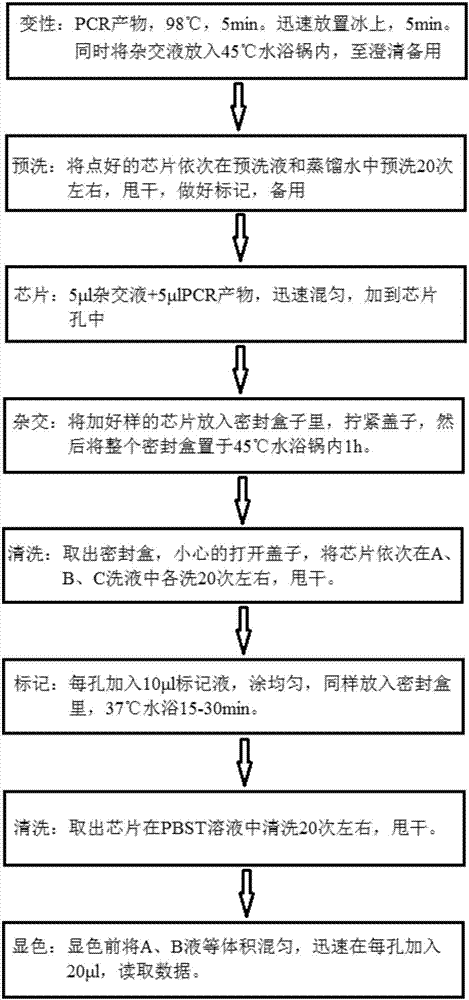
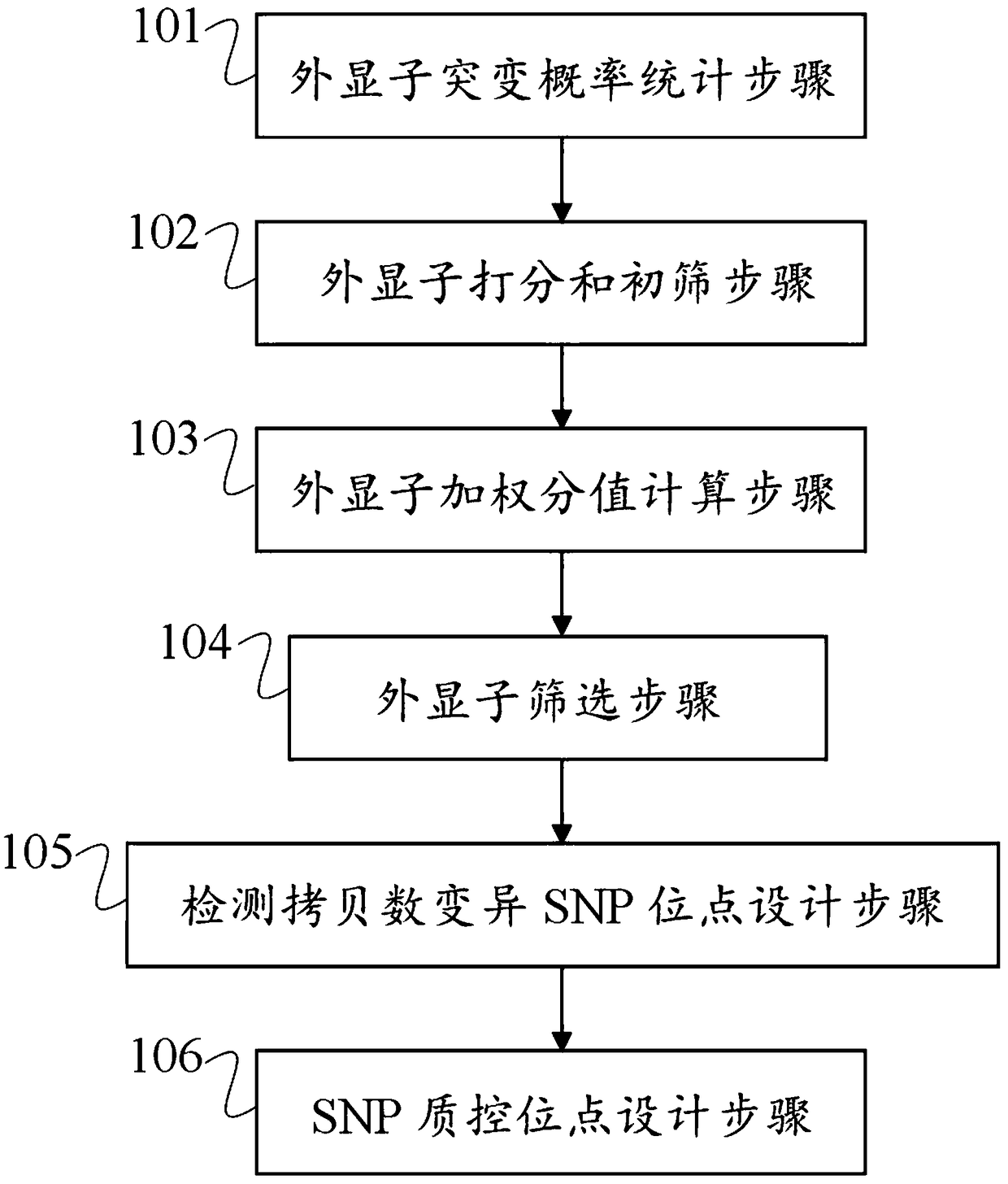
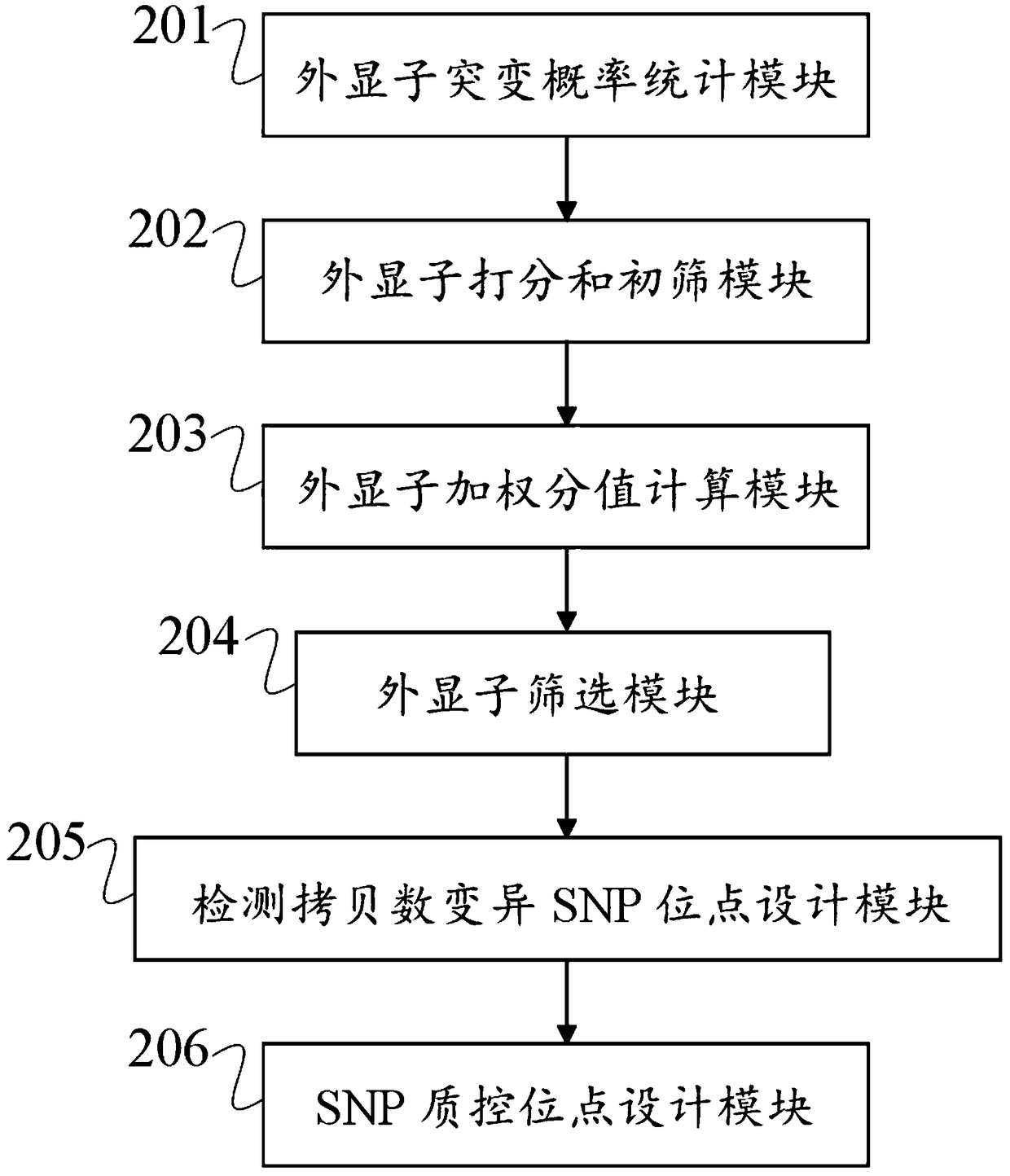
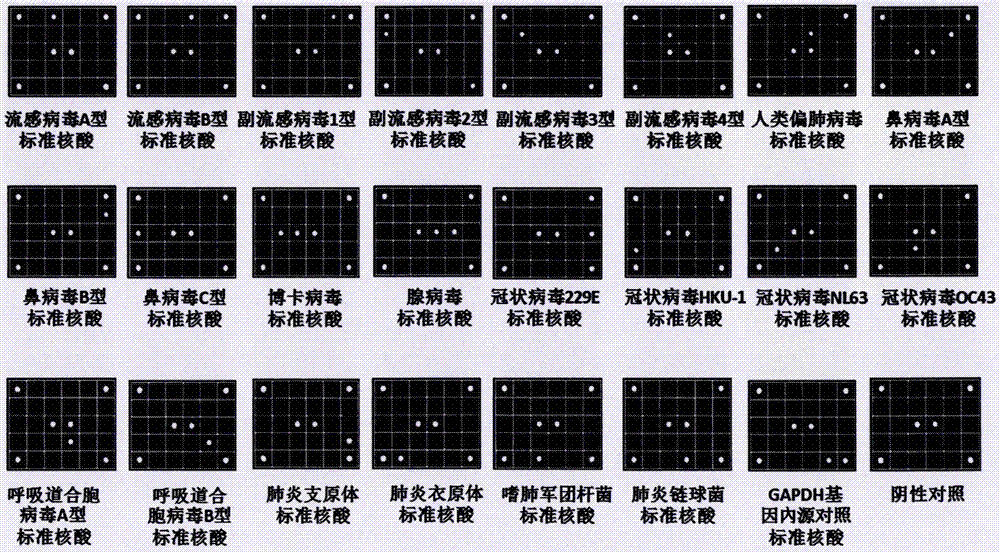
Patents
Literature
869 results about "Gene Microarray" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
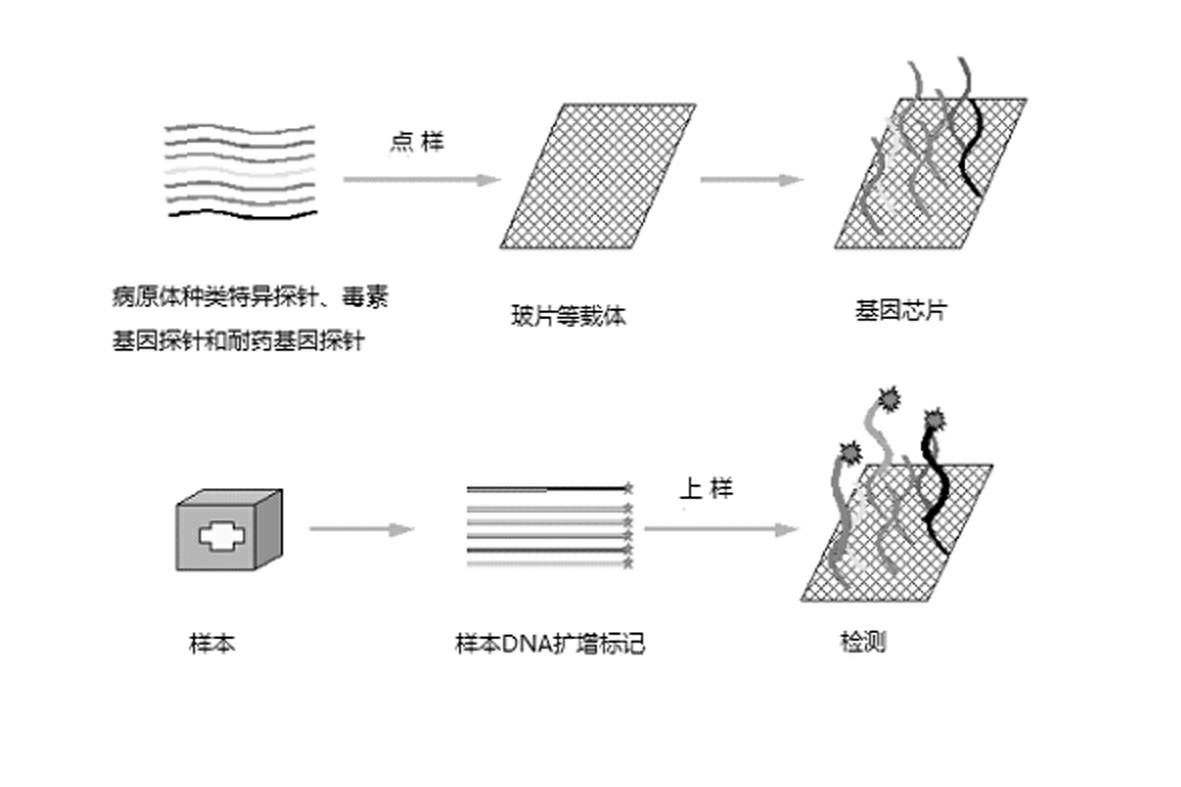
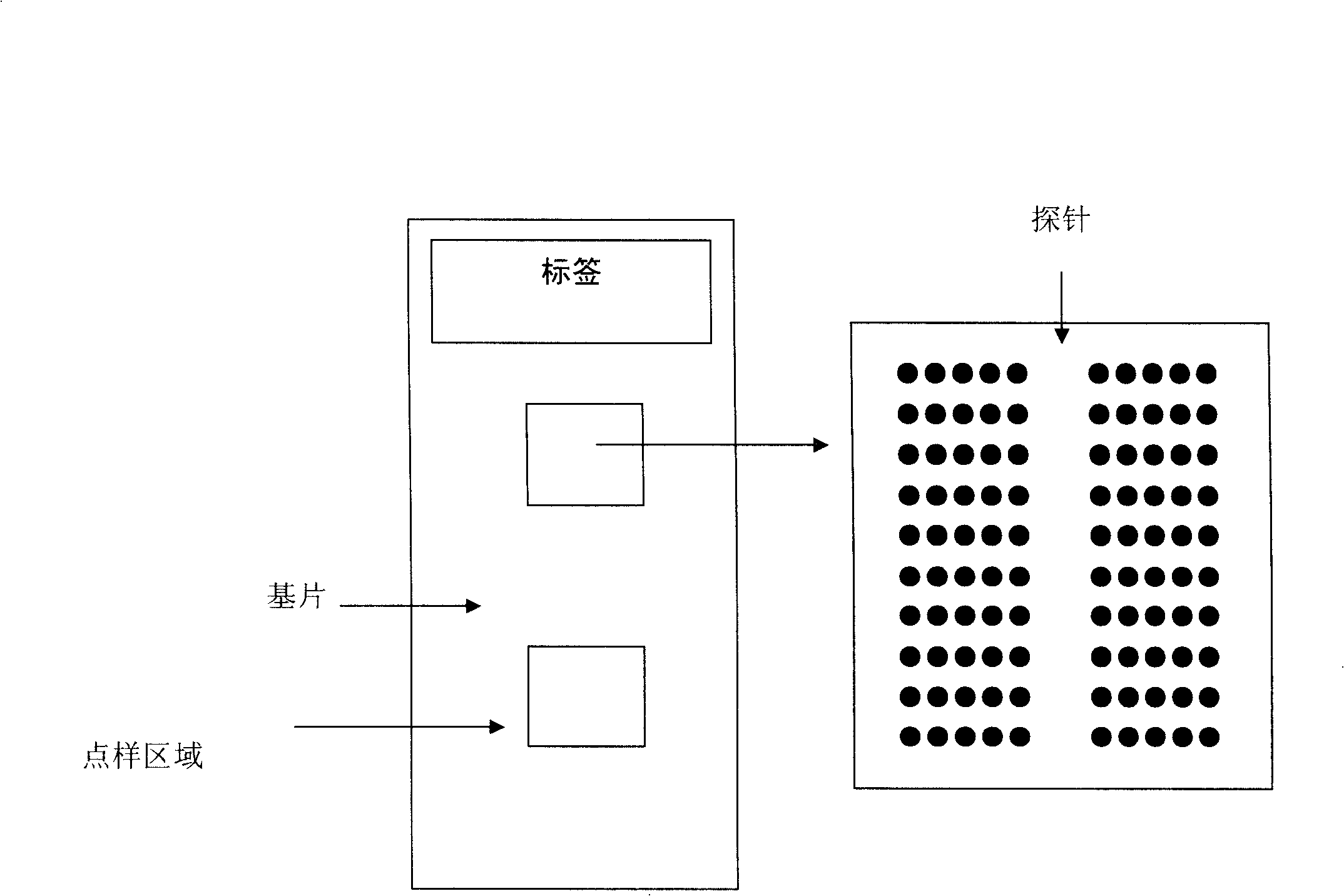
A DNA microarray is sometimes referred to as a gene chip or DNA chip. Scientists can use DNA microarrays to study the activity of several thousand genes at the same time. The microarray can be used in analyzing chromosomes.
Method for the early detection of pancreatic cancer and other gastrointestinal disease conditions
InactiveUS20080248484A1Early diagnosisSpecific and focused early diagnosisMicrobiological testing/measurementOrgan systemNeoplasm
The present invention uses peripheral blood monocyte-lymphocyte for the early diagnosis of pancreatic cancer, as well as other conditions of the pancreas and other organs. The peripheral blood lymphocytes recognize the new neoplasm in the pancreas, as well as disease processes in other organ systems. The evaluation of this specific recognition of the disease process by the peripheral blood monocyte-lymphocyte through gene microarray expression patterns constitute a successful method for the early detection of pancreatic cancer and other organ disease processes. This document describes the process used in this method of early diagnosis.
Owner:BAUER A ROBERT
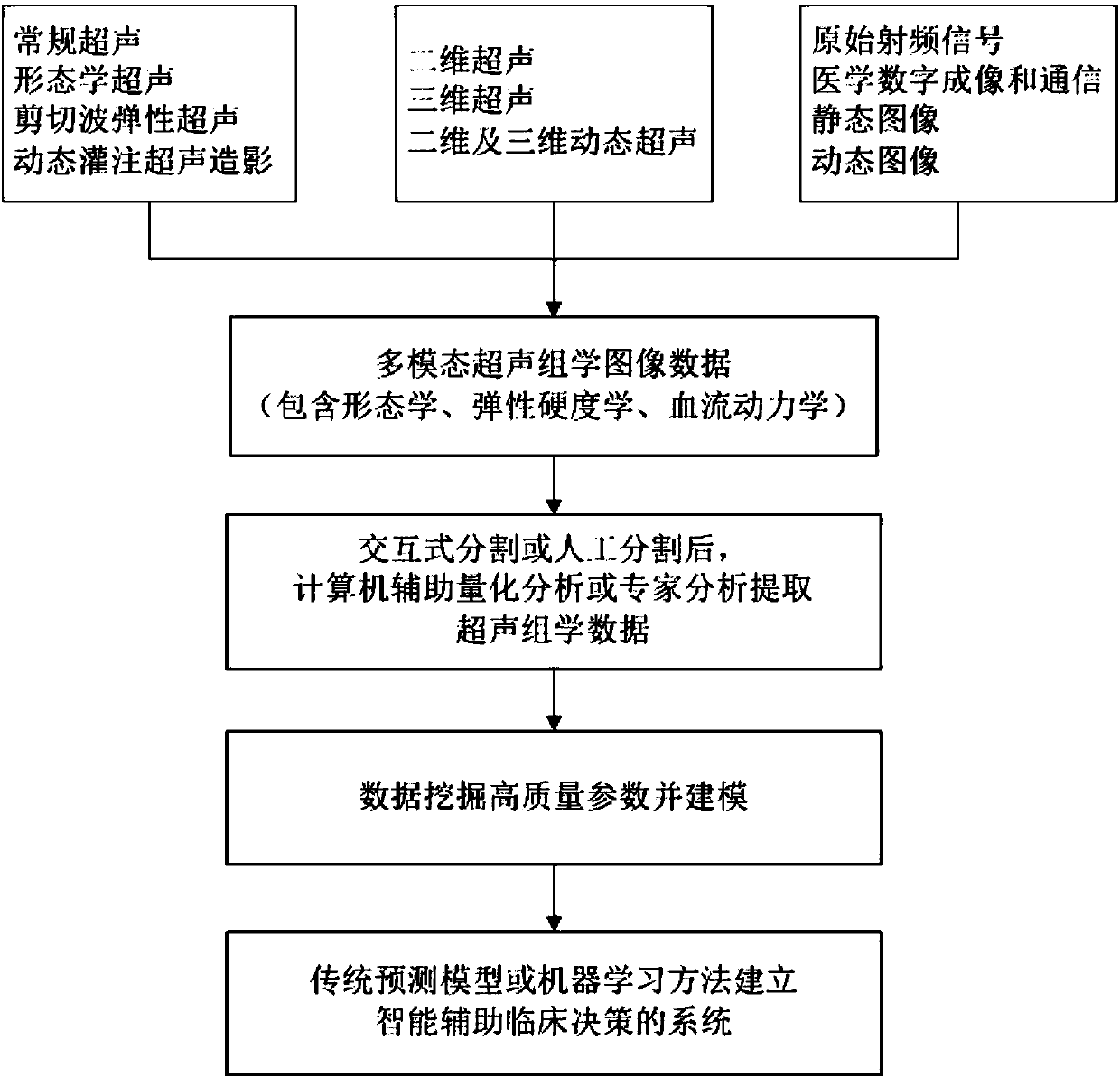
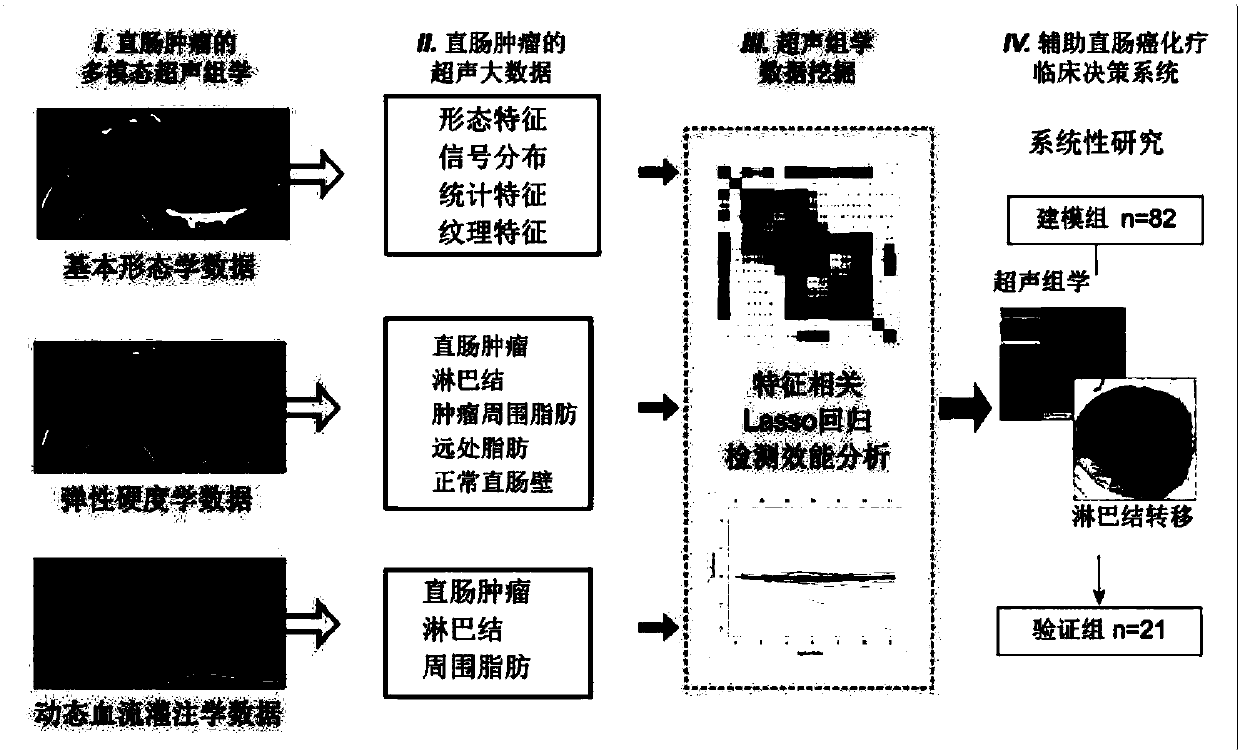
Intelligent decision-making assisting system based on multi-modal ultrasomics
InactiveCN107582097AGood segmentation resultComplete individualized precision medical adviceBlood flow measurement devicesOrgan movement/changes detectionDiseaseGenomics
The invention discloses an intelligent decision-making assisting system based on multi-modal ULTRASOMICS. Firstly, ultrasonic data from different signal sources of an instrument is acquired, an interactive or manual dividing method is adopted to locate an interested area in an ultrasonic image, data of morphology, blood perfusion and hardness science of diseases is obtained, and a multi-modal ultrasonic characteristic database is constructed; through the combination of relevant clinical information, laboratory checking information and gene chip information, ultrasomics data is mined, a machinelearning method is utilized to construct different models, the recurrence rate and the survival rate are predicted, the curative effect and the radiotherapy and chemotherapy sensitivity are judged inthe early stage, and the correlation of ultrasomics, genomics, proteomics, transcriptomics and metabonomics is analyzed. As macroscopic ultrasomics, the intelligent decision-making assisting system provides a repeatable evaluation means for noninvasive individualized precise medicine.
Owner:THE FIRST AFFILIATED HOSPITAL OF SUN YAT SEN UNIV
Kit for quickly detecting 15 pneumonia pathogenic bacteria
ActiveCN107338315AMicrobiological testing/measurementMicroorganism based processesBacteroidesStaphylococcus aureus
The invention discloses a kit for quickly detecting 15 pneumonia pathogenic bacteria. The kit can detect streptococcus pneumoniae, staphylococcus aureus, haemophilus influenzae, mycoplasma pneumoniae, pseudomonas aeruginosa, baumanii, enterococcus faecalis, enterococcus faecium, klebsiella pneumoniae, escherichia coli, enterobacter cloacae, stenotrophomonas maltophilia, burkholderia cepacia, legionella pneumophila and chlamydia pneumoniae which cover clinically common pneumonia pathogenic bacteria difficult to culture. 16S rDNA and specific gene sequences corresponding to the pneumonia pathogenic bacteria are detected by combining gene chips with multiple asymmetric PCR reactions, and the categories of the bacteria in a to-be-detected sample are identified in genus and species. The kit makes up for the defect that current clinical detection of pneumonia pathogenic bacteria is not in time or comprehensive and a novel detection means for early diagnosis and early treatment of patients suffering from pneumonia is provided.
Owner:GENERAL HOSPITAL OF PLA +1
Gene chip for detecting tumor mutation load, and preparation method and device thereof
ActiveCN109022553AReduce testing costsMeet the needs of useBioreactor/fermenter combinationsBiological substance pretreatmentsSequencing dataImmunotherapy
The invention discloses a gene chip for detecting tumor mutation load, and a preparation method and a device thereof. The gene chip disclosed by the invention contains probes capturing 811 genes. According to the gene chip disclosed by the invention, according to a tumor genome database, 811 chip capture regions are designed; by specific capture sequencing of the 811 chip capture regions, the change trend of the tumor mutation load in a whole human genome can be truly and effectively reflected, and the method is specially suitable for analysis of the tumor mutation load in Chinese people. Thegene chip can replace traditional full exons for sequencing and detecting the tumor mutation load, shrinks the sequencing data size, reduces the detection cost of the tumor mutation load, shortens thedetection cycle, and provides a relatively cheap, fast, highly efficient and accurate solution for clinical detection of the tumor mutation load. The gene chip and a tumor mutation load detection method based on the gene chip have significant clinical guiding significance for immunotherapy medication.
Owner:裕策医疗器械江苏有限公司
Gene chip for high-flux detection of pathogens and application thereof
InactiveCN102534013AStrong specificityDetermine the typeMicrobiological testing/measurementAgainst vector-borne diseasesYersinia pestisBrucella
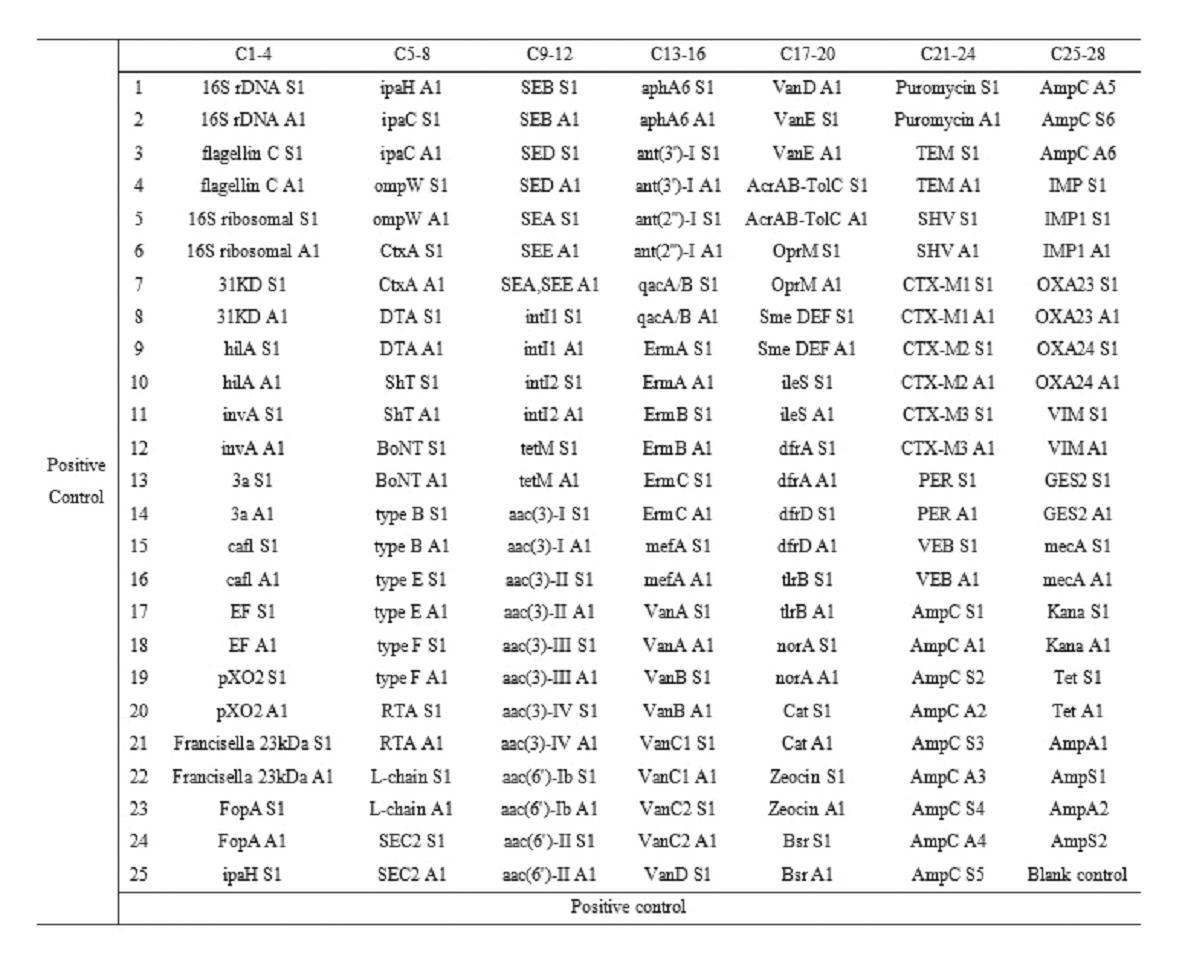
The invention relates to a gene chip for high-flux detection of pathogens and application thereof. The gene comprises (1) a combination of 174 oligonucleotide probes of pathogen variety specific genes, toxin genes and drug-resistant genes; and (2) a probe array, which is formed by curing the oligonucleotide probes on a carrier material by arm molecules. The gene chip comprises 174 gene probes, namely 32 pathogen variety specific gene probes of the following 8 pathogens of Burkholderia mallei, Burkholderia pseudomallei, Brucella, salmonella, Yersinia pestis, Bacillus anthracis, comma bacillus and the like, 25 toxin gene probe of the following 7 toxins of diphtheria toxin, Shiga toxin, staphylococcus enterotoxin, choleratoxin and the like, and 117 drug-resistant gene probes of 17 drug-resistant genes of extended-spectrum beta-lactamase, cephalosporinase, carbapenemase, integrase gene, common gene engineering carrier drug-resistant gene and the like. The gene chip can be used to detect multiple pathogen variety specific genes, toxin genes and drug-resistant genes.
Owner:李越希
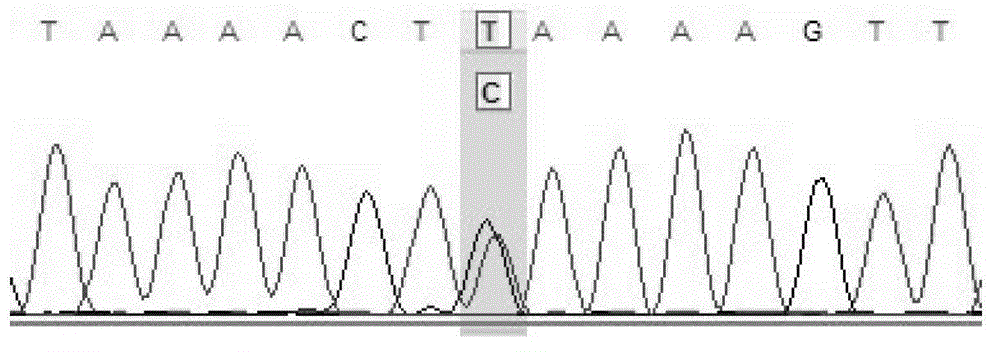
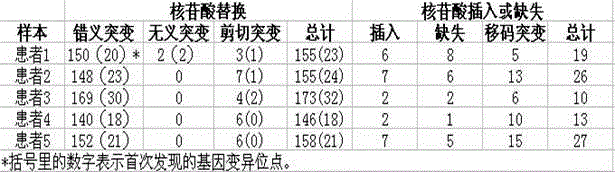
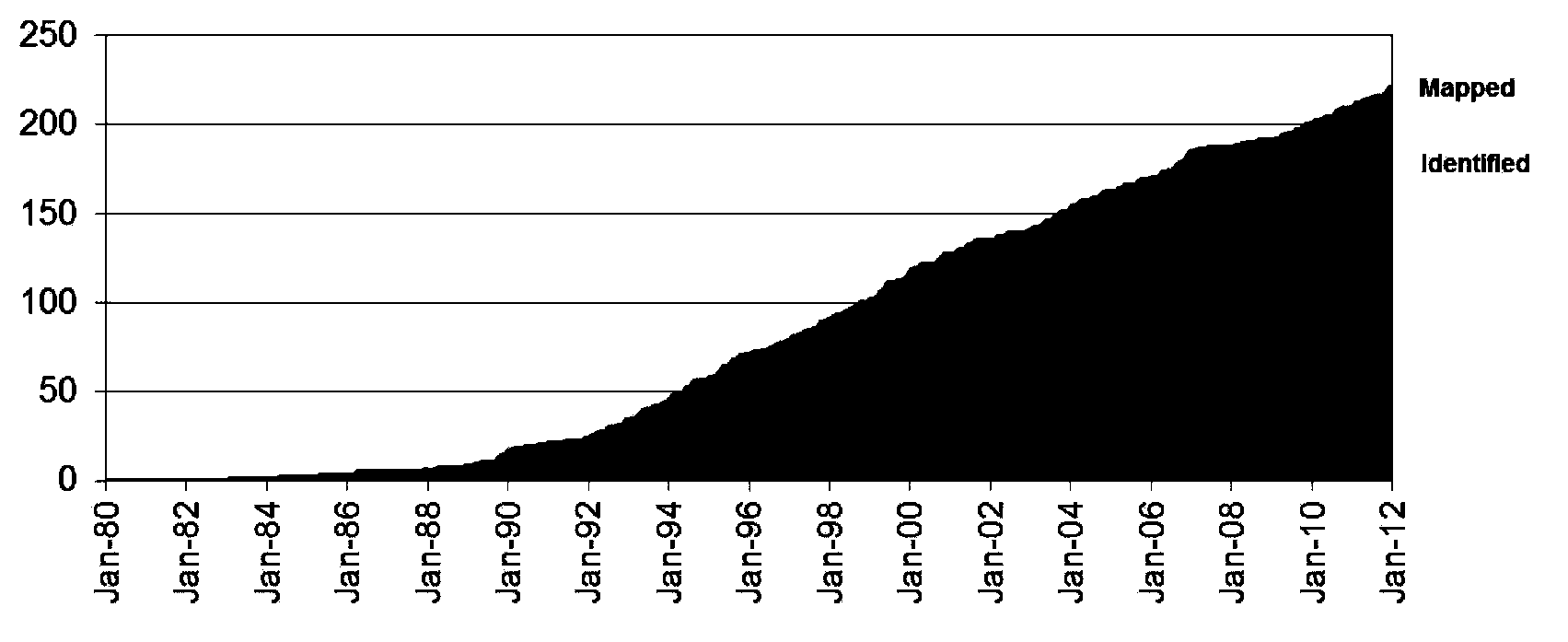
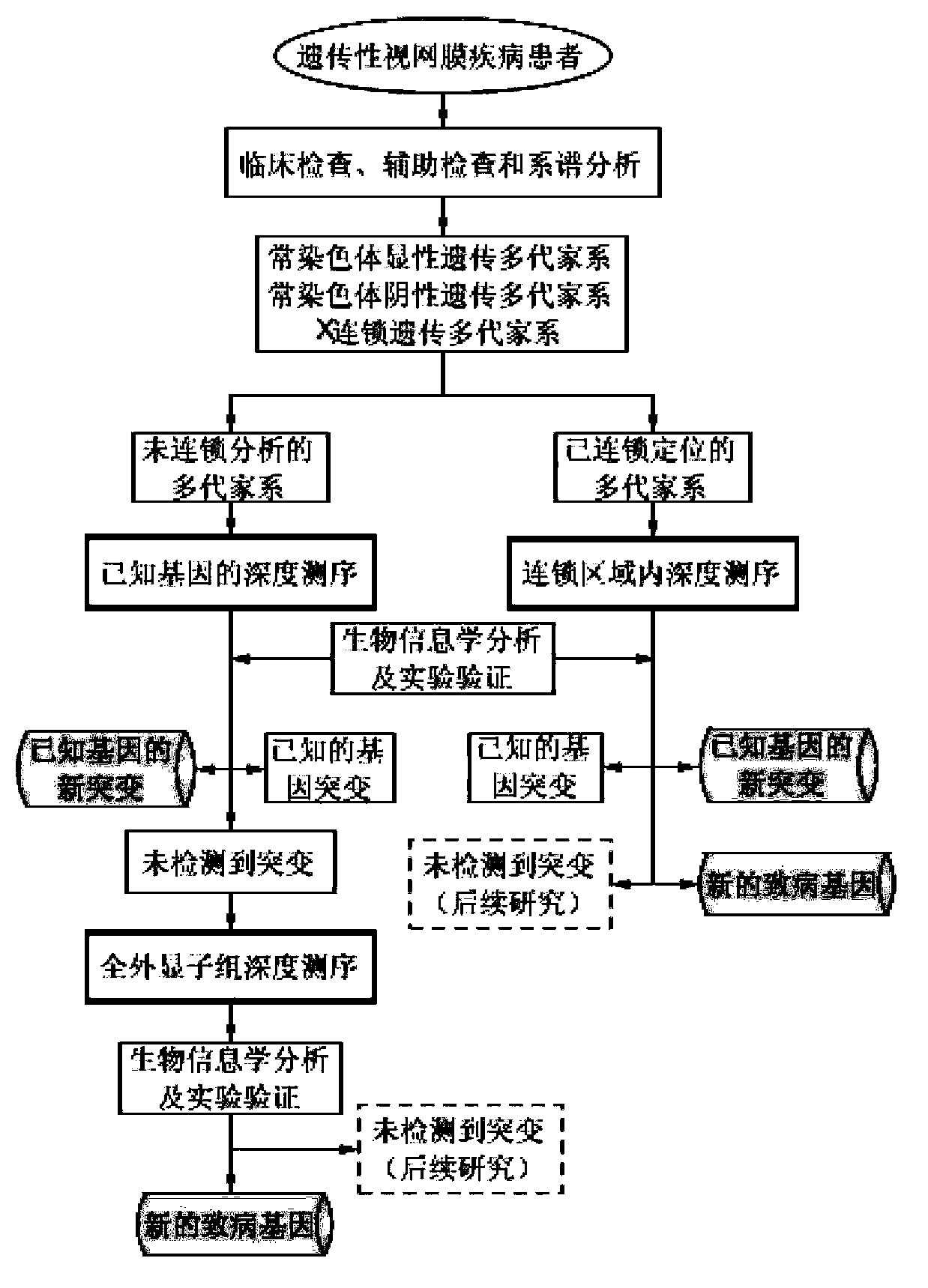
Gene chip for screening various ophthalmological hereditary diseases as well as preparation and usage method of gene chip
InactiveCN102978284AImprove efficiencyAccurate diagnosisNucleotide librariesMicrobiological testing/measurementVirulent characteristicsIntein
The invention relates to the technical field of biological gene chips, and particularly relates to a gene chip for screening various ophthalmological hereditary diseases as well as preparation and a usage method of the gene chip. Specific oligonucleotide probes of gene sequences which are related to 261 ophthalmological hereditary diseases are fixed on the surface of a carrier of the gene chip for screening various ophthalmological hereditary diseases; and the gene chip which is related to the 261 ophthalmological hereditary diseases comprises the sequences of all coding region sequences of 954 related virulence genes or disease predisposing genes and introne sequences adjacent to the coding regions. As the specific oligonucleotide probes of the gene sequences which are related to 261 ophthalmological hereditary diseases and the sequences of all the coding region sequences of the related virulence genes or disease predisposing genes and the introne sequences adjacent to the coding regions are fixed on the surface of the carrier of the gene chip, the gene chip provided by the invention is capable of screening a plurality of ophthalmological hereditary diseases at the same time and then the efficiency is greatly improved.
Owner:金子兵
Method for screening HRDs disease-causing mutation and gene chip hybridization probe designing method involved in same
ActiveCN103667438AClear relationshipScreening benefits are highMicrobiological testing/measurementDNA preparationDiseaseHybridization probe
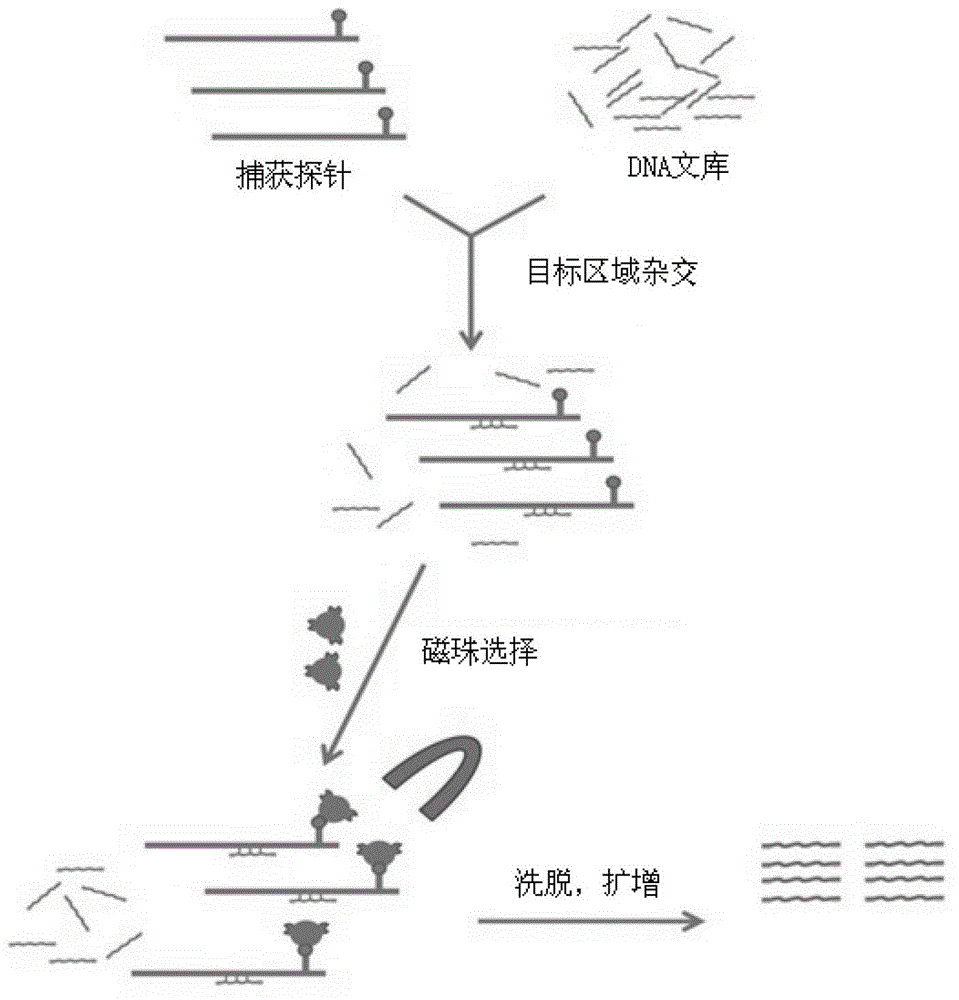
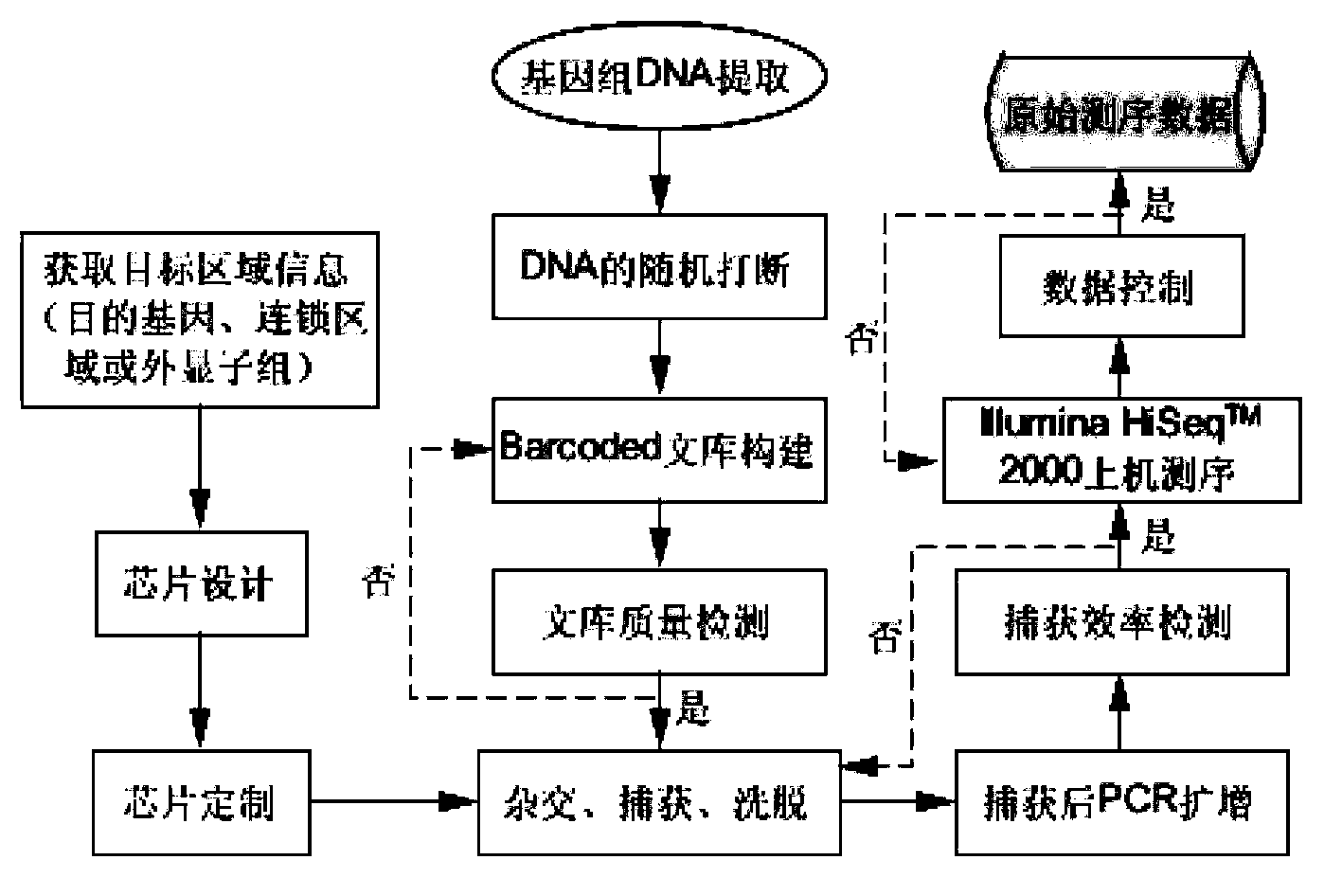
The invention belongs to the field of biological medicines, and relates to a method for screening HRDs disease-causing mutation and a gene chip hybridization probe designing method involved in the same. The method for screening the HRDs disease-causing mutation comprises the steps of (1) establishing an HRDs genetic resource repository; (2) designing and synthesizing a gene chip hybridization probe of an HRDs disease-causing gene, and integrating the gene chip hybridization probe onto a gene chip; (3) capturing a target area by utilizing the prepared gene chip and executing the depth sequencing; (4) analyzing the sequencing data on the aspect of bioinformatics, and screening the candidate disease-causing gene; (5) functionally predicting a newly-discovered splicing gene mutation site. By establishing the high-efficient HRDs target gene capturing technology, adopting the depth sequencing as a means and confirming the efficiency of the HRDs capturing chip, a high-efficient credible biological information analysis model is established.
Owner:赵晨
Virtual genome-based cryptosystem (VGC)
InactiveCN102025482ASolve the first sharing problemHuge key spaceGenetic modelsSecuring communicationPlaintextDNA Microarray Chip
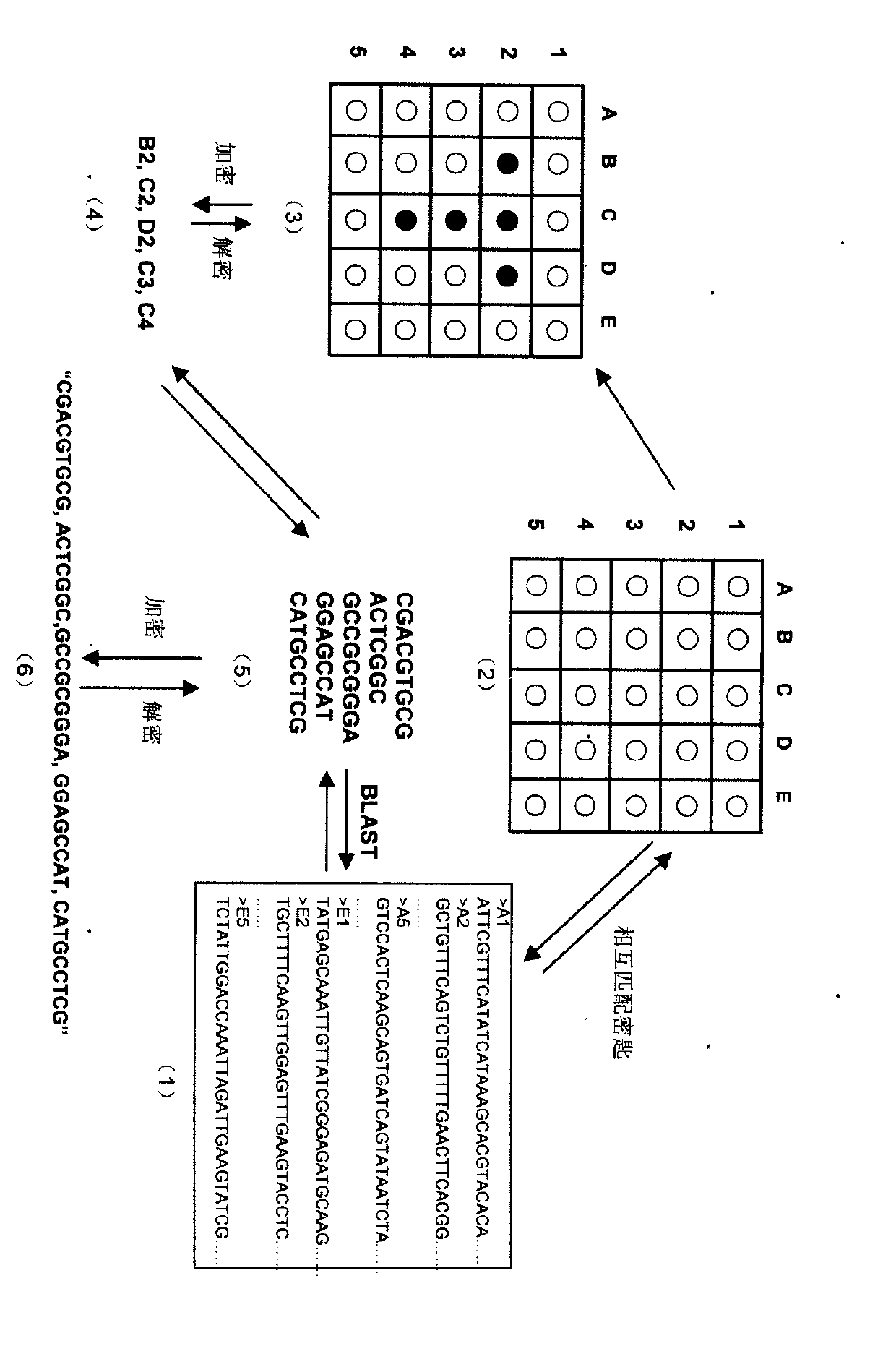
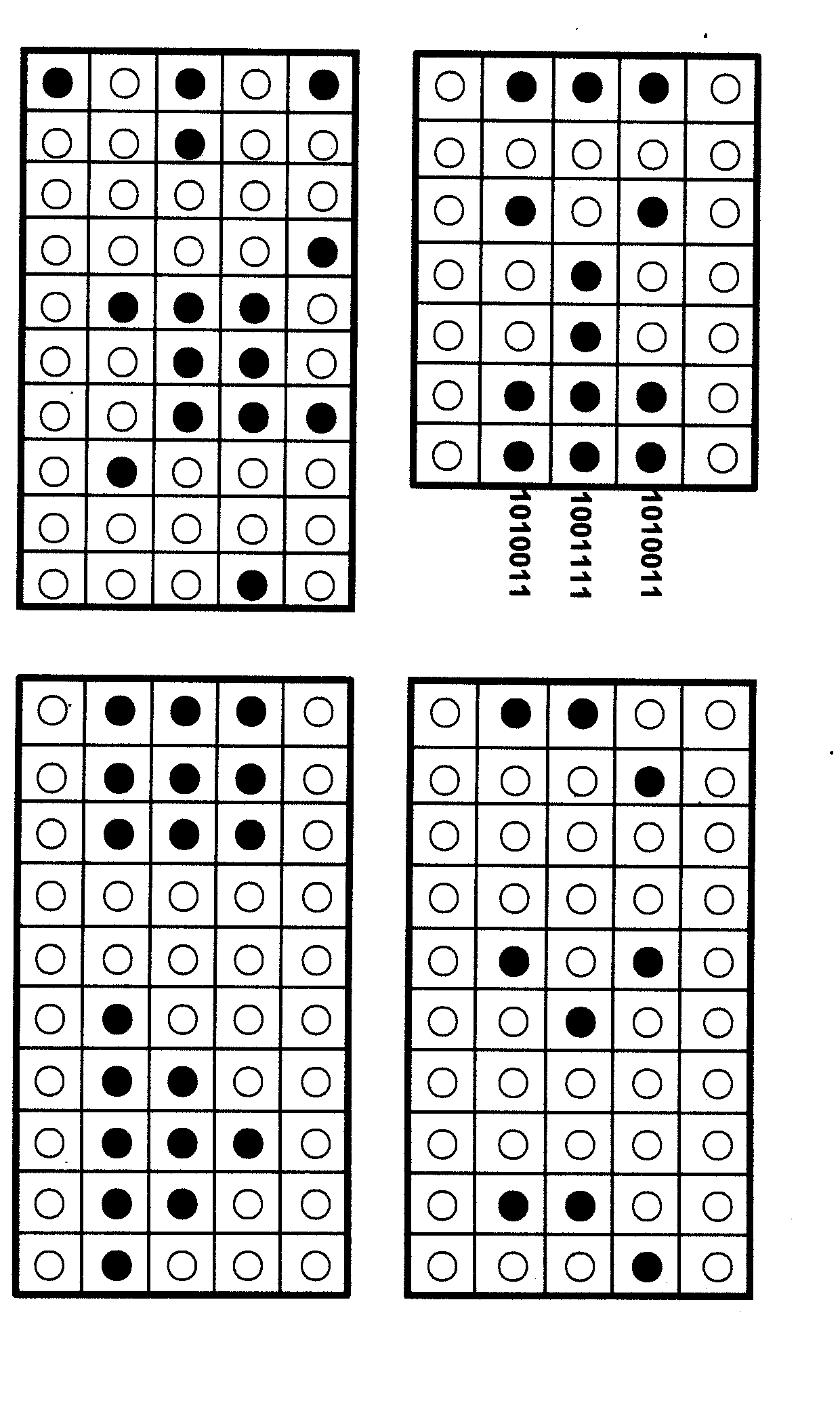
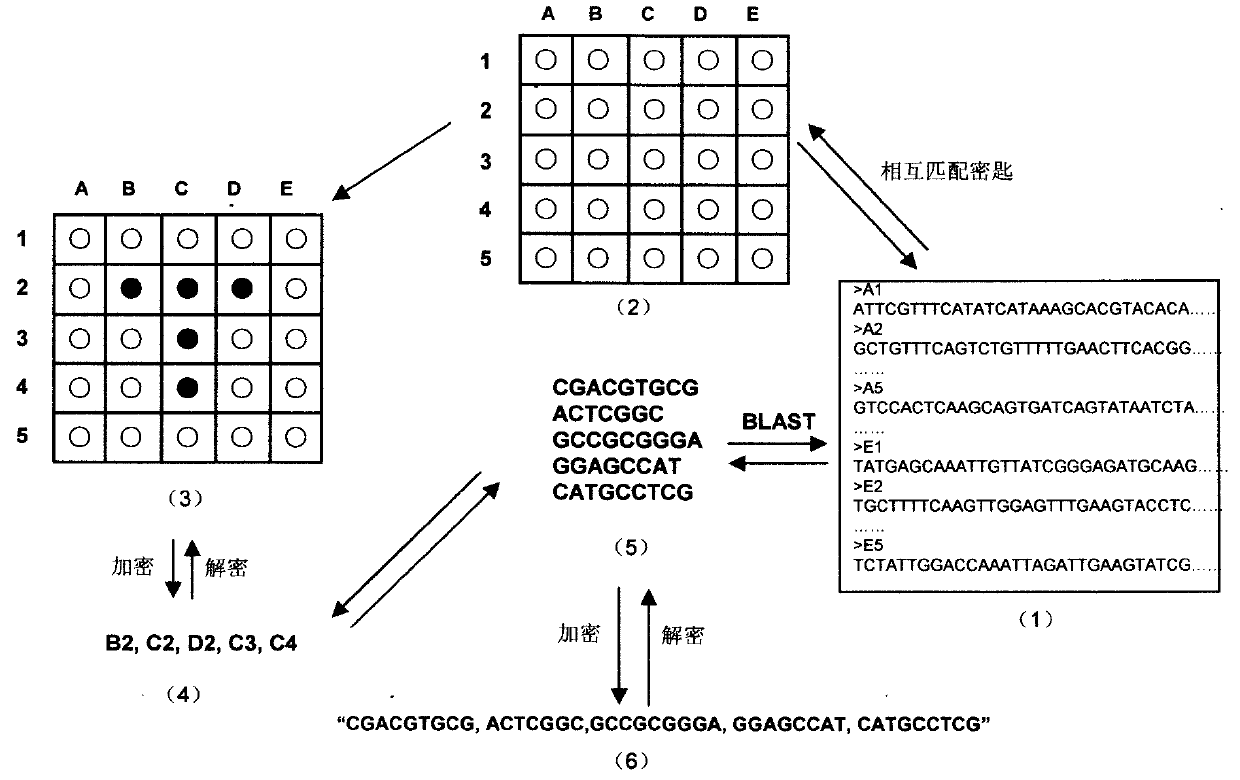
The invention relates to information security technology, in particular to a virtual genome-based cryptosystem (VGC). The cryptosystem is provided with two matched keys, of which one is a virtual genome database (VGDB) consisting of random deoxyribonucleic acid (DNA) sequences and the other one is a position table that virtual genes of the VGDB are randomly distributed in a two-dimensional microarray, namely a virtual DNA microarray chip (VDMC). Any plaintext information can be freely written on the VDMC, namely points for forming the plaintext information are selected from the VDMC microarray. The selected points correspond to the virtual genes in the VGDB; small segments of DNA sequences are randomly selected from the virtual genes; and the uniqueness of the small segments of DNA sequences in the VGDB is determined by using a common tool of the bioinformatics, namely a basic local alignment search tool (BLAST), or other character string search algorithms such as a Knuth-Morris-Pratt (KMP) algorithm and the like. A cipher text is combined by the small segments of DNA sequences. The small segments of DNA sequences need only to perform BLAST on the VGDB during decryption, namely the points for forming the plaintext information can be discovered, and the plaintext information can be restored according to the VDMC. Any non-VGDB sequence can be randomly inserted into the cipher text and does not have any influence on the encryption. Thus, the VGC is an excellent information hiding system. In addition, the VGC key can be updated automatically so as to realize an indecipherable one-time-pad system. The cryptosystem is used for real-time quick secret information communication, digital signature and identity authentication.
Owner:SOUTH CHINA BOTANICAL GARDEN CHINESE ACADEMY OF SCI
Gene chip for detection of hyperpiesis individual medicine correlated gene mutation and uses thereof
InactiveCN101343658AAccurate detectionSensitive detectionMicrobiological testing/measurementGNB3CYP2C9*3
The invention relates to a gene chip used for detecting gene mutation relating to a high blood pressure personalized medicine. The gene chip for detecting the gene mutation relating to high blood pressure personalized medicine comprises a solid phase support, a gene probe (an oligonueleotide probe) fixed in the solid phase support sequentially and a PCR primer used for amplifying the mutated gene fragment in the sample; the gene probe (the oligonueleotide probe) and the PCR primer are designed aiming at one or two points of the gene mutation in ACE (I / D) and CYP3A5*3 and / or two or more points as follows: CYP2C9*3, CYP2C9*13, AGTR1(A1166C), CYP2D6*10, ADRB1(C1165G), TSC(C1784T), ADRB3(T727C), SCNN1G_rs5729, SCNN1G_rs5723, ENOSA_rs1799983 and GNB3(C825T). The invention provides the gene chip and applications thereof for conveniently, quickly and systematically detecting the gene mutation relating to high blood pressure personalized medicine so as to determine drug reactions.
Owner:湖南宏灏基因生物科技有限公司
Respiratory tract common pathogen multiple PT-PCR combined gene chip detection kit
ActiveCN107090519AAvoid non-specific amplificationAvoid non-specific hybridizationMicrobiological testing/measurementMicroorganism based processesPositive controlReaction system
The invention relates to a respiratory tract common pathogen multiple PT-PCR combined gene chip detection kit. The kit detects one or more of 22 respiratory tract common pathogens by the usage of multiple specific conservative degenerate primer combination and probe combination and provided with endogenous control, positive control and negative control at the same time. According to the detection kit, the coverage rate for a high mutant pathogen subject sequence is increased, the problem of nonspecific cross reaction between the multiple primers and the probe is avoided, one single reaction system can detect more than 20 respiratory tract common pathogens simultaneously, and a detection tool which is simple and sensitive, rapid and large in flux and capable of carrying out multi-index parallel detection is provided for respiratory tract common pathogen detection.
Owner:SUZHOU GENEWORKS TECH CO LTD
Nucleic acid combined testing kit of respiratory tract infection pathogens
InactiveCN111378789AHigh detection throughputAvoid false negative resultsMicrobiological testing/measurementMicroorganism based processesDiseaseNucleotide
The invention discloses a nucleic acid combined testing kit of respiratory tract infection pathogens. The invention develops a set of primer-probe combinations which can detect multiple types of respiratory tract infection pathogens such as novel coronavirus, influenza virus a, influenza virus b, respiratory syncytial virus, human parainfluenza virus, adenovirus, mycoplasma pneumonia and chlamydiapneumonia through combination of a multiple fluorescence quantitative PCR technology and a flow-through hybridization and gene chip technology, wherein nucleotide sequences thereof are shown by SEQ ID NO:1-36 respectively. The nucleic acid combined testing kit of the respiratory tract infection pathogens is established. The kid can realize synchronous combined testing of the 8 respiratory tract infection pathogens, is high in detection accuracy, specificity and sensitivity, good in repeatability, low in false negativity and false positivity, short in detection time and low in cost, can realize comprehensive detection of a patient, can locate a disease source accurately, can realize treatment in time or make corresponding quarantine measures and is of important significance to effective control of respiratory tract infection and subsequent prevention of outbreak of relevant contagion and infection.
Owner:GUANGZHOU HYBRIBIO MEDICINE TECH LTD +2
Pathogenic epiphyte detection gene chip
InactiveCN101492743AQuick checkHigh sensitivityMicrobiological testing/measurementMicroorganism based processesRNA SequenceOligonucleotide
The invention relates to a gene chip for detecting pathogenic fungi, comprising a solid phase carrier, a specific oligonucleotide probe fixed on the solid phase carrier, wherein the specific oligonucleotide probe fixed on the solid phase carrier comprises oligonucleotide sequences represented by SEQ ID NO:1-SEQ ID NO:20 or DNA or RNA sequences complementary to the oligonucleotide sequences represented by SEQ ID NO:1-SEQ ID NO:20. The invention further relates to a preparation method and use method thereof. The gene chip of the invention is capable of simultaneously and rapidly detecting pathogenic fungi, has the characteristics of high sensitivity, good specificity and simple operation, does not require special instrument, and is suitable for development in clinical laboratory.
Owner:ARMY MEDICAL UNIV
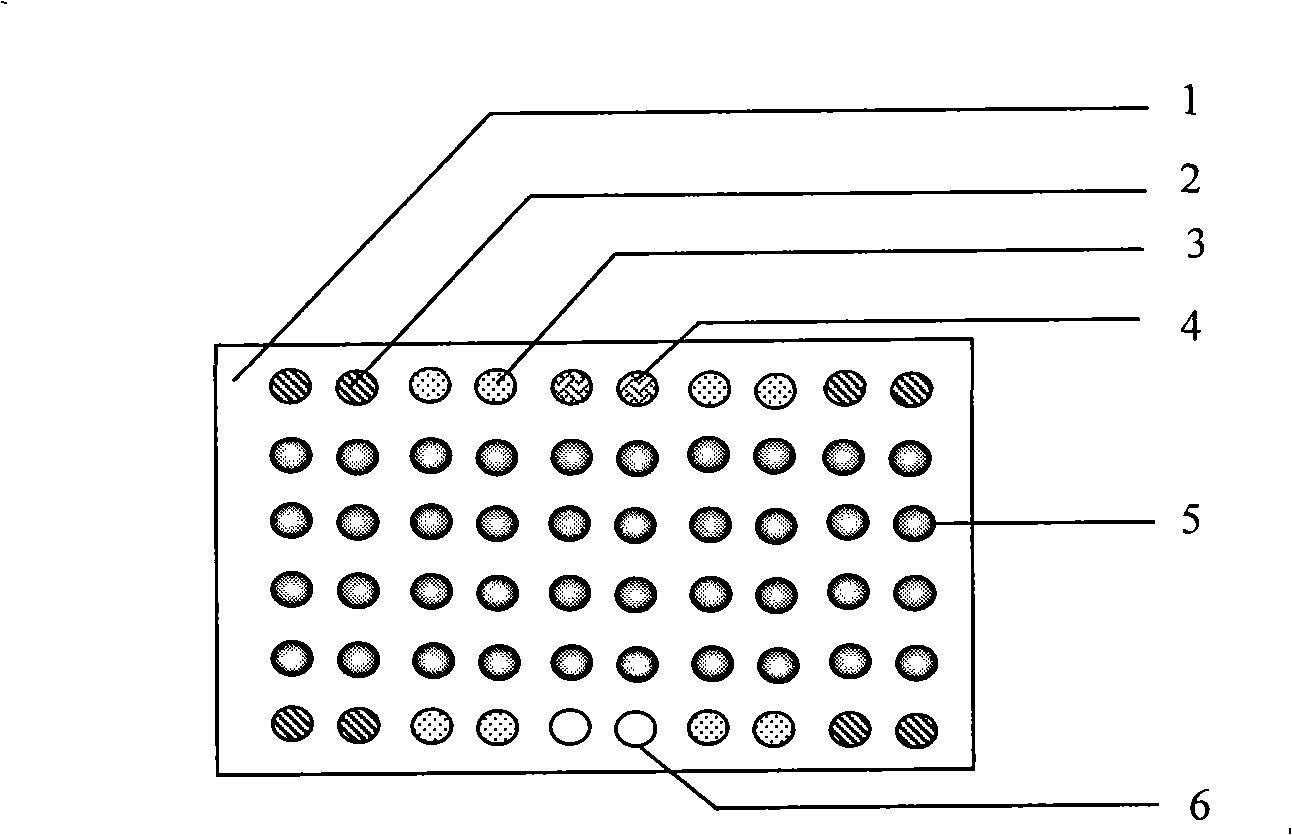
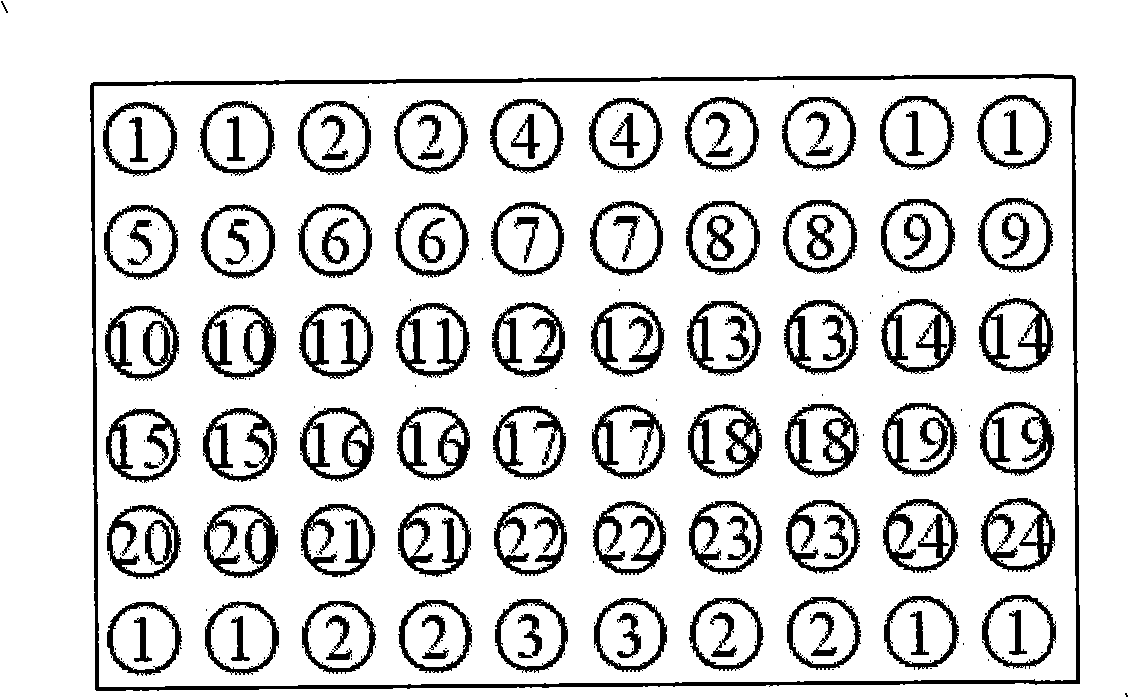
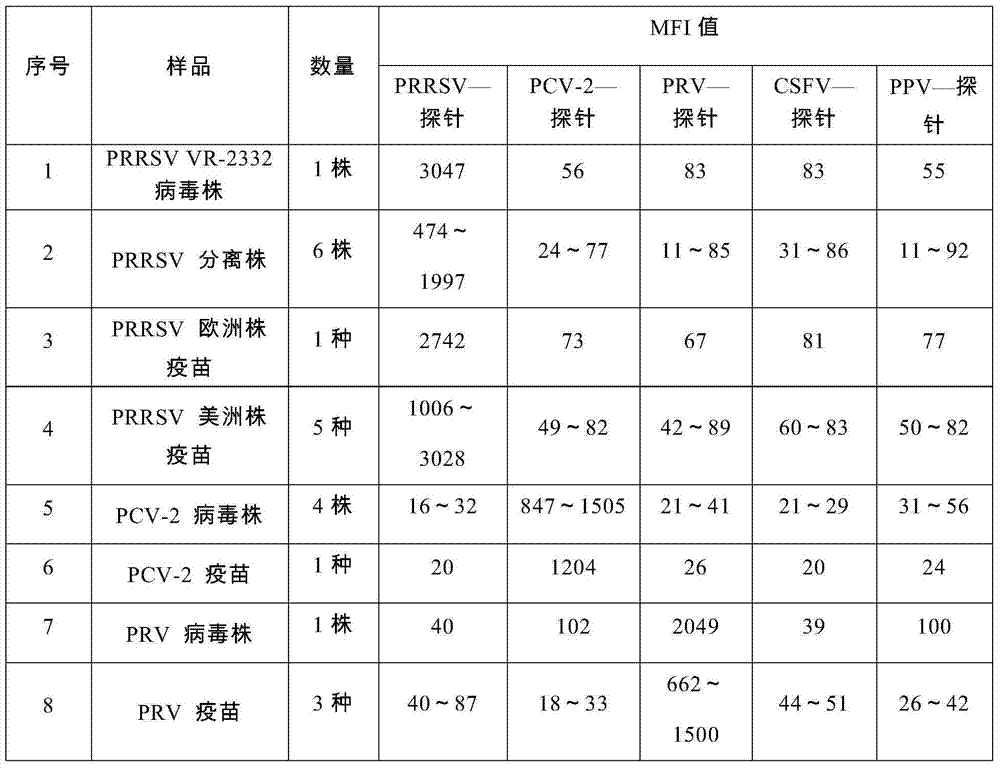
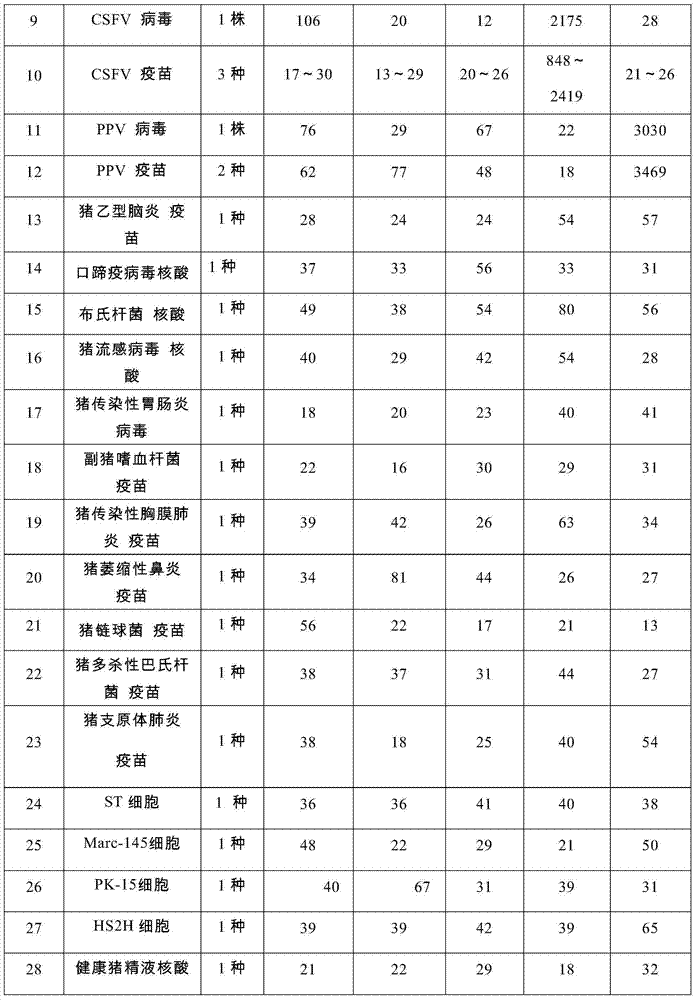
Nucleic acids of liquid-phase gene chip for synchronously detecting five porcine viruses and detection method thereof
ActiveCN104328218AHigh detection sensitivityStrong specificityMicrobiological testing/measurementMicroorganism based processesClassical swine fever virus CSFVMultiplex
The invention provides a set of nucleic acids of a liquid-phase gene chip for synchronously detecting five porcine viruses, which comprise forward and reverse primers and hybrid probes for porcine reproductive and respiratory syndrome virus (PRRSV), porcine circovirus type 2 (PCV2), porcine pseudorabies virus (PRV), classical swine fever virus (CSFV) and porcine parvovirus (PPV). The invention also provides a multiplex liquid-phase chip high-flux molecular biology detection method of the five porcine viruses. According to the method, porcine virus nucleic acids in the sample to be detected are extracted to perform multiplex unsymmetric nucleic acid amplification / multiplex liquid-phase gene chip (suspension chip) combined detection, thereby synchronously and accurately detecting and identifying the five porcine viruses in the sample to be detected. The method has the advantages of high specificity, high sensitivity, high stability, high flux and high detection speed, and is simple to operate.
Owner:INSPECTION & QUARANTINE TECH CENT OF GUANGDONG ENTRY EXIT INSPECTION & QUARANTINE BUREAU
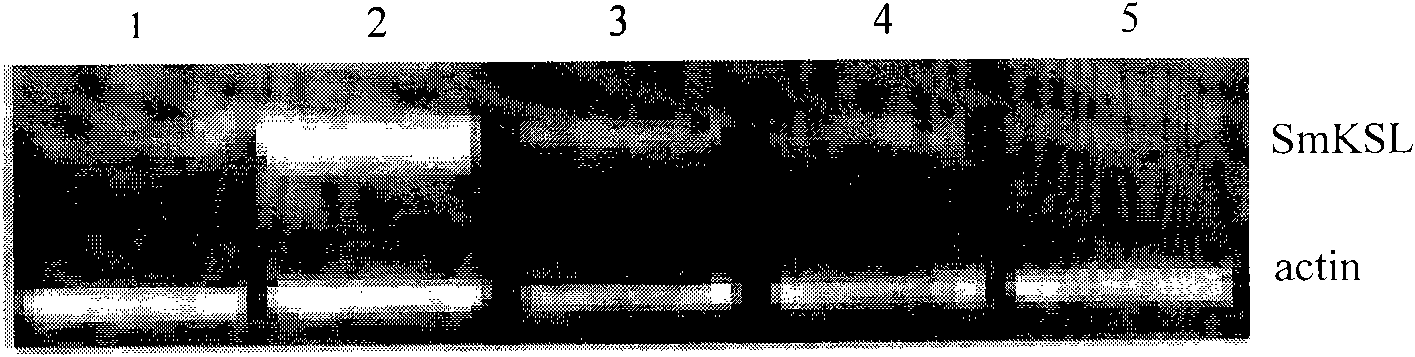
Gene for generating related diterpene synthase together with tanshinone type compound as well as encoding product and application thereof
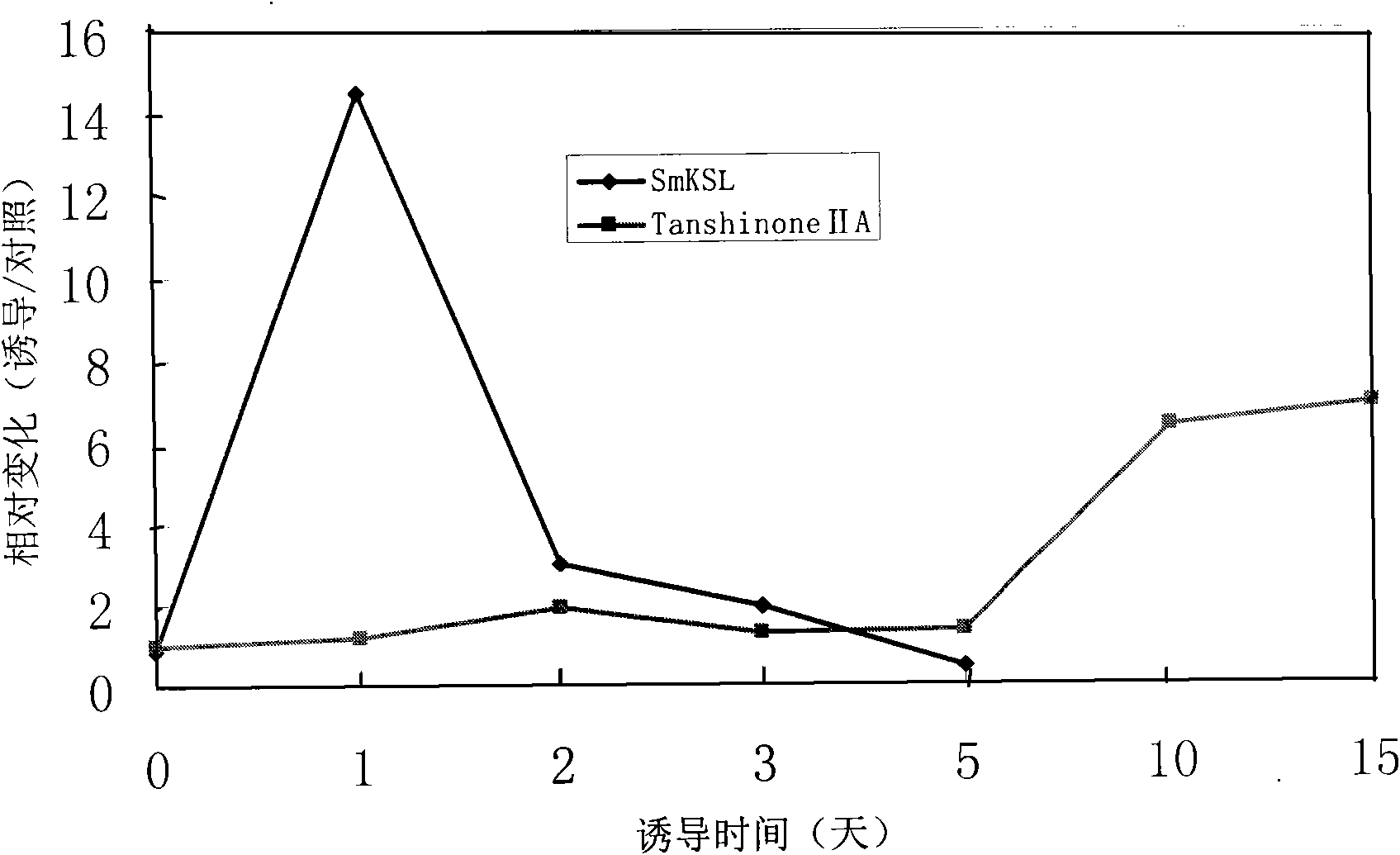
The invention discloses a gene for generating related diterpene synthase together with a tanshinone compound as well as an encoding product and an application thereof. The gene is named as the tanshin ent-kaurene synthase gene (SmKSL) and is obtained from tanshin by cloning and adopting a gene chip technology, and as for the sequence, see to SEQ ID No.1. The protein encoded by the SmKSL is the protein provided with an aminoacid residue sequence with SEQ ID No.2 or is the protein derived via the SEQ ID No. 2 by replacement, deletion or addition of one or more aminoacid residues for the aminoacid residue sequence with SEQ ID No.2 and provided with the same activity with the aminoacid residue sequence with SEQ ID No.2. The SmKSL is a key enzyme gene in a tanshin diterpene secondary metabolic pathway, the genetic expression is closely related with the generation of the tanshinone compounds, for example, tanshinone IIA, and the invention has important theoretical and practical significances for adjusting and producing plant diterpenoid compounds and cultivating the excellent medical new plant variety.
Owner:INST OF CHINESE MATERIA MEDICA CHINA ACAD OF CHINESE MEDICAL SCI
Double-probe gene mutation detecting method based on allele special amplification as well as special chip and kit thereof
ActiveCN101619352AEasy to detectAvoid the hassle of markingMicrobiological testing/measurementGenotypeOligonucleotide
The invention relates to a method for identifying gene mutation types in the field of gene analysis as well as a special chip and a kit thereof. The gene mutation detecting method comprises the following steps: taking a genome to be detected from a human tissue as a template, carrying out multiple allele special PCR amplification by a primer group that is designed aiming at special mutant sites and DNA polymerase without 3'-5' end exonclease activity, then hybridizing the obtained PCR product and an oligonucleotide probe (allele special probe) on the gene chip, and confirming mutation types of all gene sites according to the hybridizing result. The allele special probe is designed aiming at special gene types of gene mutant sites to be detected. The invention can detect gene mutations in comprehensive, systemic and high-flux ways and has light environmental pollution as well as simple and rapid operation compared with PCR-RFLP and a sequencing method.
Owner:CENT SOUTH UNIV
Food-originated pathogenic bactenium quick detection gene chip and its application
InactiveCN1536090AMicrobiological testing/measurementAgainst vector-borne diseasesFood poisoningBeta-hemolytic streptococcus
The present invention provides a gene chip for quickly detecting pathogens from food source, discloses the preparation method of said gene chip and provides 26 oligonucleotide probe sequences for detection. Said gene chip can quickly, accurately and high-effectively detect and identify the class of the pathogens in food, and its detection range includes staphylococcus aureus, Shiga's bacillus, salmonella, colibacillus 0157, bacillus proteus, mononuclear hyperplastic listerella, enterocolitis yersinia, aeruginous pseudomonads, vibrio parahaemolyticus, vibrio cholerae, bacillus cereus, beta hemolytic streptococcus, coconut fermentation pseudomonads, boticin, vibrio jejuni and bacillus perfringens, etc.
Owner:INST OF HYGIENE & ENVIRONMENTAL MEDICINE PLA ACAD OF MILITARY MEDICAL
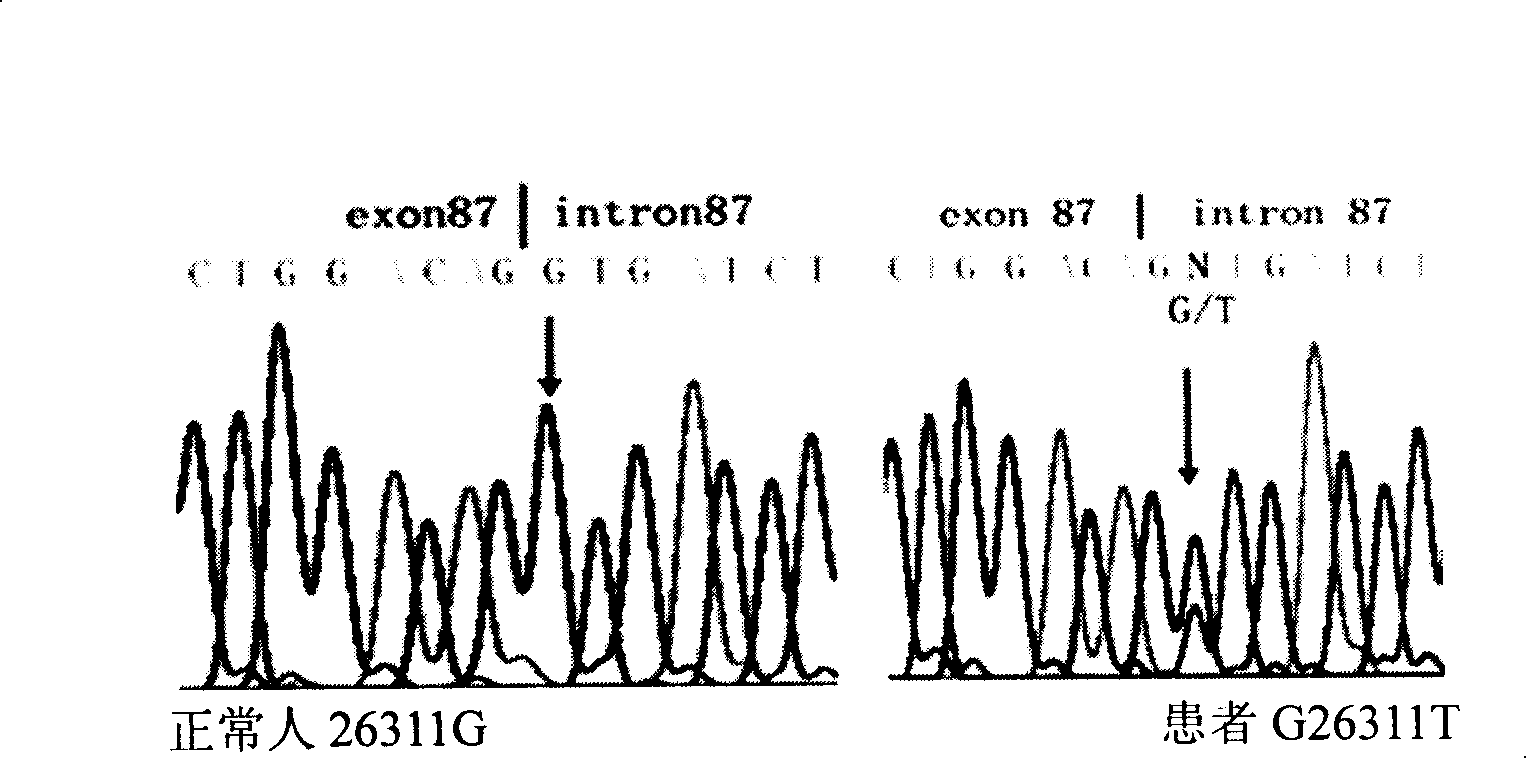
Method for detecting COL7A1 gene mutation and uses thereof
The invention discloses a method of detecting 26311 Mutation of COL7A1 gene and a usage thereof. The method adopts primers of p87F (TGGGCCTGAAATATGAGGAG) and P87R (TAGGCCACTGGAGAGACAGG) to PCR-amplify COL7A1 gene including the section of 26251-26350, implements the sequence after the purification of products, or uses BslI for the endonuclease detection after using p87Fa (TCCCACGGGGGCCCCTGGACAG) and P87Ra (TCCCCCGCCCCCACCCT GCCA) for the nest amplification of the PCR products. The usage is the application in preparations or gene chips for detecting the diagnosis of dominant dystrophic epidemolysis bullosa genealogy. The invention provides a novel way for the prenatal diagnosis of the bullosa and provides a basis for the gene therapy.
Owner:HUAZHONG UNIV OF SCI & TECH
Flu/human avian influenza virus detection gene chip and production method and use
InactiveCN101392302AShort detection timeImprove detection accuracyMicrobiological testing/measurementHighly pathogenicTyping
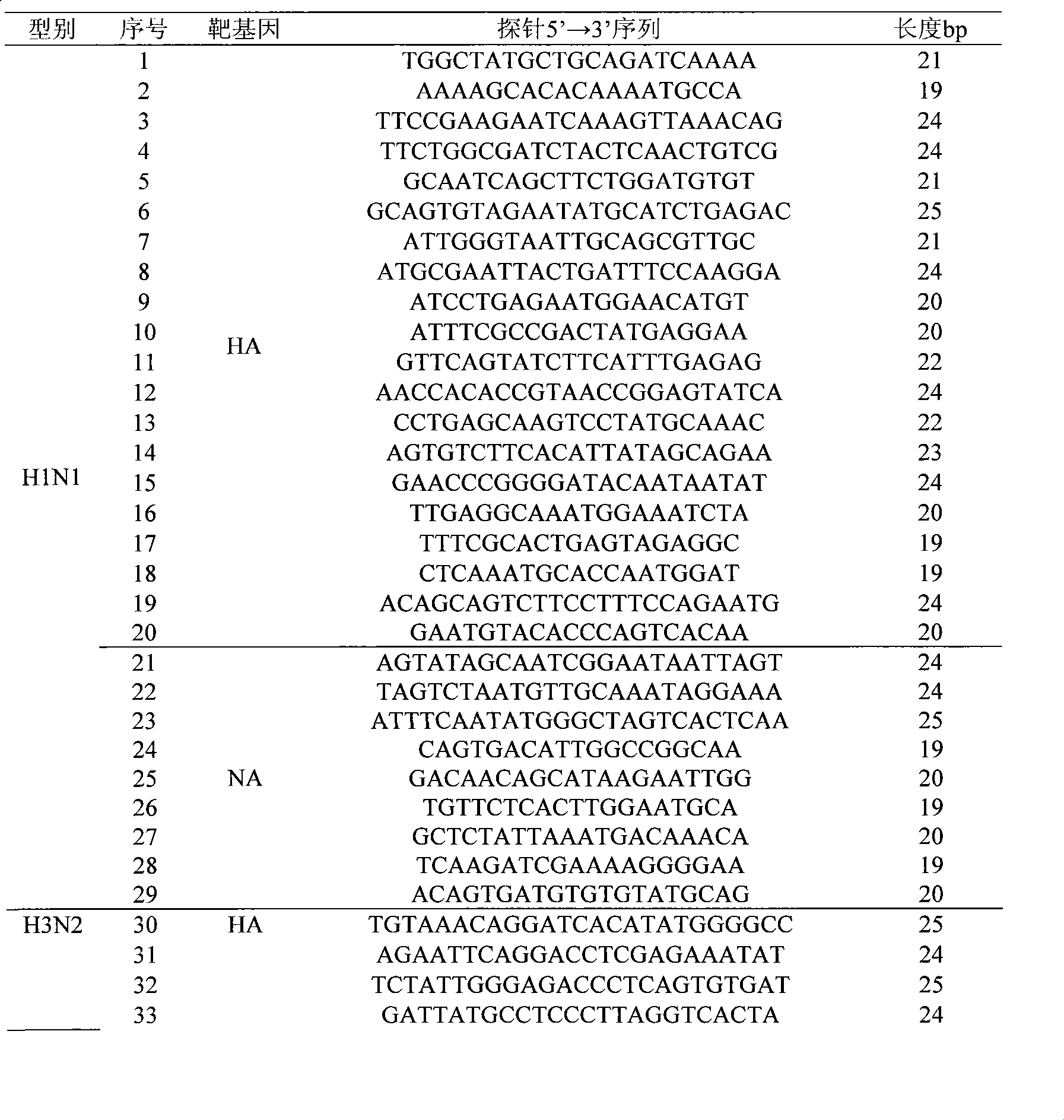
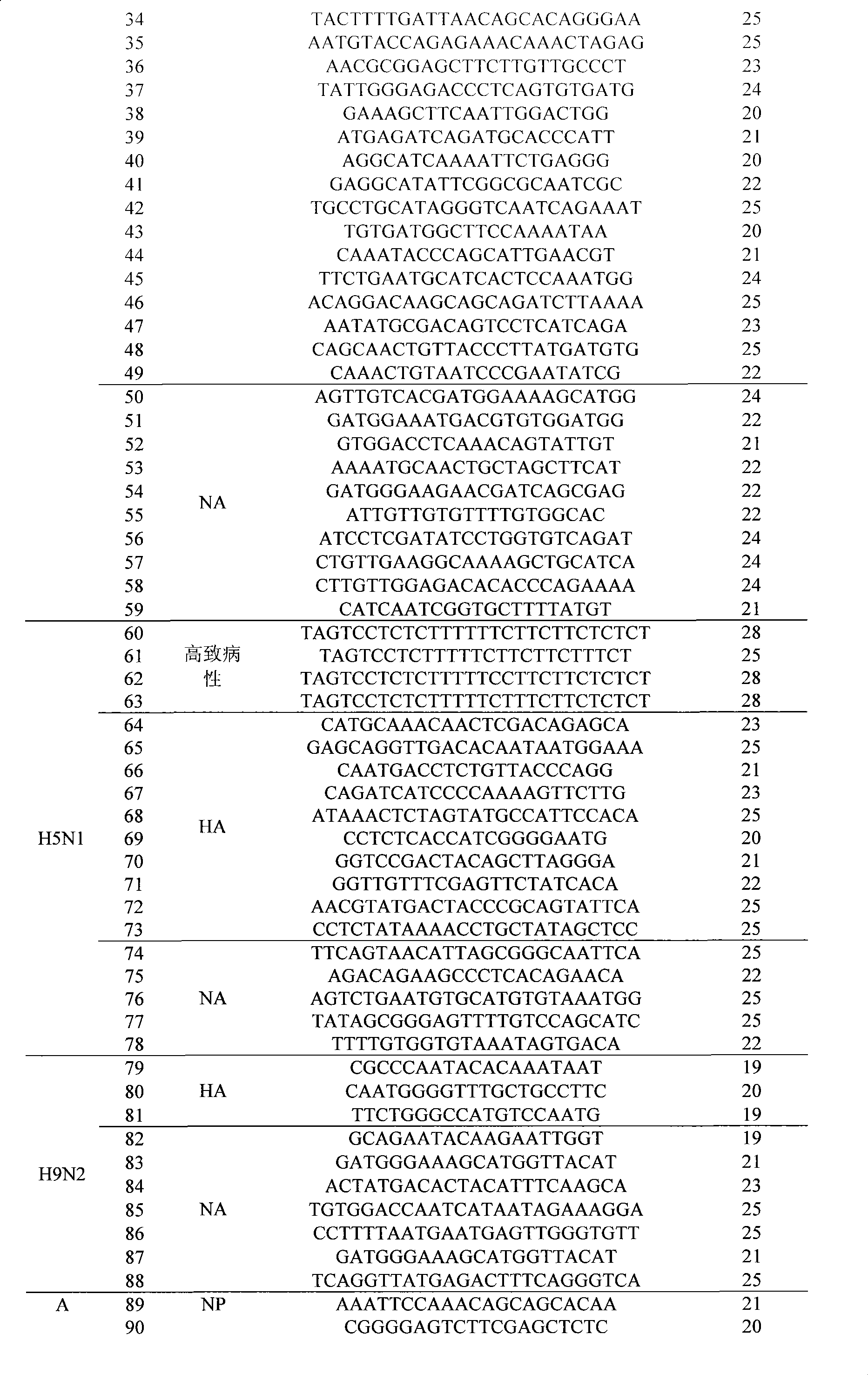
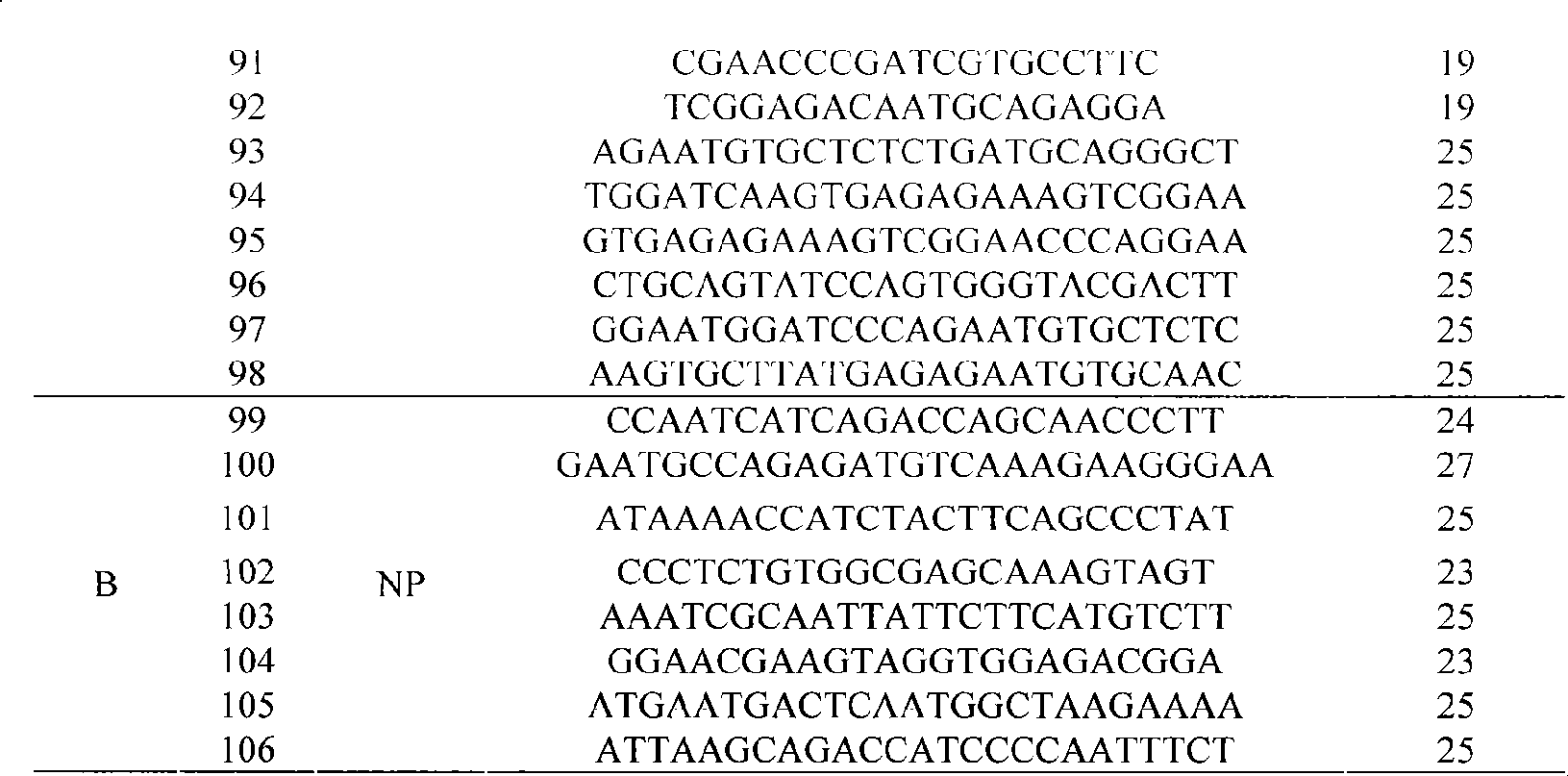
The invention discloses an influenza / human and avian influenza virus detection gene chip and a preparation method and an application thereof, 102 probes used for detecting A type, B type, H1N1 hipotype, H3N2 hipotype, H5N1 hipotype and H9N2 hipotype influenza viruses and 4 highly pathogenic differential diagnosis probes, as well as a process of sample treatment, hybridization, elution and chip identification which can carry out detection, typing or identification over the 6 types of viruses simultaneously and can identify the high pathogenicity of viruses. The invention not only greatly shortens the detection time of influenza viruses, but also improves the detection accuracy and provides a feasible technique support for the early warning mechanism of influenza epidemic situation in fields of clinical diagnosis, inspection quarantine, and the like.
Owner:中国疾病预防控制中心病毒病预防控制所 +1
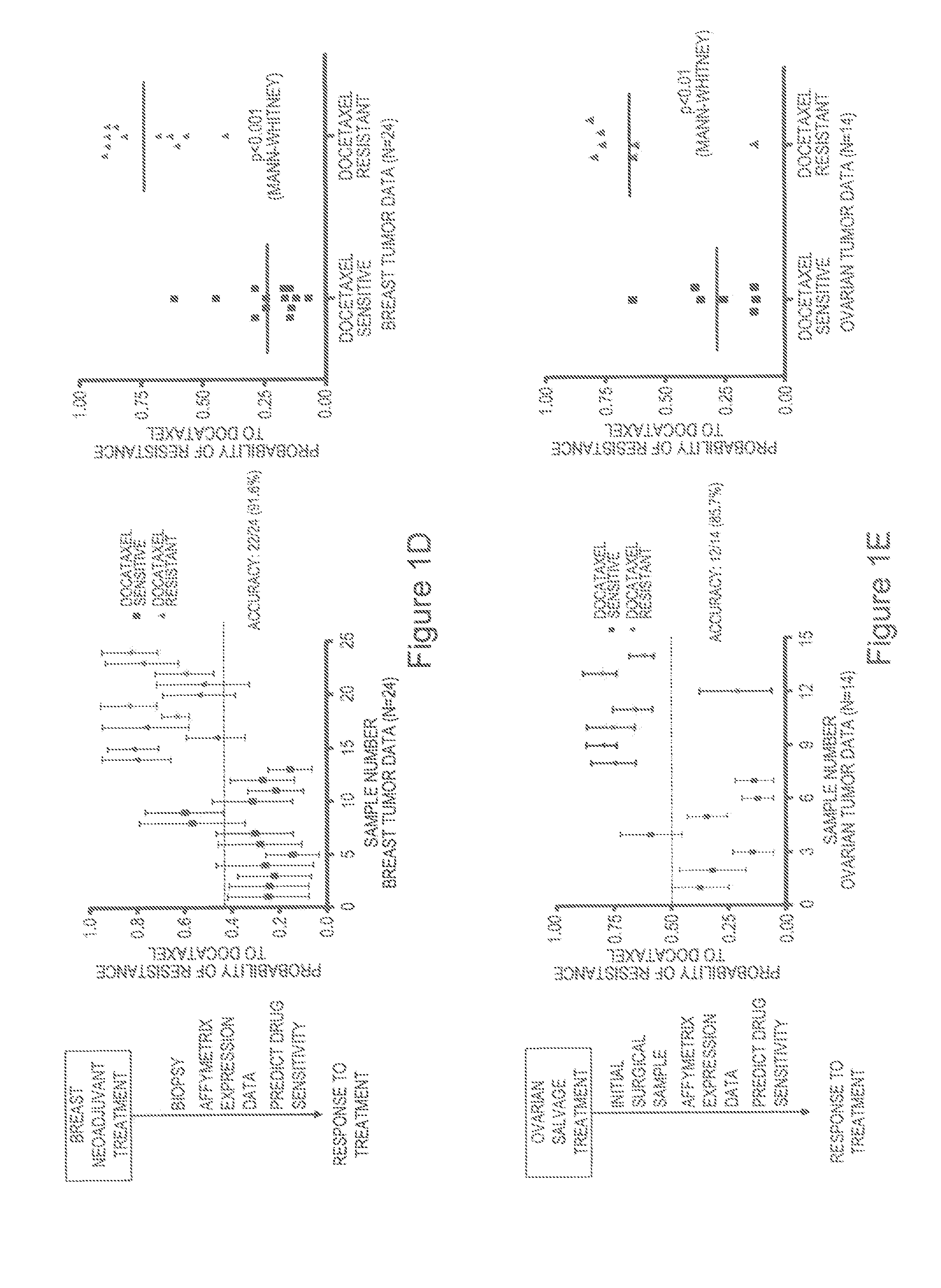
Predicting responsiveness to cancer therapeutics
Owner:UNIV OF SOUTH FLORIDA +1
Coupled two-way clustering analysis of data
InactiveUS6965831B2Lower utility and strengthValid checkData processing applicationsBiostatisticsData setFull data
Owner:YEDA RES & DEV CO LTD
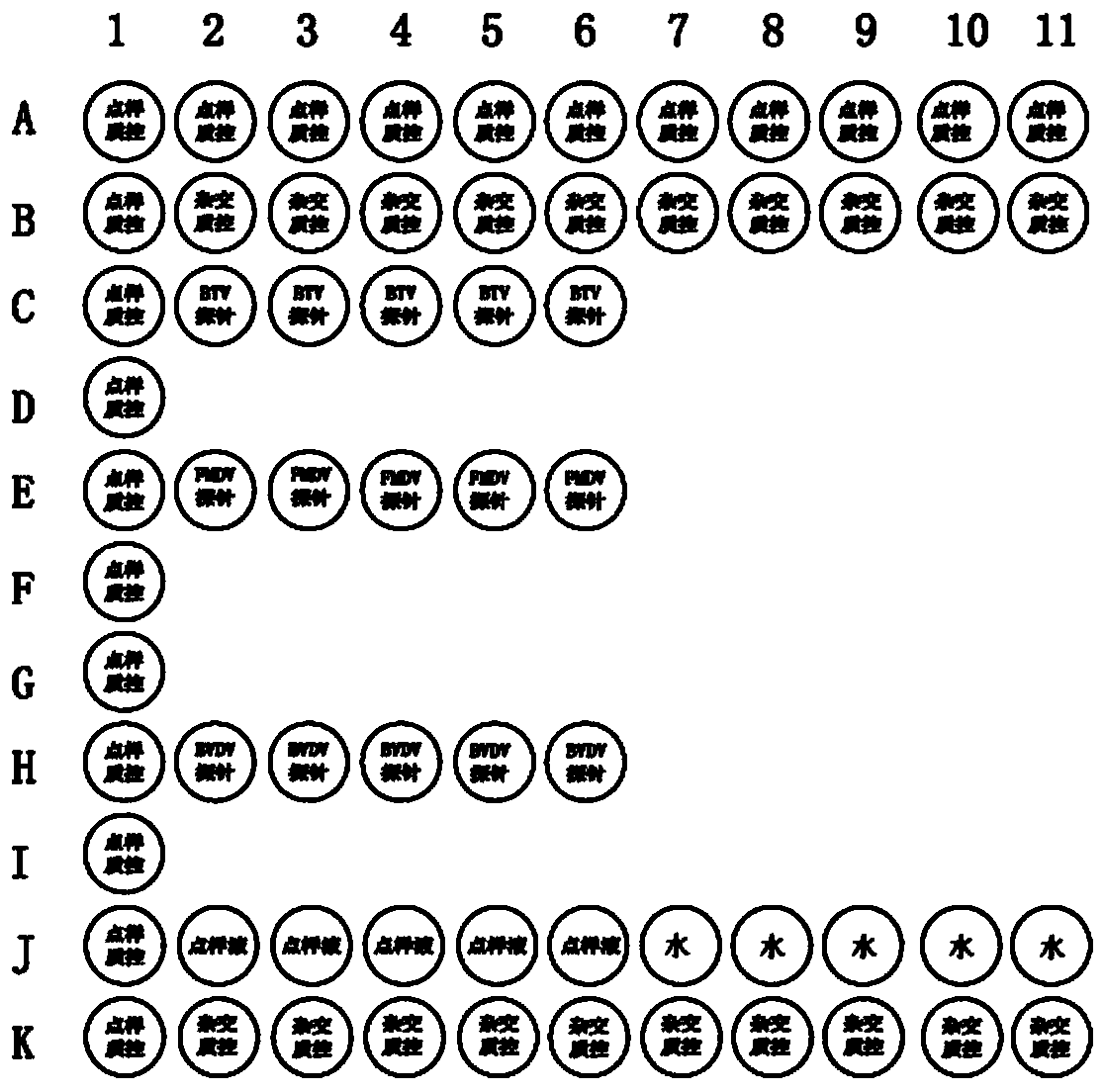
Multiplex PCR (polymerase chain reaction) primer, probe and gene chip for detecting bluetongue virus, foot and mouth disease virus and bovine viral diarrhea virus
InactiveCN103695566AImprove throughputShorten diagnostic timeMicrobiological testing/measurementDNA/RNA fragmentationForward primerMultiplex
The invention relates to a multiplex PCR (polymerase chain reaction) primer, a probe and a gene chip for detecting the bluetongue virus, foot and mouth disease virus and bovine viral diarrhea virus. The multiplex PCR primer and probe have the nucleotide sequences shown by SEQ ID No.1 to SEQ ID and No.9. The gene chip comprises a solid-phase carrier, a sample application quality control probe, a positive hybrid quality control probe and a multiplex PCR primer for detecting the bluetongue virus, foot and mouth disease virus and bovine viral diarrhea virus and the corresponding probe. In the invention, the forward primers of three viruses are marked with fluorescence, a gene chip detection technology carrying three viruses in animal fur is established based on multiplex RT-PCR (reverse transcription-polymerase chain reaction), and the RNA virus in the fur can be sensitively and specifically detected with high flux; the three viruses are screened at the same time in detection once, and the situation that a specific method is required for each virus before is changed, thereby saving the diagnosis time, meeting the needs for quick detection of mass imported / exported fur samples of the exit-entry inspection and quarantine departments and the fur import and export enterprises, and realizing relatively high application values.
Owner:徐超
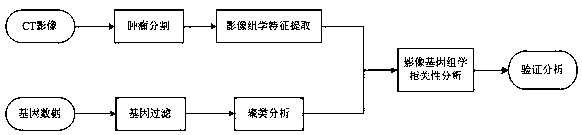
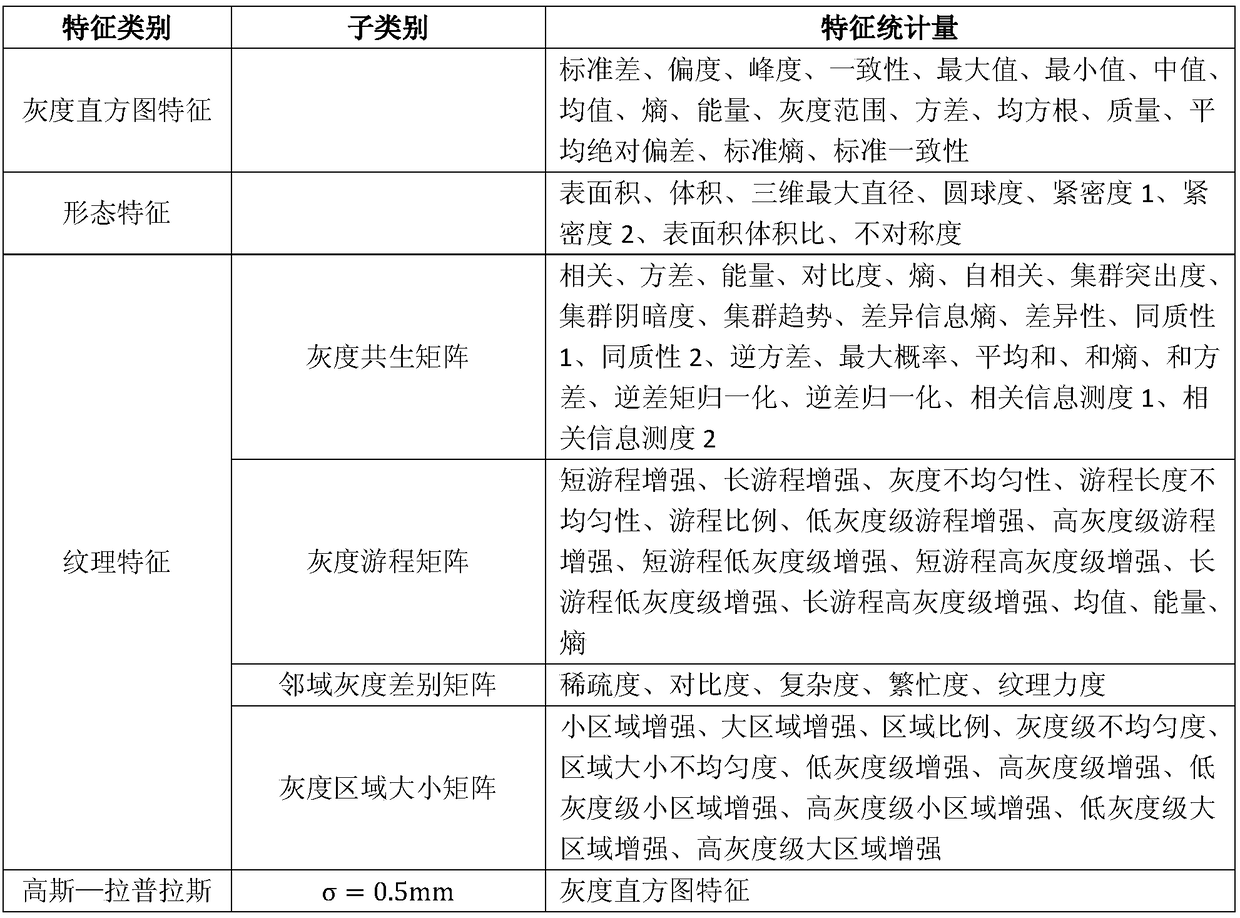
Method for analyzing correlation between CT imaging genomics characteristics and gene expression in lung cancer
InactiveCN108897984AAdjunctive Personalized TherapyImage enhancementImage analysisPersonalizationGenomics
The invention relates to a method for analyzing the correlation between CT imaging genomics characteristics and gene expression in lung cancer. The method includes: using a semi-automatic segmentationmethod to extract CT imaging genomics characteristics of a segmented tumor were; performing clustering analysis on the basis of preprocessed gene data, and taking a first principal component as a representative of a gene clustering result with a similar expression profile; and finally, using a gene chip saliency analysis algorithm to find the correlation between CT imaging genomics characteristics and gene expression in lung cancer, and verifying and analyzing the result. The invention provides a novel scheme for exploring the relationship between the image characteristics and the gene data,and attempts to find an imaging substitute of the gene, interprets the image characteristics from the gene level, and assists the personalized treatment of the tumor well.
Owner:UNIV OF SHANGHAI FOR SCI & TECH
Method related to DNA (Deoxyribose Nucleic Acid) folded paper and structure and application thereof
InactiveCN102559891ABioreactor/fermenter combinationsBiological substance pretreatmentsEngineeringA-DNA
The invention discloses a method related to DNA folded paper and a structure and an application thereof. The method comprises the following steps of: (1) preparing a DNA folded paper staple chain of a tail end modification reactant I, and arranging a connector between the tail end of the staple chain and the reactant I; (2) performing self-assembly on the staple chain obtained in the step (1) and a scaffold chain; (3) adding a reactant II which is specifically combined with the reactant I; and (4) detecting information on the combination of the reactant I and the reactant II. The connector is arranged between the reactant I and a staple short chain, so that the reaction locus of the reactant I which is effective to the reactant II on the DNA folded paper can be accurately controlled in particular to the DNA folded paper absorbed on a solid-liquid interface. The method can be taken as a detection method for a gene chip, and be used for specifically distributing reactants. The method can be used for detecting monomolecular reactions, and has extremely high sensitivity and specificity.
Owner:SHANGHAI INST OF APPLIED PHYSICS - CHINESE ACAD OF SCI
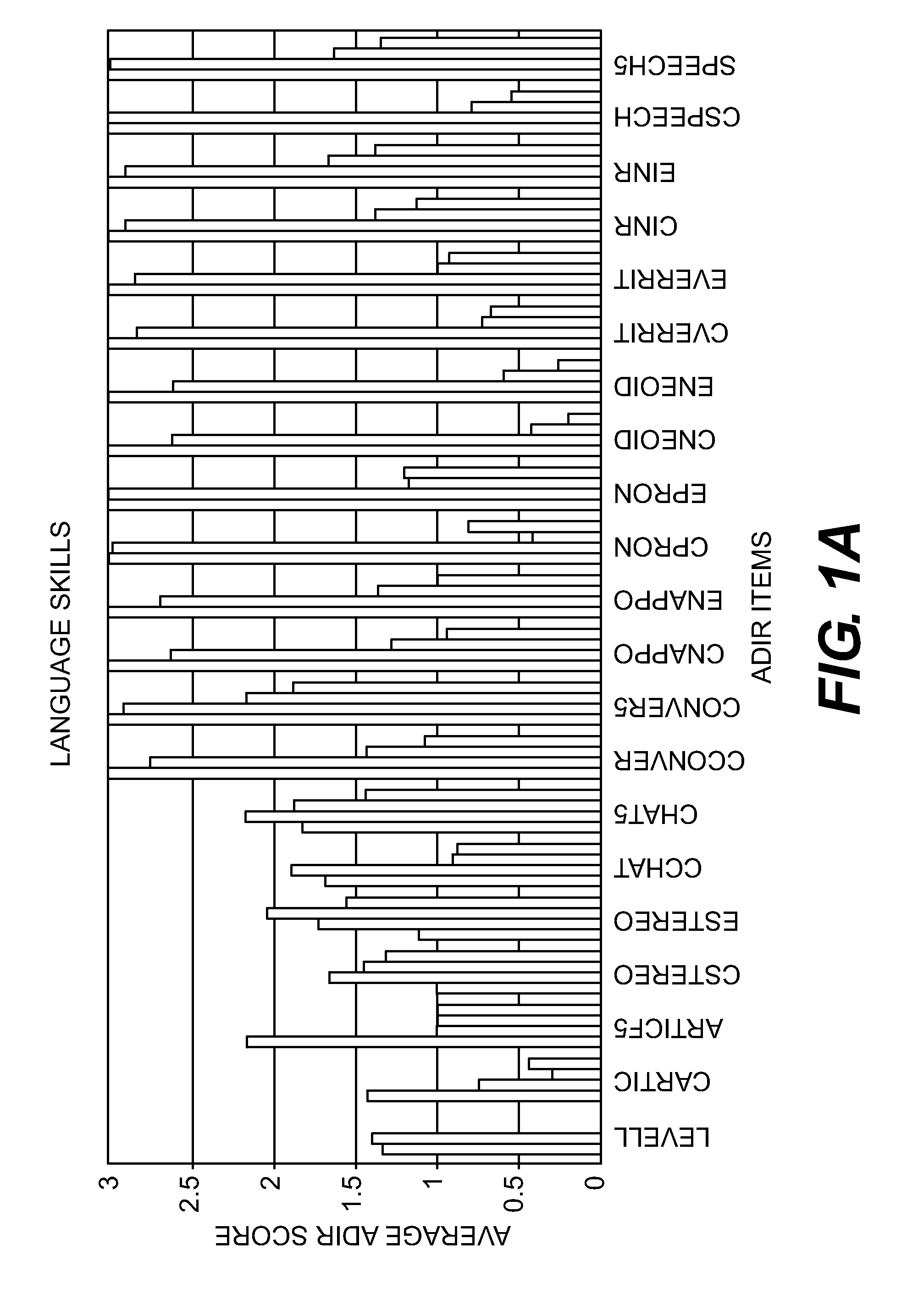
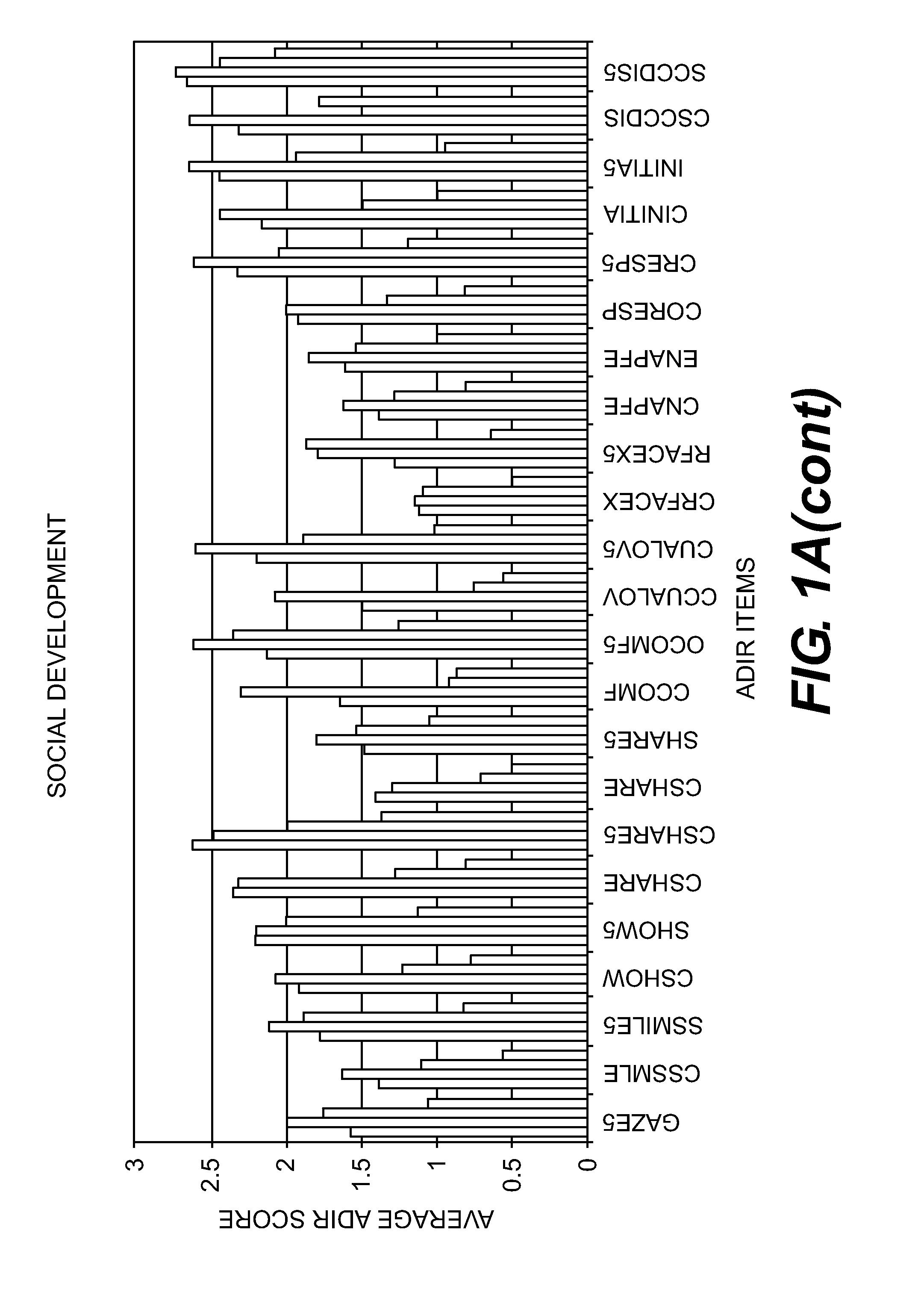
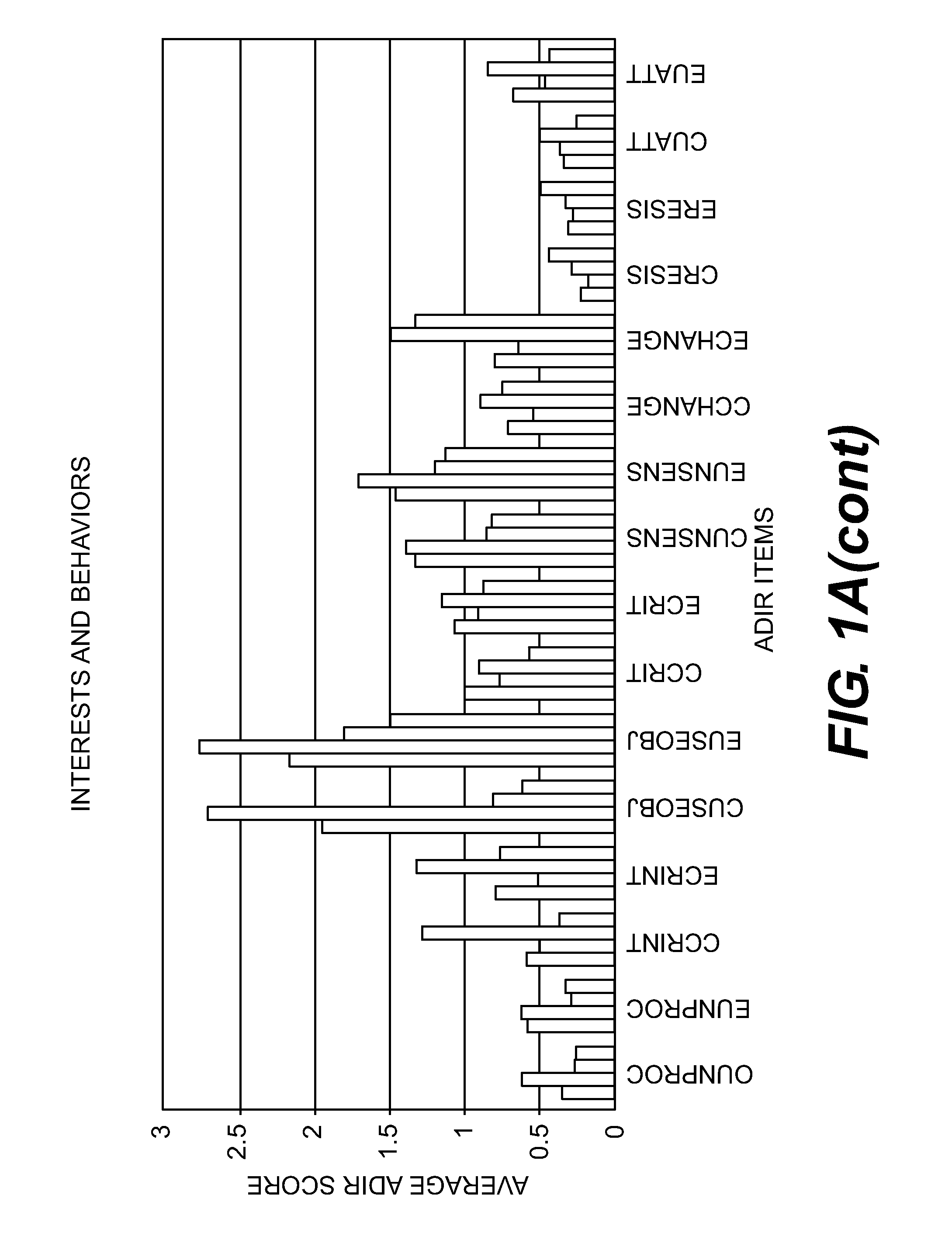
Compositions and Methods for Identifying Autism Spectrum Disorders
The compositions and methods described are directed to gene chips having a plurality of different oligonucleotides with specificity for genes associated with autism spectrum disorders. The invention further provides methods of identifying gene profiles for neurological and psychiatric conditions including autism spectrum disorders, methods of treating such conditions, and methods of identifying therapeutics for the treatment of such neurological and psychiatric conditions.
Owner:GEORGE WASHINGTON UNIVERSITY
Diagnosis and treatment methods related to aging, especially of liver
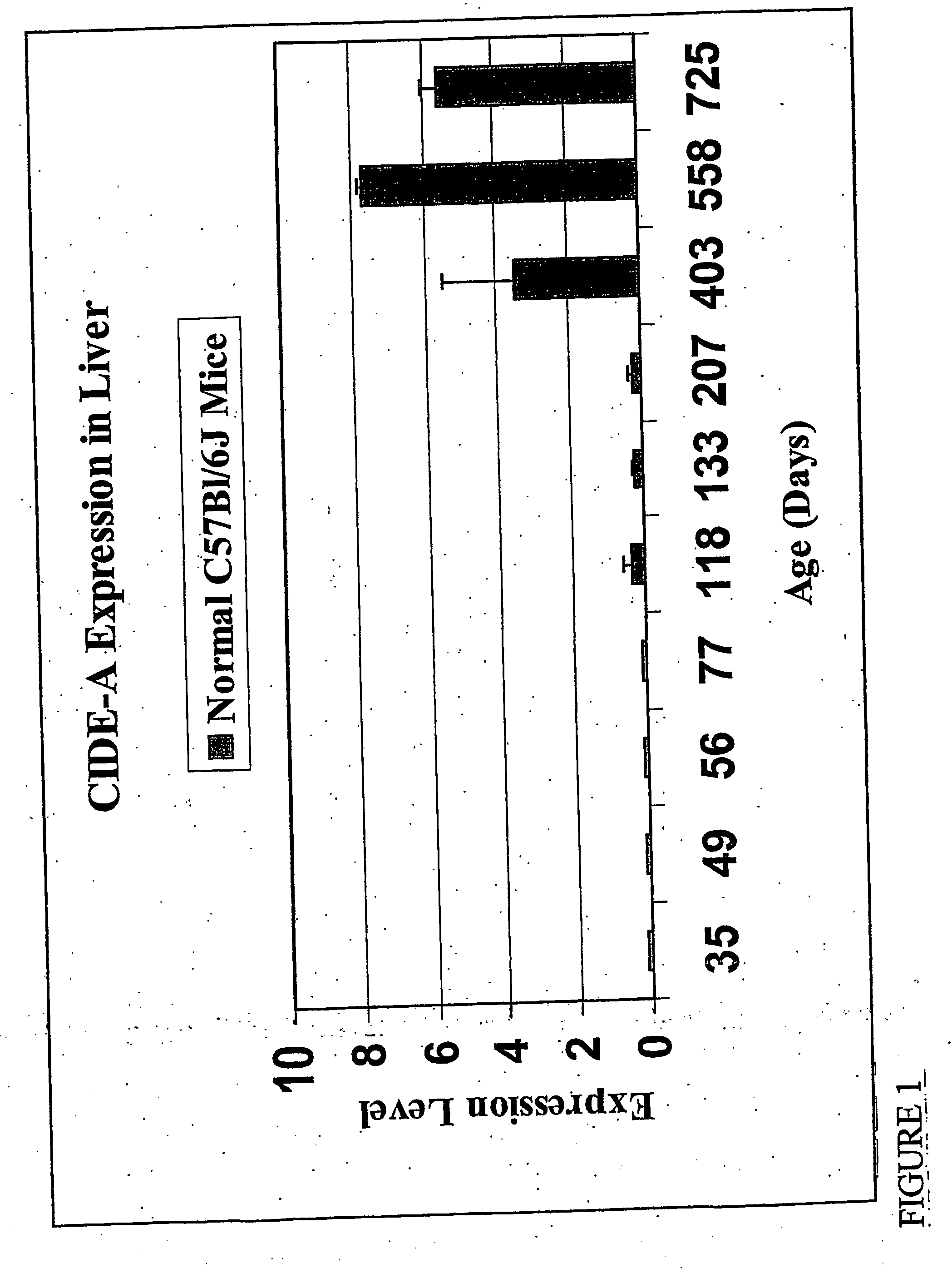
InactiveUS20070111933A1Reduce probabilityReduce severityPeptide/protein ingredientsGenetic material ingredientsHepaticaGene
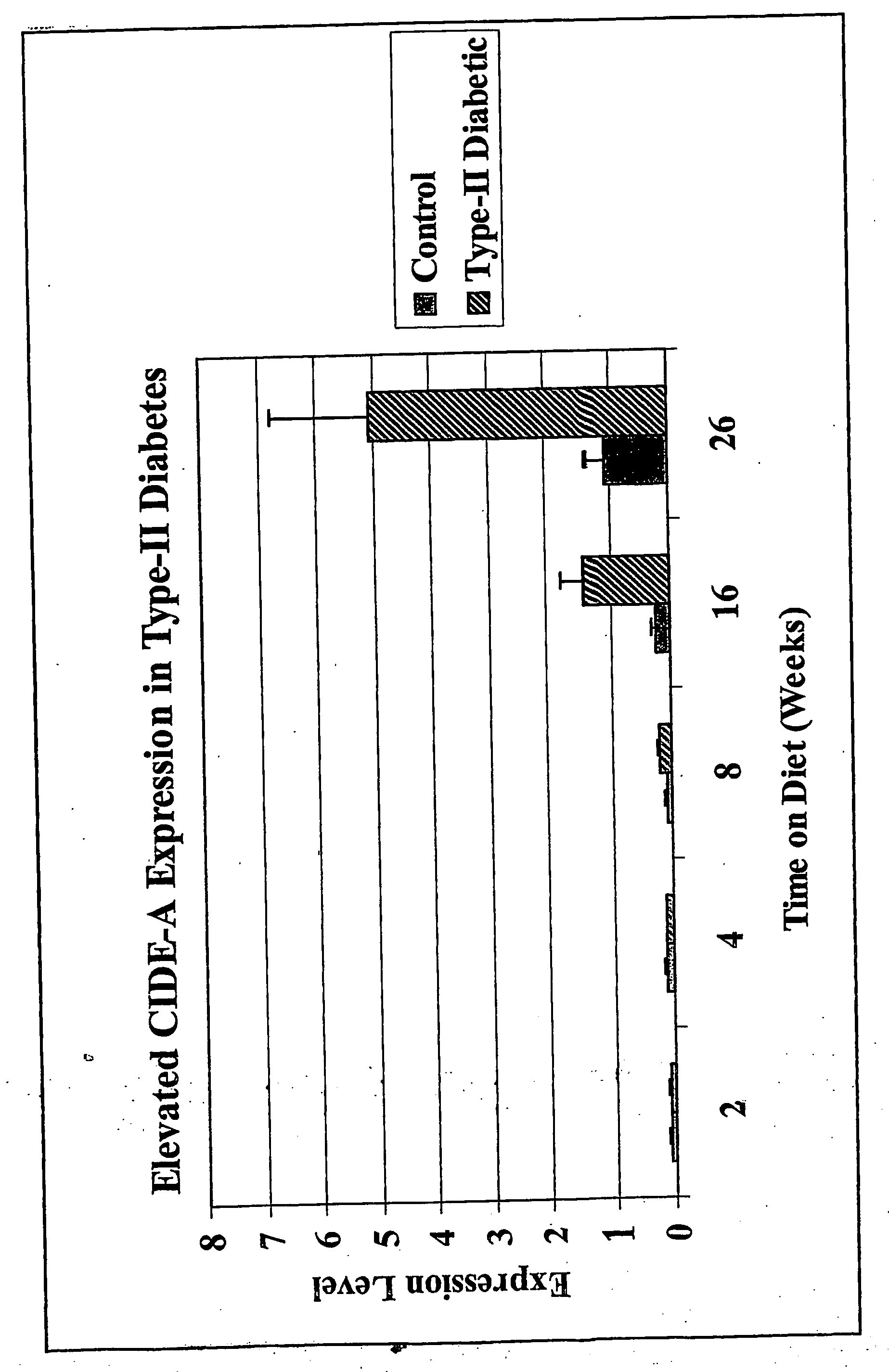
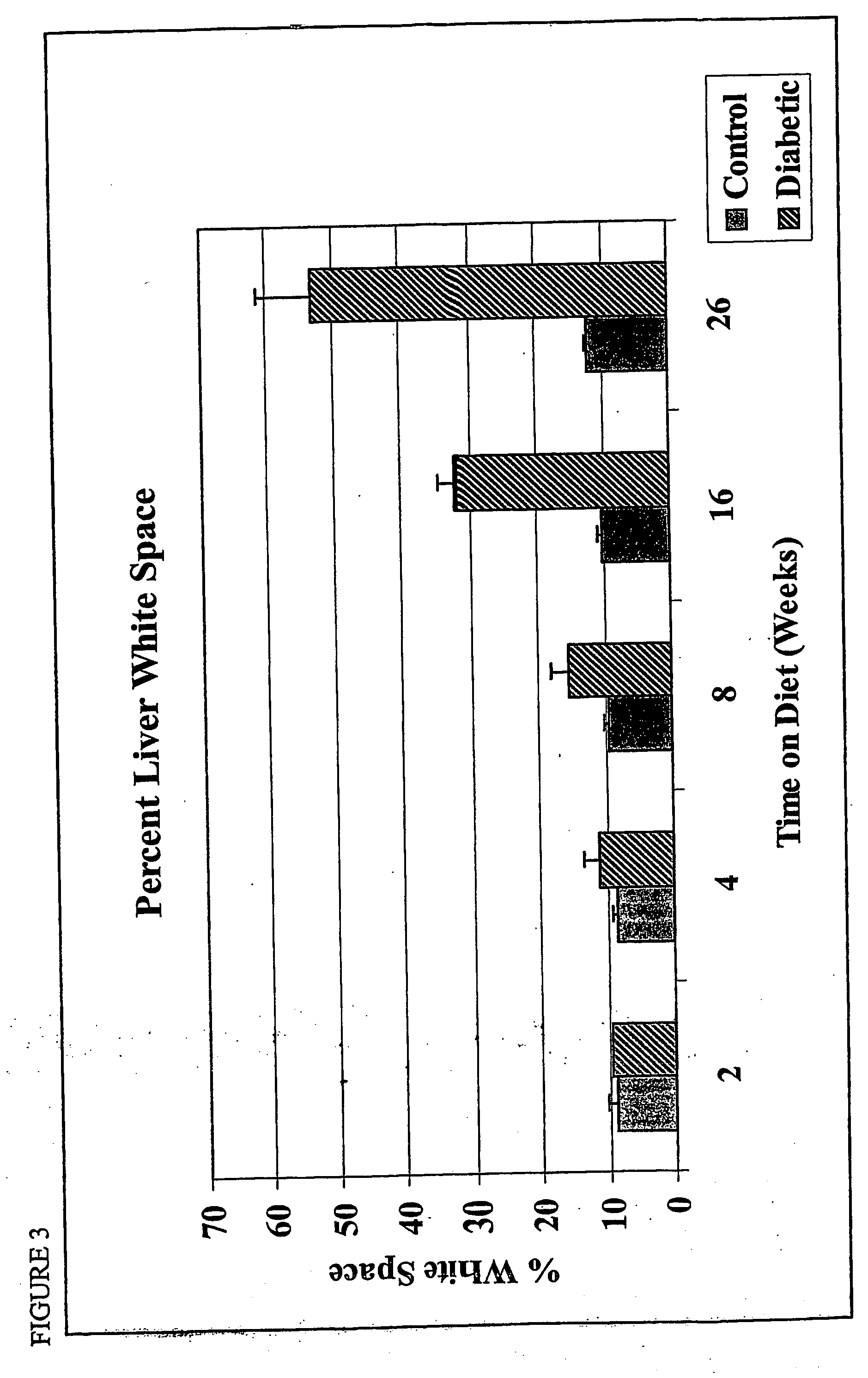
Mouse genes differentially expressed in comparisons of older and younger livers by gene chip analysis have been identified, as have corresponding human genes and proteins. The human molecules, or antagonists thereof, may be used for protection against faster-than-normal biological aging, or to achieve slower-than-normal biological aging. The human molecules may also be used as markers of biological aging.
Owner:OHIO UNIV
Methods for enhanced detection & analysis of differentially expressed genes using gene chip microarrays
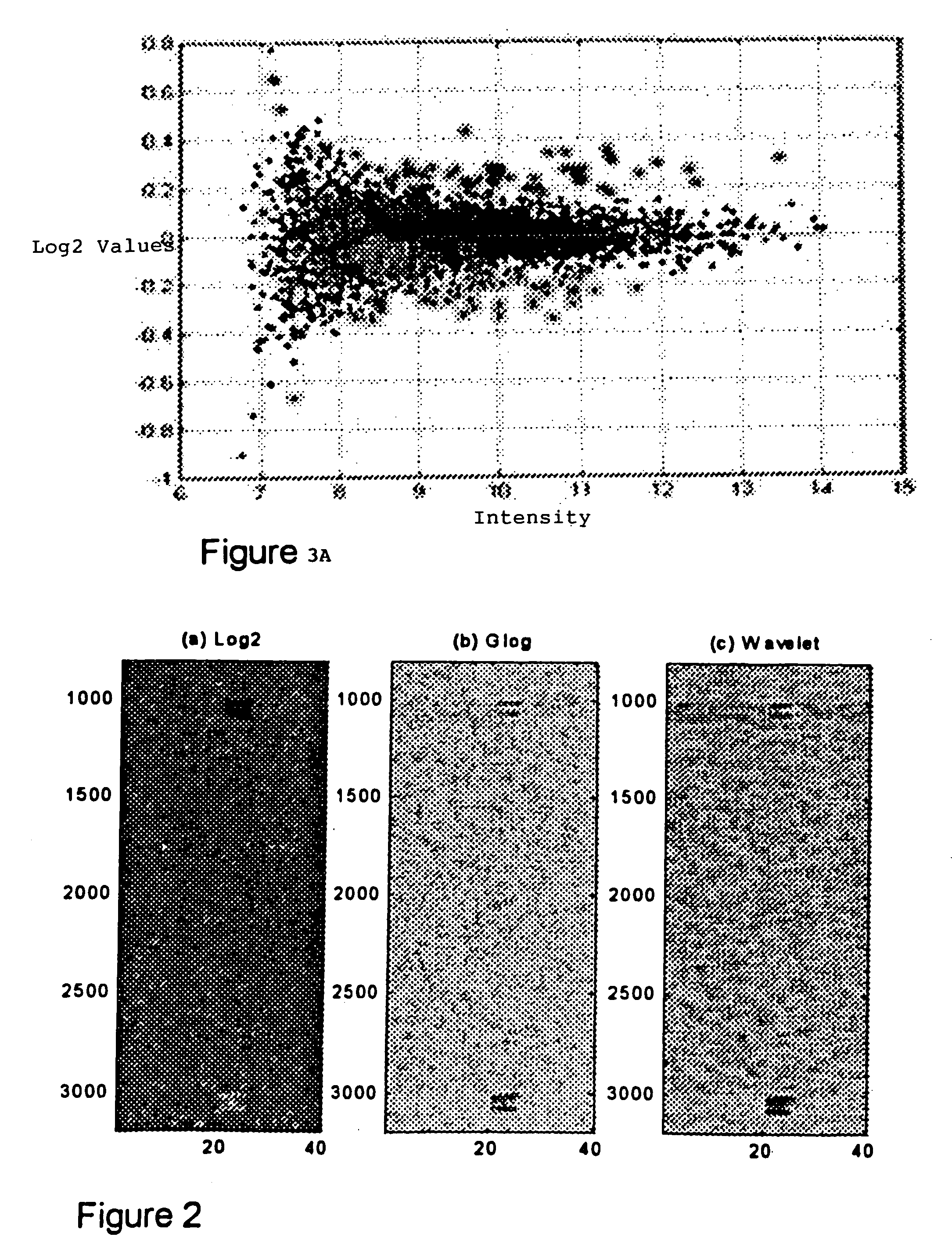
InactiveUS20050181399A1Improve signal-to-noise ratioImprove concentrationMicrobiological testing/measurementBiological testingSingular value decompositionWavelet denoising
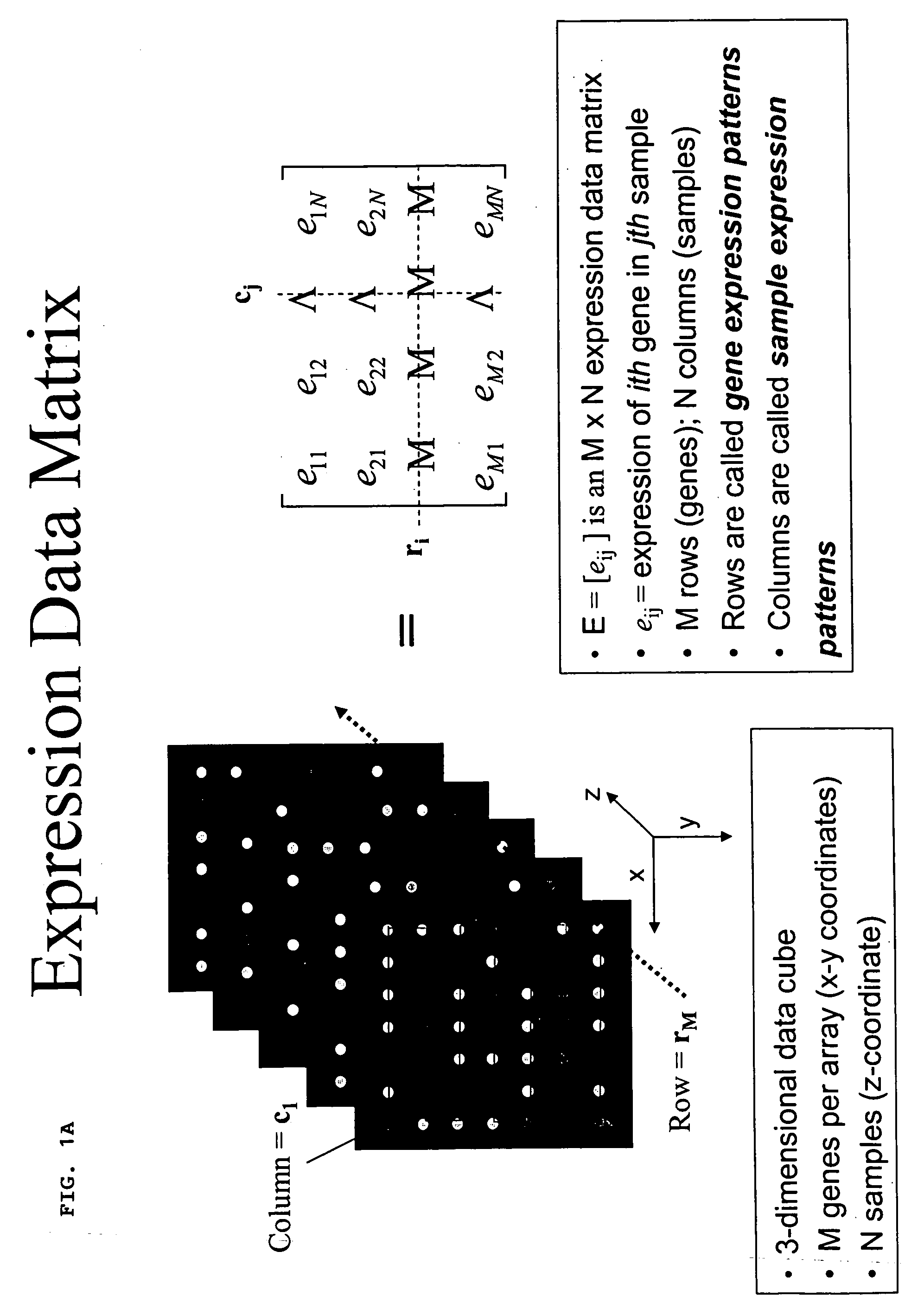
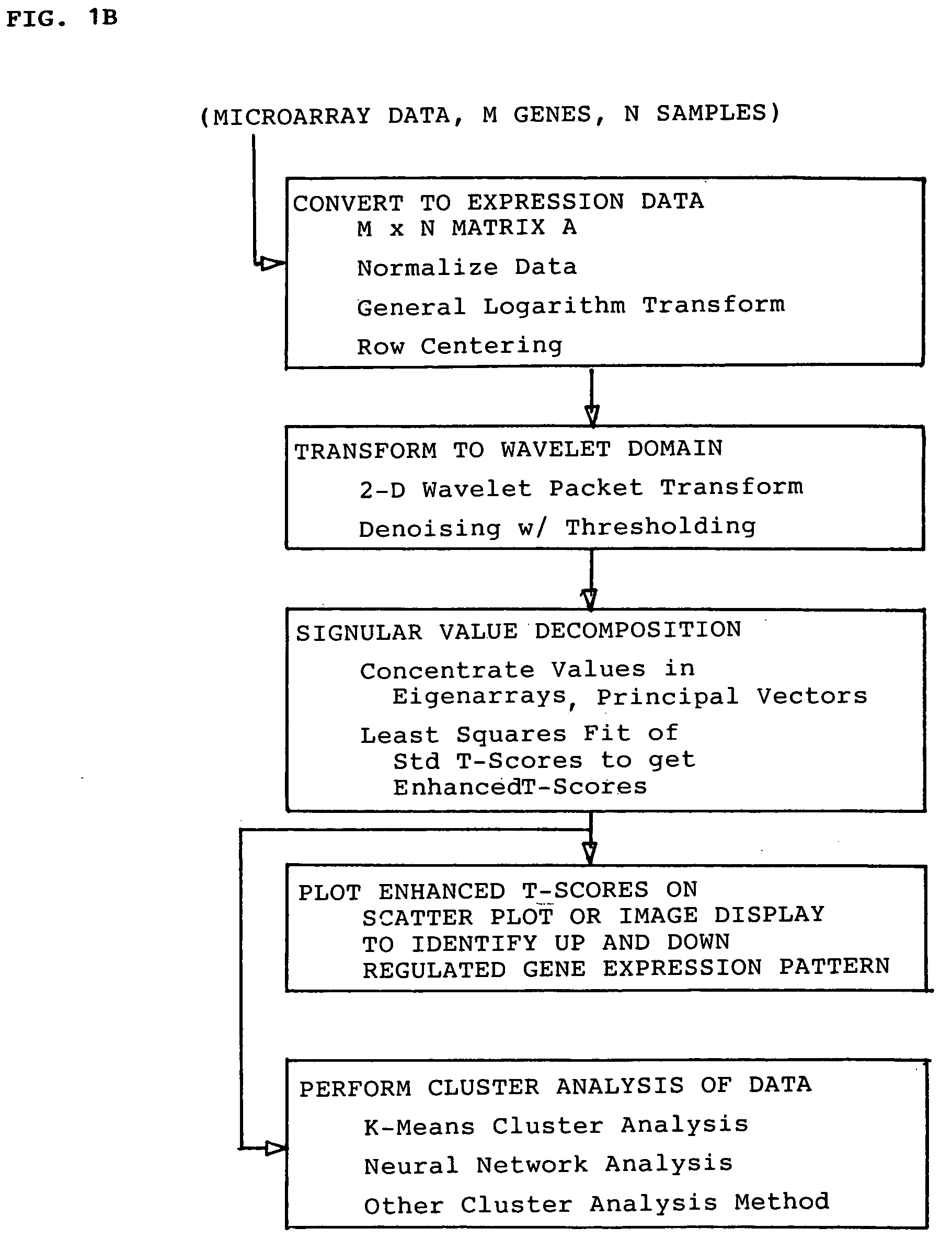
A method for enhanced detection and statistical analysis of differentially expressed genes in gene chip microarrays employs: (a) transformation of gene expression data into an expression data matrix (image data paradigm); (b) wavelet denoising of expression data matrix values to enhance their signal-to-noise ratio; and (c) singular value decomposition (SVD) of the wavelet-denoised expression data matrix to concentrate most of the gene expression signal in primary matrix eigenarrays to enhance the separation of true gene expression values from background noise. The transformation of gene chip data into an image data paradigm facilitates the use of powerful image data processing techniques, including a generalized logarithm (g-log) function to stabilize variance over intensity, and the WSVD combination of wavelet packet transform and denoising and SVD to clearly enhance separation of the truly changed genes from background noise. Detection performance can be assessed using a true false discovery rate (tFDR) computed for simulated gene expression data, and comparing it to estimated FDR (eFDR) rates based on permutations of the available data. Where a small number (N) of samples in a group is involved, a pair of specific WSVD algorithms are employed complementarily if N>5 and if N<6.
Owner:UNIV OF HAWAII
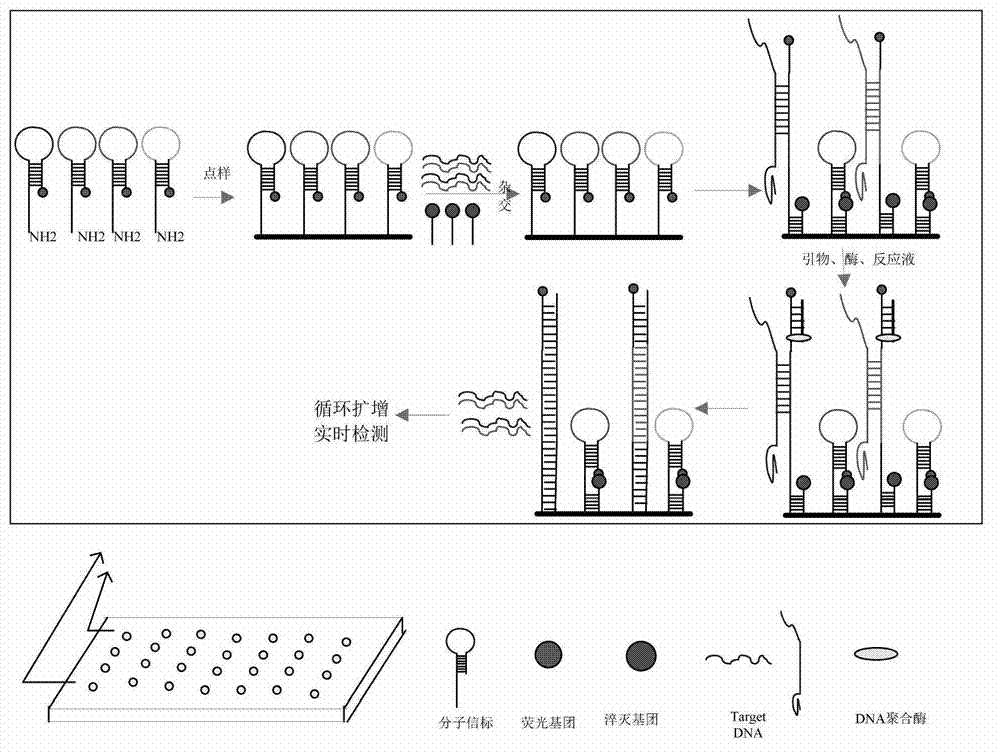
Detection technology of molecular beacon strand displacement isothermal amplifying gene chip and kit
ActiveCN102888457AReduce usageAvoid the influence of steric hindranceMicrobiological testing/measurementFluorescence/phosphorescenceDeoxyriboseGenomic clone
The invention discloses a detection technology of molecular beacon strand displacement isothermal amplifying gene chip and a kit. The detection technology disclosed by the invention combines the advantages of strand displacement isothermal amplifying technology and molecular beacon technology. According to the detection technology, DNA (Deoxyribose Nucleic Acid) is amplified under the isothermal condition; fluorescence signals of the molecular beacon are continuously released along with the amplification of the DNA, and the released target DNA are combined to new molecular beacon, so that the amplification and releasing of the fluorescence signals are carried out in turn. According to the detection technology, the purpose of amplifying the signals is achieved based on the strand displacement reaction of DNA polymerase; and the purpose of detecting signals is achieved based on open loop and closed loop of the molecular beacon. With the adoption of the detection technology disclosed by the invention, the amplification of the DNA / RNA (Ribose Nucleic Acid) and the detection of signals are integrated, the processes such as additional marketing, hybridizing and washing are saved, so that, the operation steps are simplified, the efficiency is improved; and moreover, the chip can be detected on real time, and accurate and reliable results are obtained.
Owner:武汉中科志康生物科技有限公司
Nucleotide specific to Vibrio fluvialis O2, O4, O13, O15 and O18 and application thereof
ActiveCN103898108APracticalImprove accuracyMicrobiological testing/measurementDNA/RNA fragmentationNucleotideSerotype
The invention relates to a nucleotide specific to Vibrio fluvialis O2, O4, O13, O15 and O18 serotypes and an application thereof. The nucleotide comprises 1) at least one of nucleotides shown in SEQIDNO: 1-14; and 2) at least one of nucleotides shown in SEQIDNO: 1-14. The nucleotides can be used for preparing PCR (polymerase chain reaction) kits and gene chips for detecting Vibrio fluvialis. The nucleotide specific to Vibrio fluvialis O2, O4, O13, O15 and O18 serotypes disclosed by the invention, as well as a PCR kit and a gene chip which contain the nucleotide, are strong in practicability, and the PCR kit is simple in preparation method, short in detection cycle, rapid in speed, strong in maneuverability, convenient for industrialized production, and low in detection cost; the accuracy is high; and the sensitivity is high.
Owner:NANKAI UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com