Method for detecting COL7A1 gene mutation and uses thereof
A genetic and functional technology, applied in the field of genetic testing, can solve problems such as indistinguishability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Next, the COL7A1 gene mutation was detected by gene sequencing.
[0044] 1. PCR amplification of COL7A1 gene coding region
[0045] Objects: The families with dystrophic epidermolysis bullosa collected in outpatient clinics included 9 patients, 11 healthy people and 200 normal controls. Detailed physical examination was performed on all participants, and 5ml blood samples were collected from each participant after signing the informed consent form.
[0046] Genomic DNA extraction: using phenol chloroform extraction method.
[0047] 1. Anticoagulant blood was diluted 1-fold with PBS.
[0048] 2. Add 2 times the volume of lymphatic separation solution (18°C-28°C) into the centrifuge tube, spread a layer of 1 times the volume of diluted blood on top, and centrifuge at 1000*g for 20 minutes at room temperature.
[0049] 3. Discard the supernatant, carefully suck out the nucleated cell layer in the middle, transfer to a 5ml Ep tube, 5000g, 10 minutes, and then wash once w...
Embodiment 2
[0096] Next, the change of said base is detected by restriction fragment length polymorphism.
[0097] 1. PCR amplification of intron 86 / exon 87 of COL7A1 gene
[0098] 1. Use primers p87F and p87R and other reagents to PCR amplify 86 / exon 87 of the sample DNA. For the steps, see the first part of Example 1.
[0099] 2. After the PCR product was diluted 200 times, take 1 μl as a template for the second round of PCR. The primers are as follows: p87Fa: 5'-TCCCACGGGGGCCCCTGGACAG-3' and p87Ra: 5'-TCCCCCGCCCCCACCCTGCCA-3'. For other reagents and steps, see Example 1 first part.
[0100] 3. For the purification of PCR products, refer to steps 1-5 in the second part of Example 1 for the steps.
[0101] 4. Add 1 U of Bsl I (NEB Company) to 20 μl of PCR product, 2.5 μl of 10*buffer, add 25 μl of water to make up, and incubate at 55°C for 2 hours.
[0102] 5. Use 1.5% agarose gel for electrophoresis, 1*TAE electrophoresis buffer, voltage 5V / cm, electrophoresis for 30 minutes, observe u...
Embodiment 3
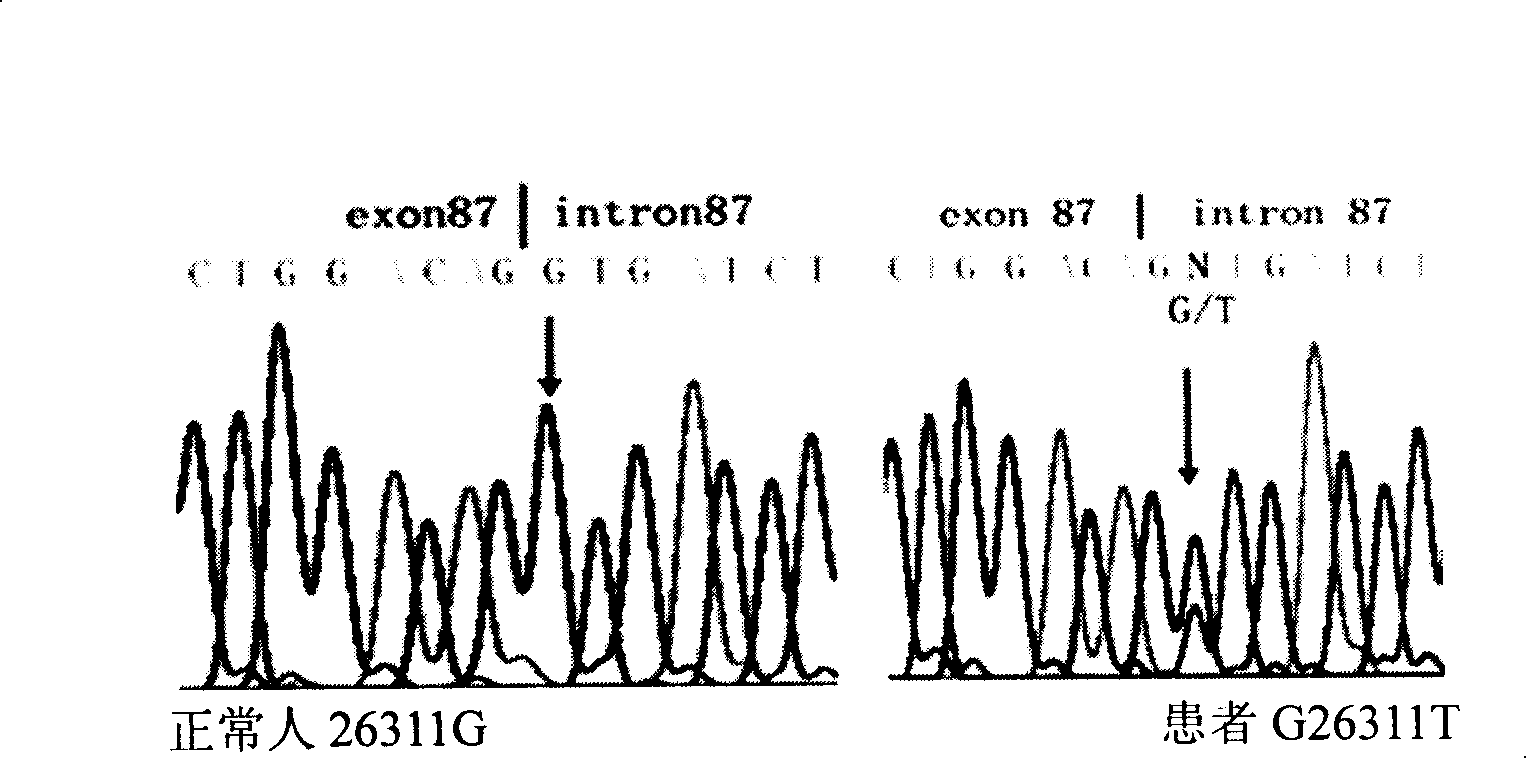
[0104] Kit for G26311T point mutation of the causative gene COL7A1 of dystrophic epidermolysis bullosa and its application
[0105] 1 kit contains:
[0106] Primers for amplification: p87F: TGGGCCTGAAATATGAGGAG
[0107] P87R: TAGGCCACTGGAGAGACAGG
[0108] Taq enzyme 5u / ul for PCR amplification
[0109] 10* buffer (containing 15ml MgCl 2 . )
[0110] dNTP2mM
[0111] Big Dyemix
[0112] 2 How to use:
[0113] It mainly includes the following steps:
[0114] A. extract DNA, utilize above-mentioned primer, carry out PCR reaction;
[0115] B: The PCR reaction product is directly sequenced after purification, and the obtained sequence is compared with the standard sequence in Genebank to determine the existence of the mutation site.
[0116] The specific method is referring to embodiment 1.
[0117] The base change can also be detected by specific hybridization of the COL7A1 gene isolated from the tissue with the COL7A1 allele.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com