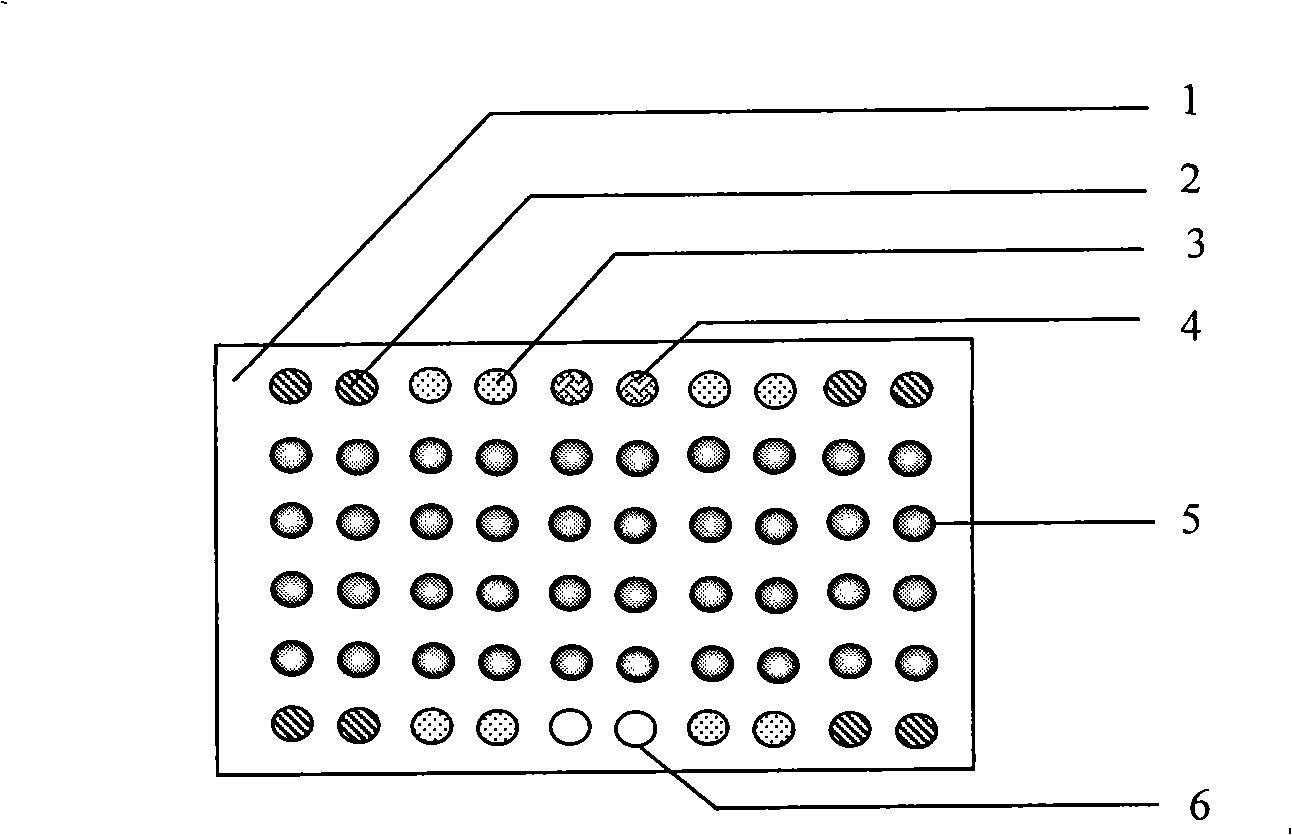
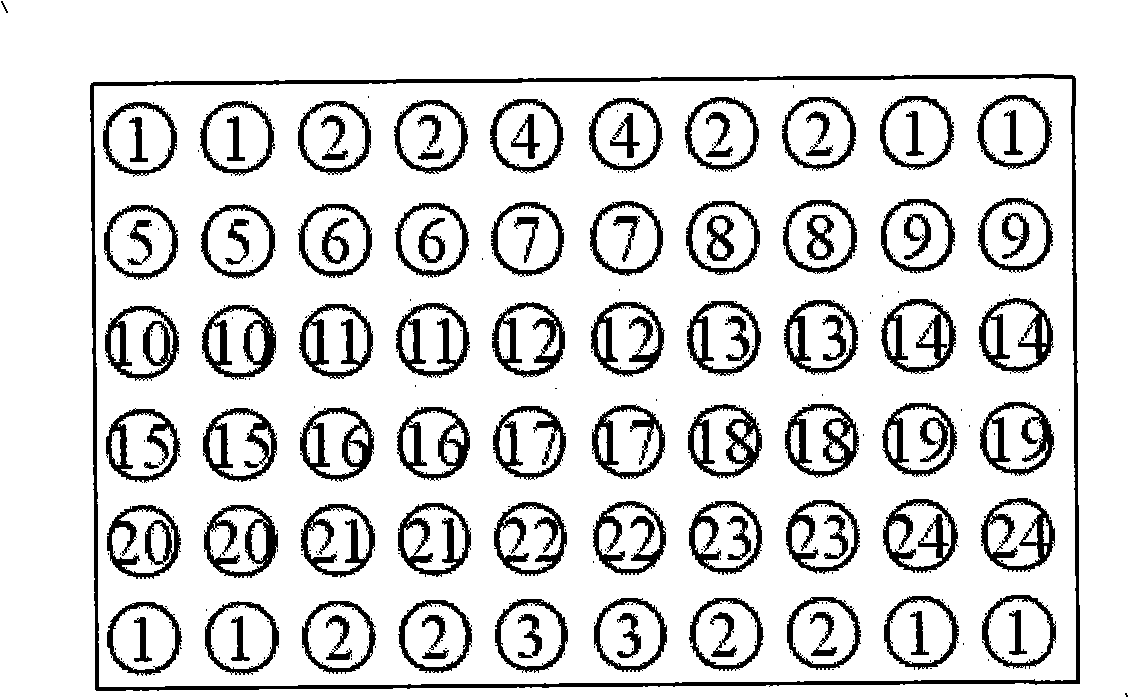
Pathogenic epiphyte detection gene chip
A technology for detecting gene chips and pathogenicity, applied in the field of gene chips, can solve the problems of strict operation requirements, easy quenching of fluorescent signals, expensive fluorescent dyes and detection equipment, and achieve the effects of high sensitivity, good specificity and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1 Universal PCR Amplification Method of Pathogenic Fungal Ribosomal Genes
[0041] 1. Design of universal primers: use the clonemanager 8.0 software to compare the DNA sequences from 5.8s to 28s of the rRNA gene of the fungus to be tested, and use the primer premier5.0 software to design common primers in the highly conserved sequence regions of 5.8S rDNA and 28S rDNA The amplification primers can amplify the partial sequence of fungal 5.8S rDNA, the whole sequence of ITS2 and the partial sequence of 28S rDNA. The primer sequences are as follows:
[0042] Oligonucleotide 1: 5'-CAACGGATCTCTTGGTTC-3' (SEQ ID NO: 24)
[0043] Oligonucleotide 2: 5'-ATGCTTAAGTTCAGCGGG-3' (SEQ ID NO: 25)
[0044] The 5' end of oligonucleotide 1 was modified with biotin.
[0045] 2. General PCR reaction system: including
[0046] 10×PCR buffer 5μl
[0047] 2.5mmol / L dNTPs 4μl
[0048] 25nmol / L MgCl 2 4μl
[0049] 25μmol / L upstream primer 0.4μl
[0050] 25μmol / L dow...
Embodiment 2
[0058] Example 2 Preparation of samples to be tested
[0059] 1. Specimen Processing
[0060] Culture: 1) For the solid medium, scrape an appropriate amount of colony or hyphae on the medium, and place it in a sterile 1.5ml EP tube;
[0061] 2) For the liquid medium, take 500 μl of the bacterial culture and put it into a sterile 1.5ml EP tube, centrifuge at 10000r / min for 15 minutes, and wash the sediment twice with PBS.
[0062] Blood: Take 500μl of blood sample containing bacteria, put it in a sterile 1.5ml EP tube, add 1ml of red blood cell lysate (10mM Tris (pH7.6), 5mM MgCl 2 , 10mMNaCl), incubated at 37°C for 10 minutes and then centrifuged at 10000r / min for 15 minutes. Add 500 μl of white blood cell lysate (10 mM Tris (pH 7.6), 10 mM EDTA (pH 8.0), 50 mM NaCl, 0.2% SDS, 200 μg proteinase K / ml) to the precipitation, and bathe in water at 65° C. for 2 hours. Centrifuge at 10000r / min for 15 minutes, and wash the sediment twice with PBS. .
[0063] Sputum: add an equ...
Embodiment 3
[0067] Example 3 Design of fungal-specific probes
[0068] The fungal rRNA gene is a tandem repeat gene, namely: 18S rRNA-ITS1-5.8SrRNA-ITS2-28SrRNA transcription unit, the sequence separating 18S, 5.8S, and 28S rRNA gene subunits is called the internal transcribed spacer (internal transcribed space , ITS), is a non-coding region. Since the ITS region does not join mature ribosomes, it suffers less selection pressure and a faster evolutionary rate, showing extremely extensive sequence polymorphisms in most eukaryotes. At the same time, the length of the ITS sequence is moderate, and there is enough information in the sequence, which can be widely used in the systematic research of interspecies or intraspecific groups within a genus.
[0069] 1. Search the rRNA gene sequence of the selected target bacteria in the Genbank nucleic acid sequence database, and use clonemanager 8.0 software for sequence comparison.
[0070] 2. Use AlleleID 6.0 software to design species-specific...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com