Gene chip for high-depth sequencing of gene mutation, preparation method and applications thereof
A gene chip, deep sequencing technology, applied in biochemical equipment and methods, microbial determination/inspection, chemical library, etc., can solve the problems of low content, low mutation frequency of tumor markers, low DNA content, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
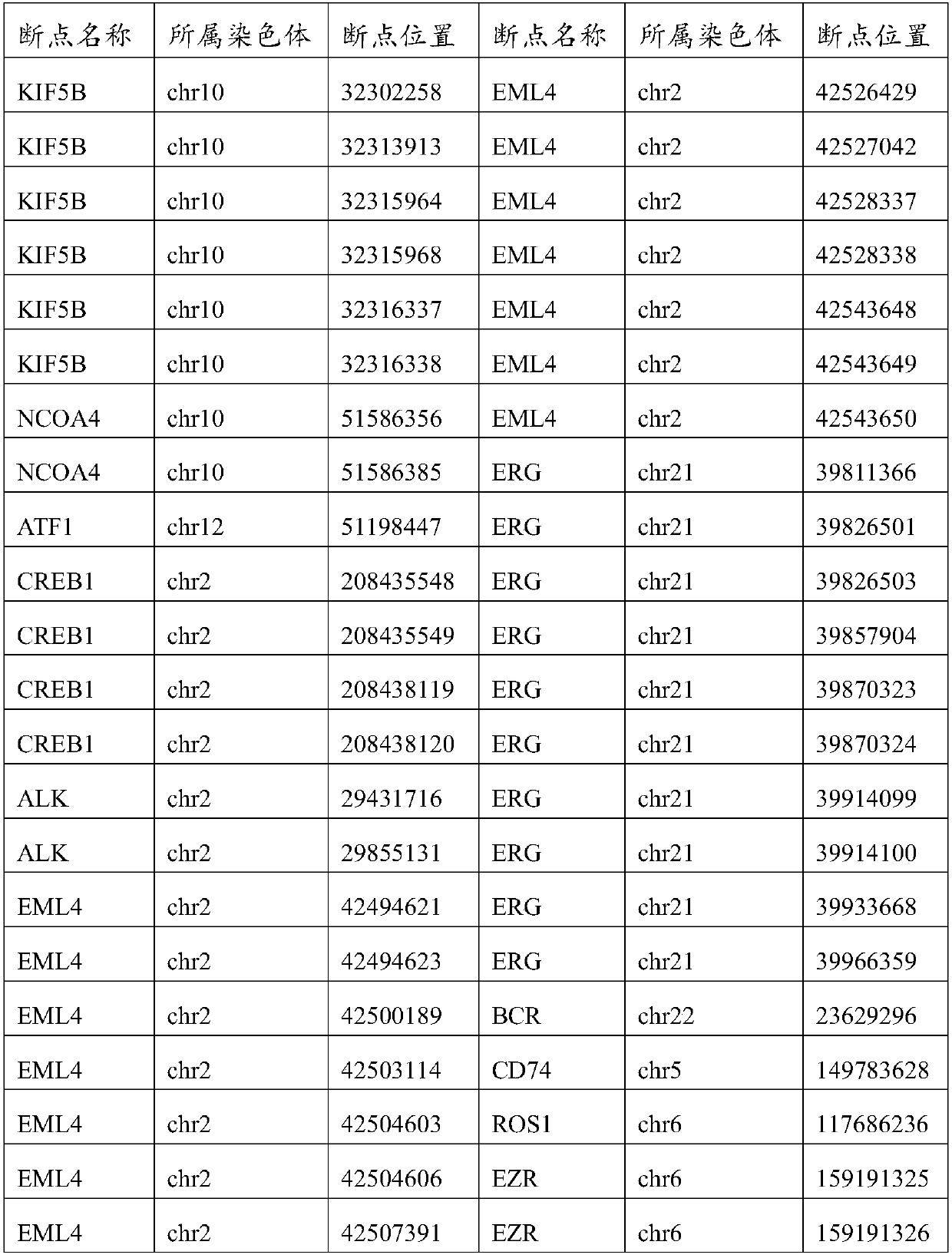
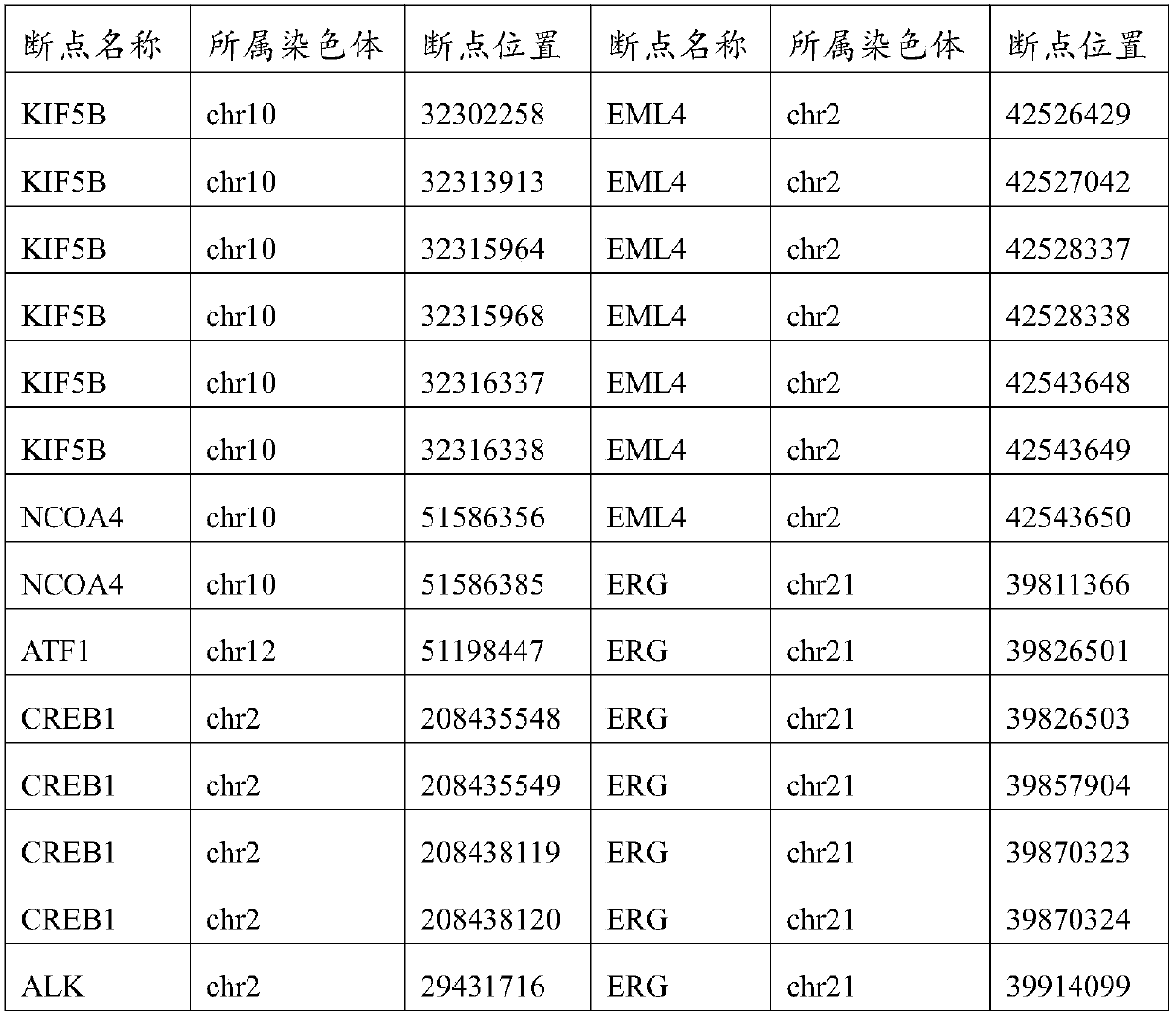
[0050] (1) The preparation method of this application constructs a capture interval library adapted to it according to different mutation types, and then designs a gene chip suitable for different mutation types. For example, for CNV hotspot genes, this application adds these hotspots The heterozygous SNV locus of the gene, of course, the selection of the number of heterozygous SNV loci is related to the size of the gene, and the appropriate number of heterozygous SNV loci is selected for each gene, and the final capture interval library is the union of the above-mentioned intervals , thus ensuring the detection of CNV.
[0051] (2) The preparation method of this application not only considers the individual needs of the product, but also comprehensively considers TCGA, ICGC, COSMIC and other databases, so that the gene chip can cover the relevant Driver Gene, high-frequency mutation gene, High-frequency mutation sites, important genes in 12 cancer-related signaling pathways, ...
Embodiment
[0056]The preparation method of gene chip for high-depth sequencing of gene mutation in this example includes different mutation types such as single nucleotide variation, insertion-deletion mutation, copy number mutation and structural variation, combined with the mutation information of the gene mutation online database and the local database, Build a capture interval library; design a gene chip based on the capture interval library, improve the capture quality and efficiency of the gene chip, and achieve high-depth sequencing. According to different mutation types, the capture interval library is divided into single nucleotide variation and insertion-deletion mutation capture interval library, copy number mutation capture interval library, and structural variation capture interval library. This example uses different samples to explain in detail the construction of the indel mutation capture interval library, the construction of the copy number mutation capture interval libr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com