Genome editing using targeting endonucleases and single-stranded nucleic acids
一种单链核酸、靶向核酸的技术,应用在基因治疗、基因工程、植物基因改良等方向,能够解决低自发重组率、阻碍进展等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0121] Example 1: Modification of RSK2 Kinase
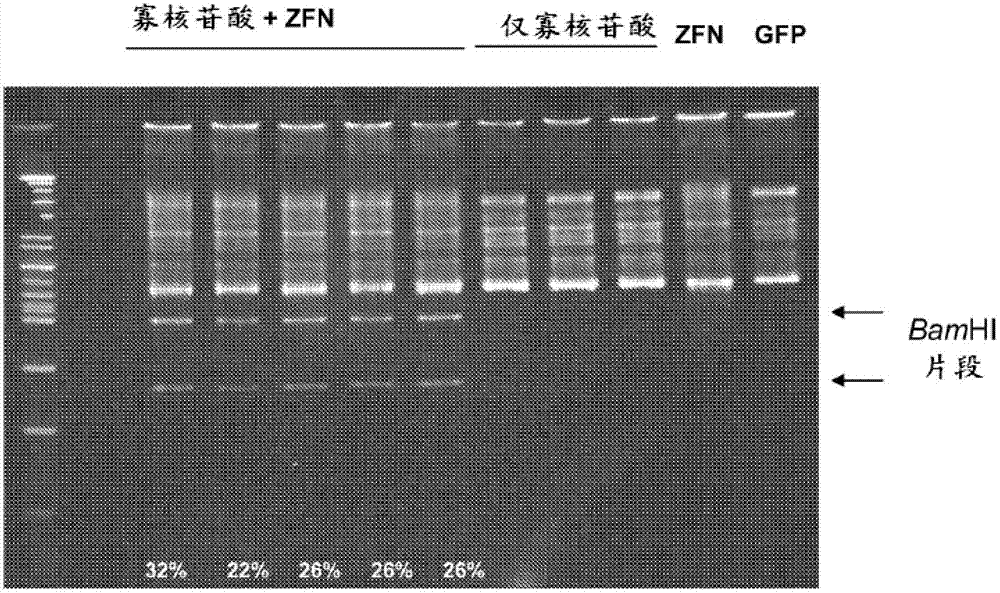
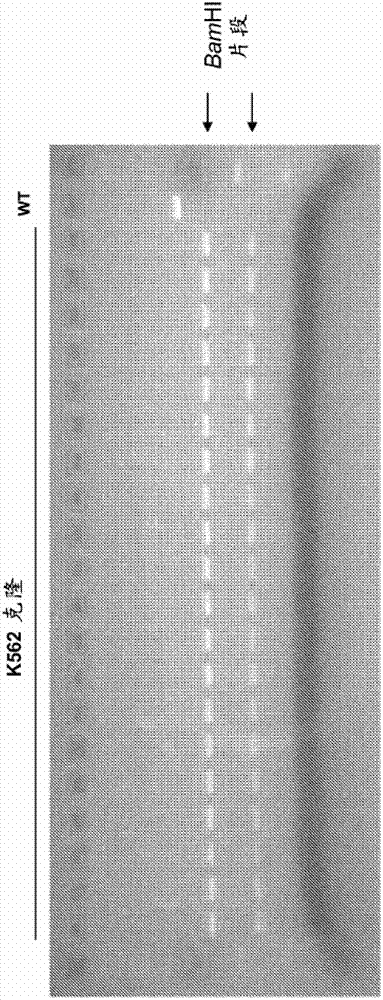
[0122] The following examples detail the modification of the RSK2 kinase locus. Oligonucleotides (125nt) were designed to incorporate 3 different mutations into the RSK2 kinase chromosomal sequence. Oligonucleotides contain: (1) two point mutations in the ZFN binding site that prevent subsequent non-homologous end joining (NHEJ), (2) TGC to GTT change to switch Cys to Val, and (3) silencing Sex C to A change to generate a single BamHI site for clone screening ( figure 1 ). Oligonucleotides are generated using standard synthetic methods (eg, without chemical modification) and purified by PAGE. A pair of zinc finger nucleases (ZFNs) were designed to target the RSK2 kinase locus. A ZFN was designed to bind the sequence 5'-GTATACATAAAGCTA-3' (SEQ ID NO: 6; figure 1 left binding site shown in ), and another ZFN was designed to bind the sequence 5'-GGAGTTTGCAGTGAAGGTA-3' (SEQ ID NO: 7; figure 1 right binding site shown in ).
[...
Embodiment 2
[0125] Example 2: Modification of the AAVS1 locus
[0126] The following examples detail the use of oligonucleotides to introduce HindIII sites into the AAVS1 locus. Figure 4 The wild type sequence of the AAVS1 locus and the sequence (108 nt) of sense and antisense oligonucleotides containing HindIII sites are shown. These oligonucleotides were generated using standard methods (eg, without chemical modification) and purified by PAGE.
[0127] A pair of ZFNs were designed to target the AAVS1 locus. A ZFN was designed to bind the sequence 5'-ACCCACACAGTGG-3' (SEQ ID NO: 8; Figure 4 left binding site shown in ), and another ZFN was designed to bind the sequence 5'-TAGGGACAGGAT-3' (SEQ ID NO: 9; Figure 4 right binding site shown in ). Capped, polyadenylated mRNAs encoding ZFNs were prepared using standard methods. ZFN mRNA and HindIII site containing oligonucleotides were nucleoffected into K562, HCT116, U2OS, A549, HEK293, HepG2 or MSF7 cells. After a period of incubatio...
Embodiment 3
[0129] Example 3: Modification of the AAVS1 locus - length of oligonucleotides
[0130] To determine whether shorter oligonucleotides might be used to deliver HindIII sites to the AAVS1 locus, oligonucleotides ranging in length from 36 to 106 nt were prepared. For example, a 106 nt oligonucleotide has 50 nt sequence identity (ie, has 50 nt homology arms) on either side of the ZFN cleavage site, with a HindIII site (6 nt) between the homology arms. The sequences of the oligonucleotides are shown in Table 1 below.
[0131]
[0132] K562 cells were nucleoffected with 2.5 μg of plasmid DNA encoding each ZFN directed to AAVS1 (5 μg total) and 3 μl of a 100 μM oligonucleotide stock (sense or antisense). Cells were harvested 2 days after nucleofection. Genomic DNA was PCR amplified and digested with HindIII. Figure 7 Integration of sense strand AAVS1-HindIII oligonucleotides of different lengths is demonstrated. Delivery of the ZFN and either oligonucleotide results in integr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com