Method related to DNA (Deoxyribose Nucleic Acid) folded paper and structure and application thereof
A kind of origami and multi-purpose technology, applied in the field of detection, can solve the problem of low sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0050] In the method for preparing the single-chain staple of the end-modified reactant I described in step 1) of the present invention, the method of connecting the reactant I to the end of the staple chain adopts the prior art. For example, staple strands (unmodified biotin) and biotin-modified single strands of staples are both commercially available. It can also be joined by chemical methods. Other kinds of reactant I also staple the 5' end of the single-stranded nucleotide according to the prior art.
[0051] In step 2) of the present invention, the staple chain and scaffold chain obtained in step 1) are self-assembled, and the scaffold chain and staple single chain (including staple single chain and biotin-modified staple single chain) are self-assembled. Mixing them allows them to self-assemble into DNA origami. The method of self-assembly of scaffold chains and staple single strands to form DNA origami is prior art. It is generally better to mix the scaffolding chai...
Embodiment 1
[0065] Example 1 Modified biotin without dT
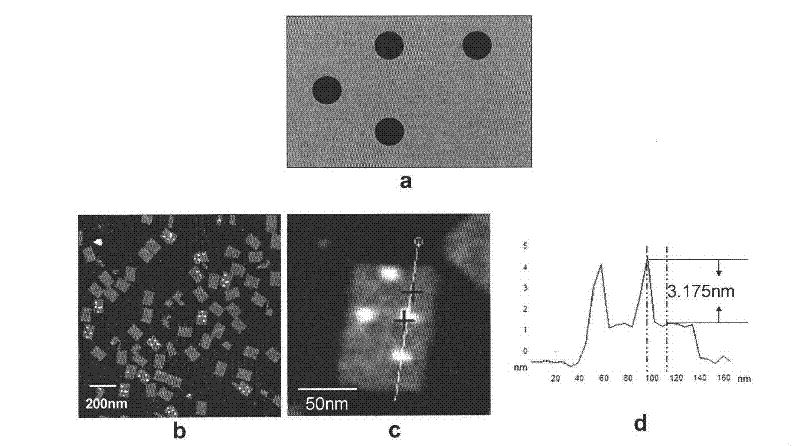
[0066] Drop 2 μL of DNA origami modified with biotin (dT-free) (such as figure 1 a) On freshly dissociated mica, after 5 minutes of adsorption, in 50 μL 1×TAE / Mg 2+ In the buffer solution, use AFM’s liquid phase tap mode for imaging. After the imaging is stable, slowly inject 50 μL of 7.6nM streptavidin solution into the liquid tank, and continuously scan to observe the streptavidin and biotin on the origami site binding process, the observations are recorded in figure 1 b. Among them, the atomic force microscope is a NanoScope IIIa system (Digital Intrument, the United States), the scanning head of the atomic force microscope is a J-type scanning head, and the AFM probe is a NP-S silicon nitride tip (elasticity coefficient 0.58Nm -1 , Veeco Company), the imaging conditions were temperature 25°C, humidity 40%-50%, imaging parameters were scanning rate 2Hz, driving amplitude 400mV, voltage 0.3V, number of scanning lines 256, and ...
Embodiment 2
[0068] Embodiment 2 Modified biotin, containing multiple dT
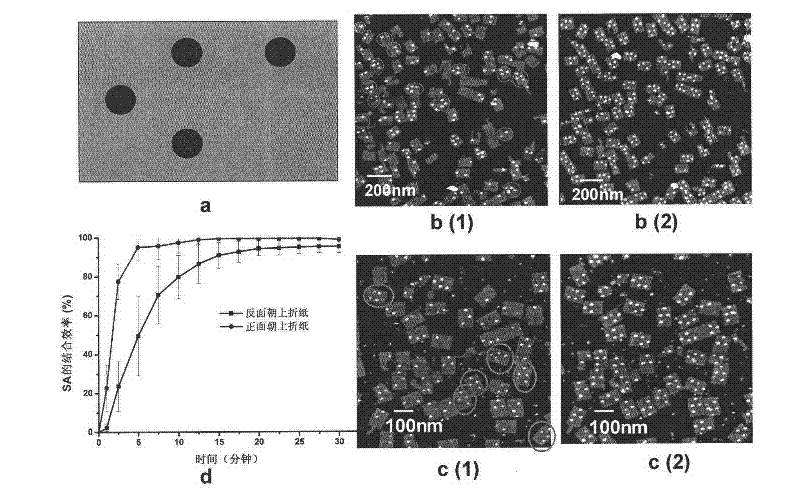
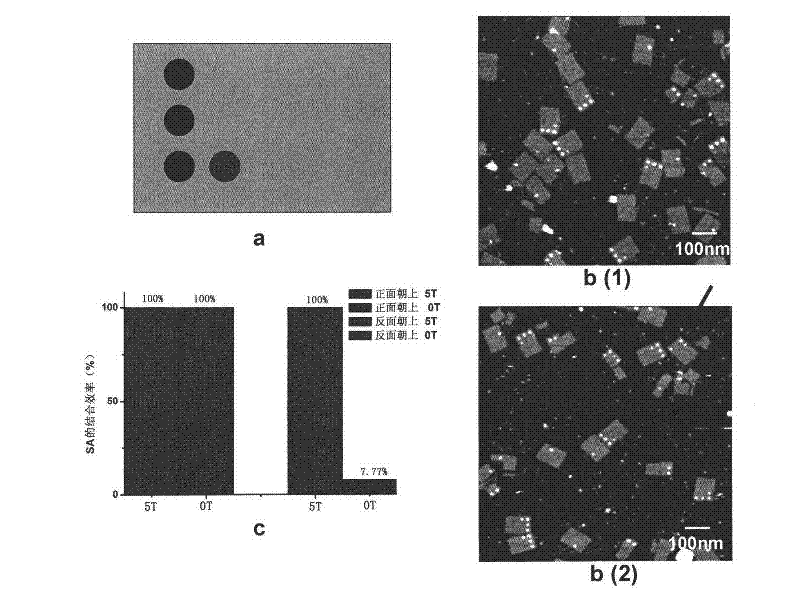
[0069] The experimental procedure is the same as in Example 1, and the origami design pattern is as figure 2 a, the design site is the same as that in Example 1, except that it contains 5 dTs. When there are 5 dTs, there is only one case of biotin and avidin reaction on origami ( figure 2 b). Before the reaction, the origami had the same appearance, the length, width and height were 100nm, 70nm and 2.5nm, respectively. The surface is flat. After adding avidin, changes appeared on the origami surface over time, and after 30 minutes, almost all of the biotin-modified sites appeared circular bumps. The height is 2.8-3.3nm, which is the bound avidin molecule. from figure 2 The avidin binding site pattern on b can clearly determine the orientation of the origami on the mica surface. figure 2 b(1) is the image obtained after adding streptavidin and reacting for 5 minutes, figure 2 b(2) is the image obtained a...
PUM
| Property | Measurement | Unit |
|---|---|---|
| length | aaaaa | aaaaa |
| height | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com