Method and prediction model for detecting tumour mutation burden
A mutation load and tumor technology, applied in the field of biomedicine, can solve the problems of high cost of WES, difficulty in conventional clinical methods, and cumbersome operations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
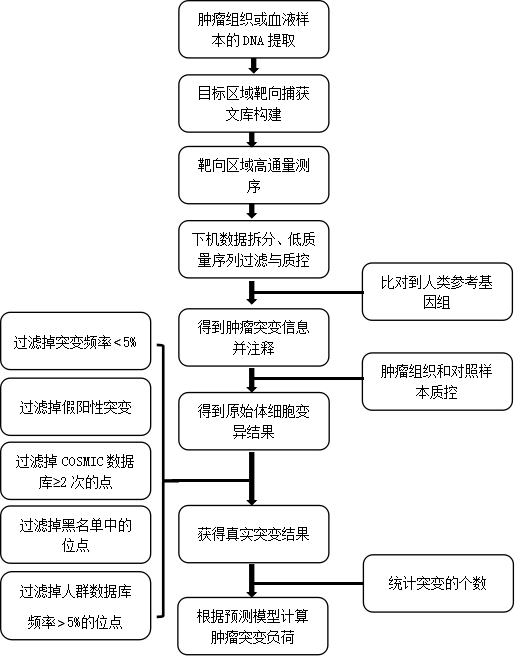
[0052] In an embodiment, the samples to be tested are paraffin-embedded tissue samples known to have a high tumor mutation burden and corresponding blood control samples.
[0053] The test steps are as follows:
[0054] 1. Sample extraction and fragmentation: Genomic DNA was extracted from blood samples and paraffin-embedded tissue samples using a nucleic acid extraction kit (Magen or Promega), and quantified using Qubit; the sample DNA was fragmented using a Covaris M220 instrument to make The size of the DNA fragment is between 100 and 500 bp, and the QIAxcel (QIAGEN) instrument is used to detect whether the fragment size meets the requirements.
[0055] 2. Double UMI library construction:
[0056] 1) End repair and "A" addition: The fragmented sample DNA is subjected to end repair and "A" addition. The reaction system is shown in Table 1 below. Vortex and mix well, then centrifuge, and place on a thermal cycler. ℃ for 30 minutes, then 65 ℃ for 30 minutes.
[0057] Table ...
Embodiment 2
[0094] Whole exome sequencing and ChosenOne599 were performed on 1453 lung cancer samples Ⓡ Targeted sequencing, analyzed according to the above process, and counted ChosenOne599 Ⓡ The number of SNV synonymous mutations, the number of SNV non-synonymous mutations, the number of frameshift insertion mutations, the number of non-frameshift insertion mutations, the number of frameshift deletion mutations, and the number of non-frameshift deletion mutations in the The TMB value of the panel is used as the gold standard, and the TMB of the panel is calculated according to the multiple linear model.
[0095] Obtain the mutation data of the sample to be tested through the detection process, and calculate ChosenOne599 Ⓡ The number of SNV synonymous mutations Nsys, the number of SNV non-synonymous mutations Nnon, the number of frameshift insertion mutations Nis, the number of non-frameshift insertion mutations Nns, the number of frameshift deletion mutations Nds and the number of non-...
Embodiment 3
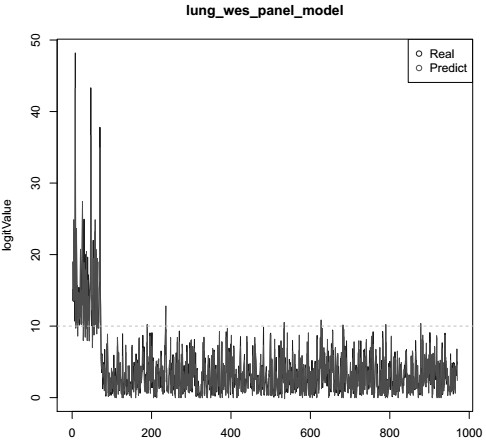
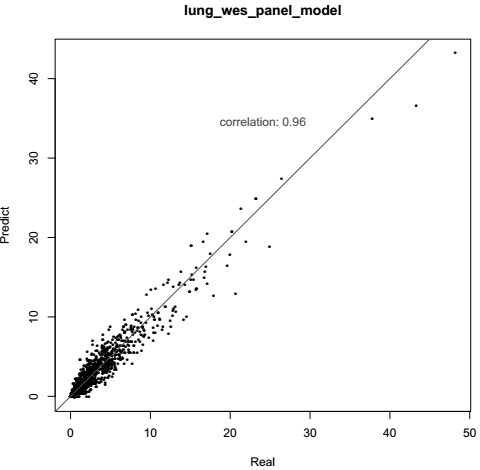
[0098] This embodiment carries out ChosenOne599 to 6 routine TMB standard items Ⓡ For targeted region sequencing, predict the results according to the TMB model, and compare the results with the gold standard as shown in Table 9 and Figure 6 shown.
[0099] Table 9 Results prediction and comparison results with gold standard
[0100]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com