Method for non-invasive prenatal screening for aneuploidy
A technology for prenatal screening and aneuploidy, applied in biochemical equipment and methods, instruments, library member identification, etc., can solve problems such as false positives
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0112] Example 1: Assay Development
[0113] The following sections describe the materials and methods used to perform the NIPS assay of the present invention.
[0114] 1. Patient sample collection
[0115] In one example, for assay development, validation, and validation studies, applicants obtained samples from pregnant women from Sequenom (San Diego, CA), Precision Medicine, and consenting volunteers. For singleton pregnancies, applicants obtained 3,750 samples from Sequenom, 165 samples from Precision Medicine, and 10 samples from volunteers; Sequenom also provided samples from 115 pairs of twin pregnancies. The Sequenom samples are scheduled to be discarded and de-identified before being sent to applicants. Samples from Precision Medicine were obtained with informed consent using their protocol. Volunteers provided written informed consent through signed forms approved by the Western Institutional Review Board, which specifically reviewed and approved the study. Thi...
Embodiment 2
[0145] Example 2: Assay verification and verification
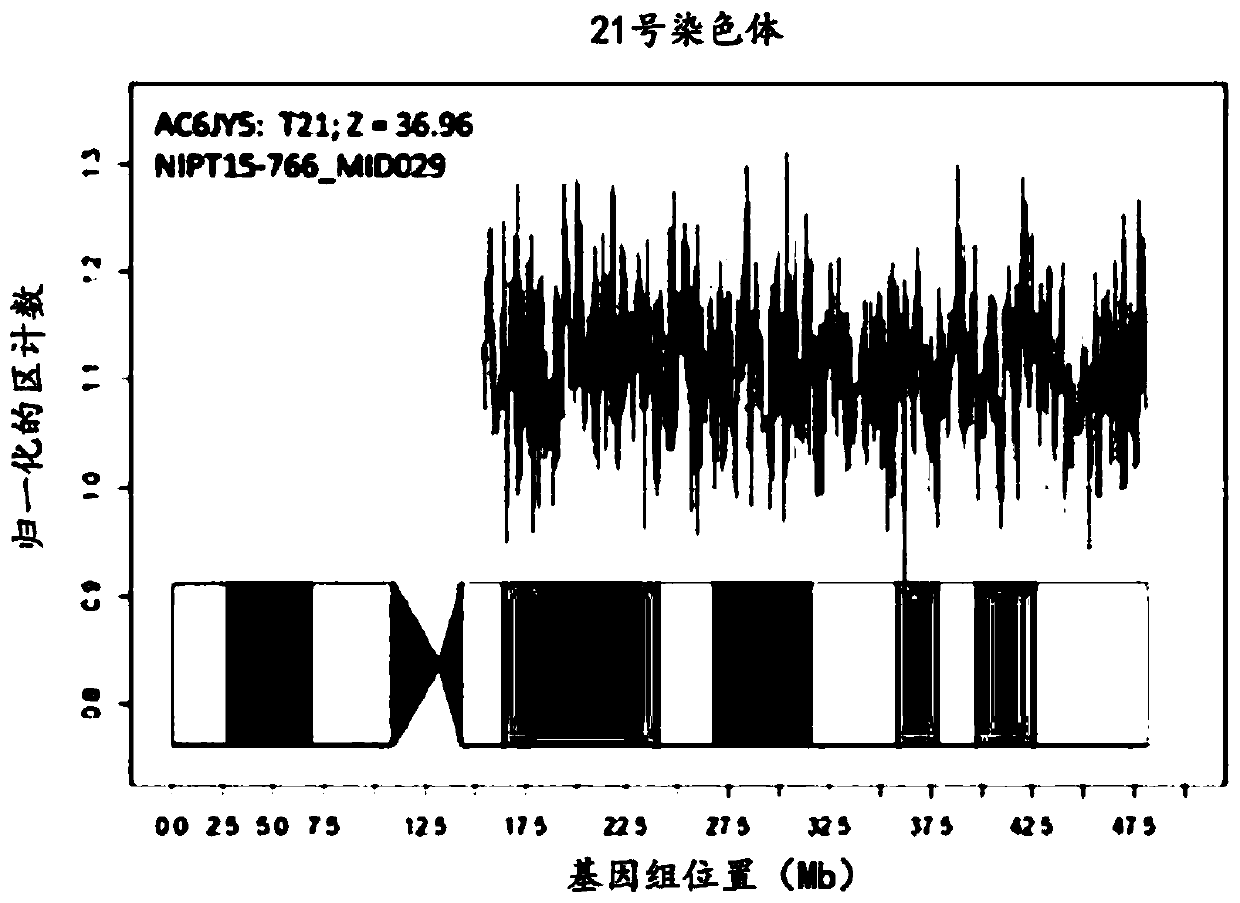
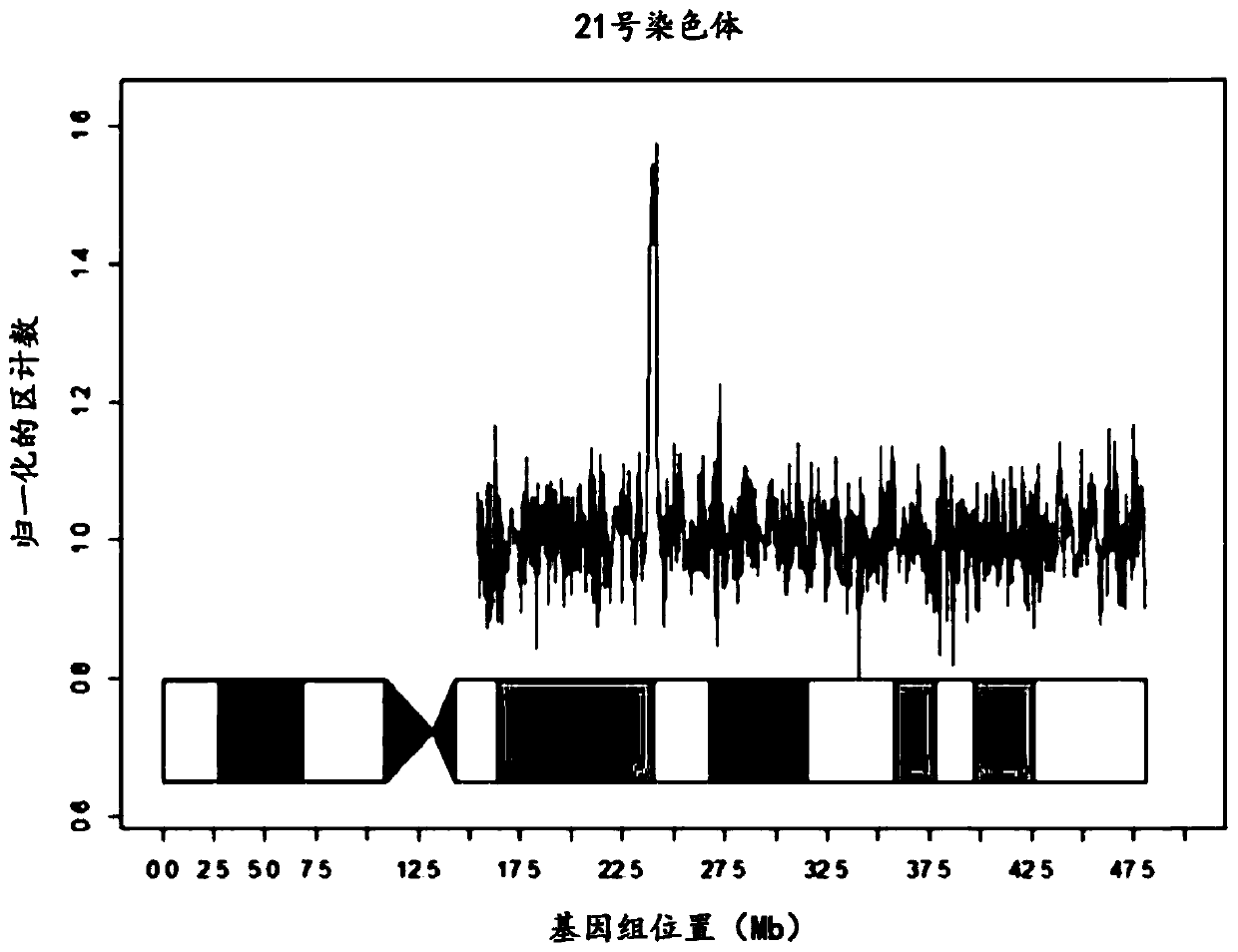
[0146] Once assay performance parameters were established, a series of assay samples were tested, including known unaffected pregnancies and known aneuploid pregnancies. The series of 2,085 samples included trisomy 21 (n=69), trisomy 18 (n=20) and trisomy 13 (n=17). There were no unaffected pregnancies with a Z-score >4 and no affected pregnancies with a Z-score 4 and no affected pregnancies with a Z-score <8.
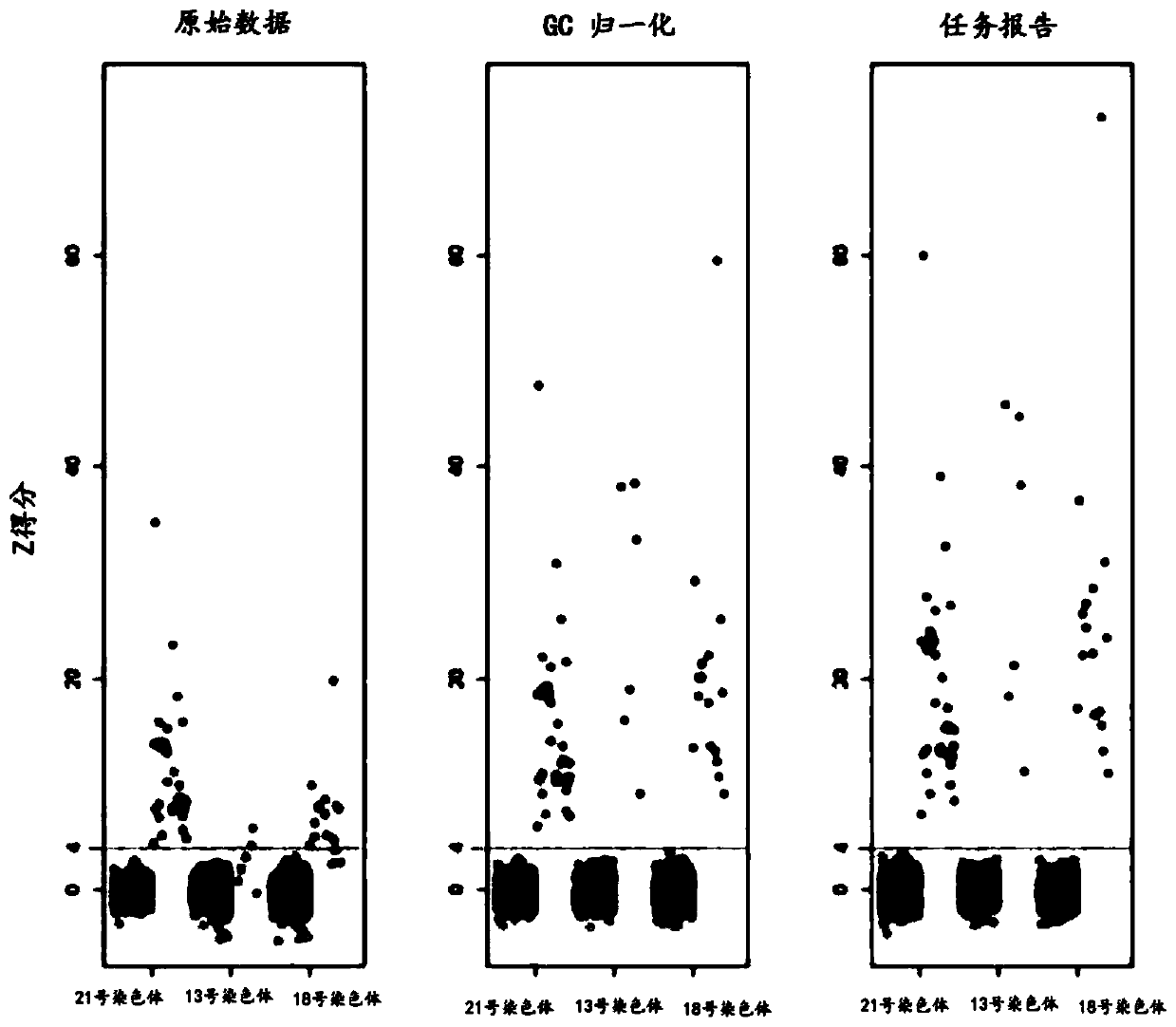
[0147] As there was no difference in performance between the testing and validation studies, the results were pooled for analysis. The effect of GC correction was minimal for chromosome 21 with normal GC content, intermediate for chromosome 18 with moderate gain and GC content, and greatest for chromosome 13 with highest GC content ( figure 1 ). Using raw data, a Z-score threshold of 4 yielded absolute discrimination between 2,498 unaffected pregnancies and 90 trisomy 21 samples; no unaffected pregnancies had...
Embodiment 3
[0152] Embodiment 3: clinical implementation
[0153] The following section describes the results of the NIPS assay of the invention in an exemplary clinical implementation. In particular, samples from the 10th gestational week were accepted. More than 90% of the samples received were from between the 10th and 15th week of gestation.
[0154] Based on the test and validation results above, for clinical implementation, a Z-score cutoff of ≤4 was used for unaffected pregnancies and >8 for affected pregnancies. A Z score >3 but <8 suggests further investigation. A review of the first 10,000 clinical samples revealed abnormal NIPS results in 180 (1.8%) (Table 1). The overall positive rate of trisomy 21 was 1.0%, trisomy 18 was 0.36%, trisomy 13 was 0.21%, and sex aneuploidy was 0.17%. One sample was positive for the DiGeorge microdeletion, and 2 cases had 2 abnormalities. Of the first 10,713 samples tested, results were unreportable in 94 (0.88%); due to low fetal fraction ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com