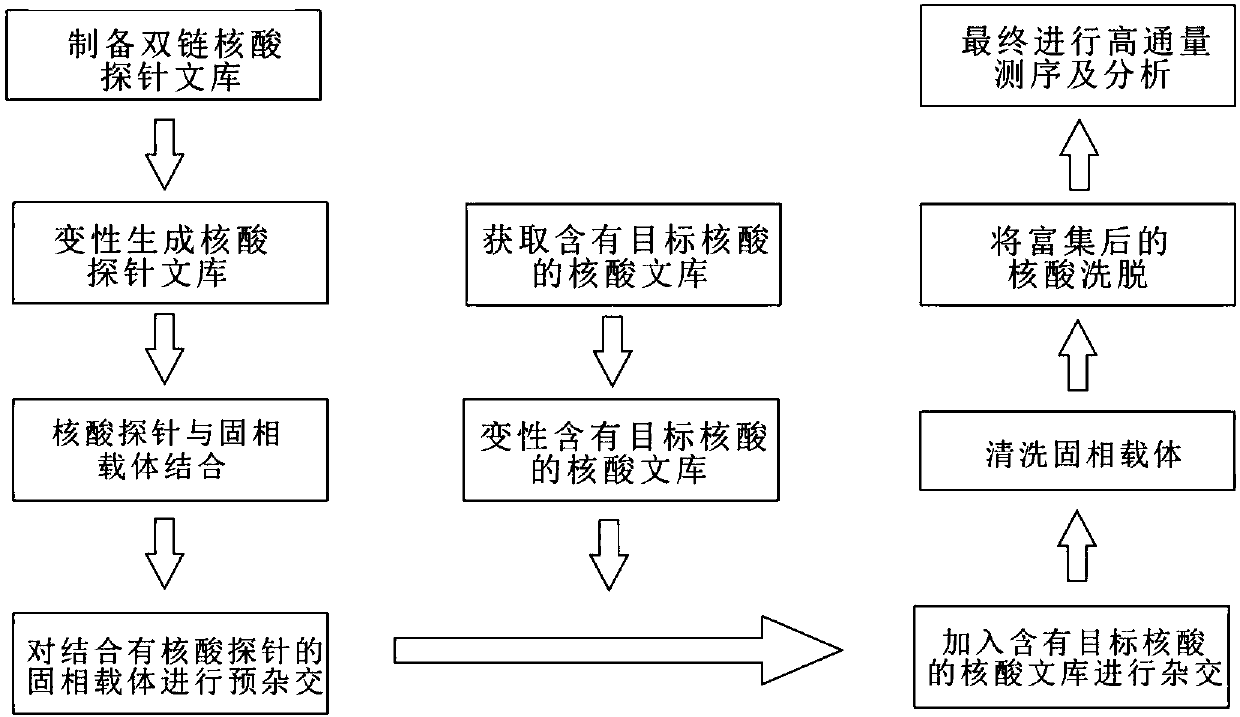
Solid-phase-carrier-membrane-based target nucleic acid enrichment method for high-throughput sequencing
A technology of solid phase carrier and target nucleic acid, which is applied in the direction of biochemical equipment and methods, measurement/testing of microorganisms, chemical library, etc., can solve the problem of unreported, narrowed applicable range, hybridization time and conditions can not have a good regulatory issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
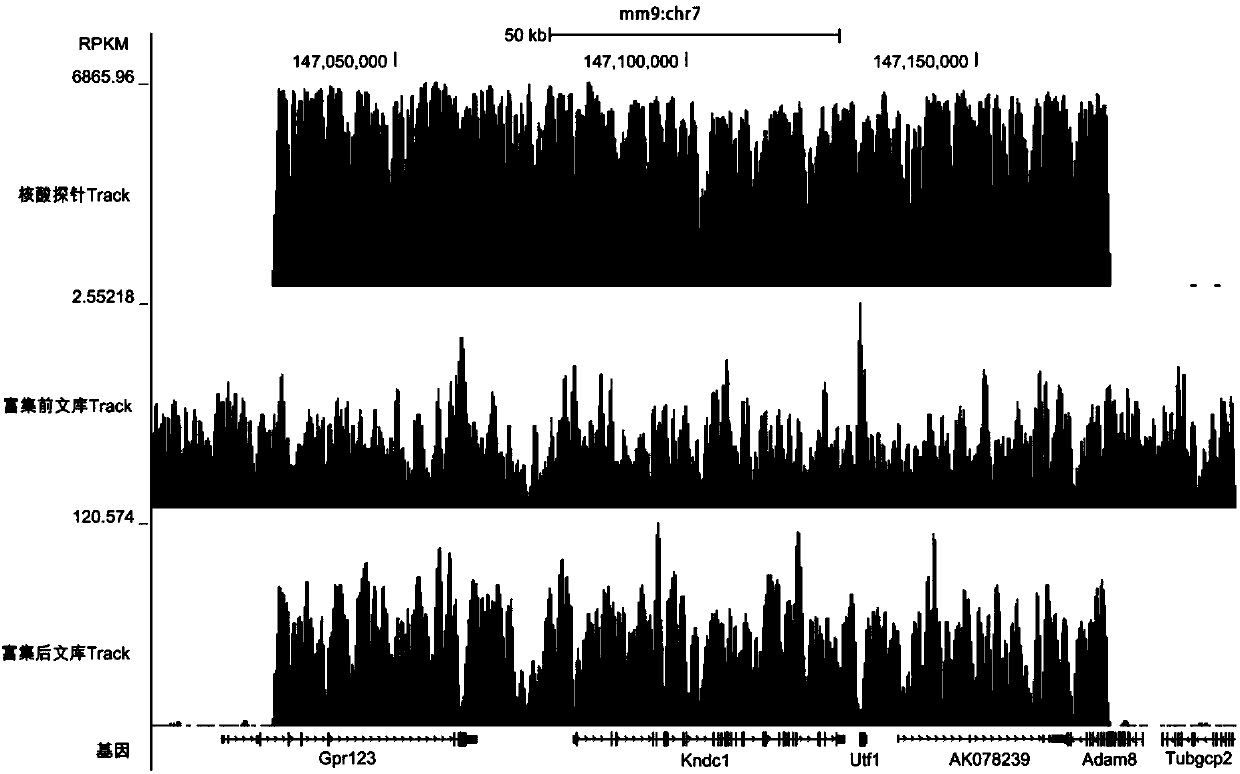
[0104] Example 1 Enrichment of Chromosome 7 Sequences of Mouse Genome 147,029,000-147,172,901
[0105] 1. Preparation of double-stranded nucleic acid library:
[0106] 1. The patent applicant has RP24-292M18 bacterial artificial chromosome (BAC), which contains the sequence of chromosome 7 of the mouse genome 147,029,000-147,172,901;
[0107] 2. Shake 10ml of chloramphenicol-containing LB medium transformed with RP24-292M18 DH5a overnight, and extract the BAC plasmid by alkaline lysis;
[0108] 3. Dissolve 5 μg of BAC in 100ul 1xTE buffer, and use an ultrasonic instrument (Bioruptor) to fragment the BAC. The conditions are 30s on / 90s off, 6 cycles, and the fragments produced range from 150bp to 800bp;
[0109] 4. Purify the sonicated solution with QIAquick PCR Purification Kit (Qiagen), and elute with 70 μl TE buffer;
[0110] 5. Take 5 μl of the solution after sonication, add 1 μl of 6x loading Buffer, and check the size of the fragment after sonication by agarose gel elect...
Embodiment 2
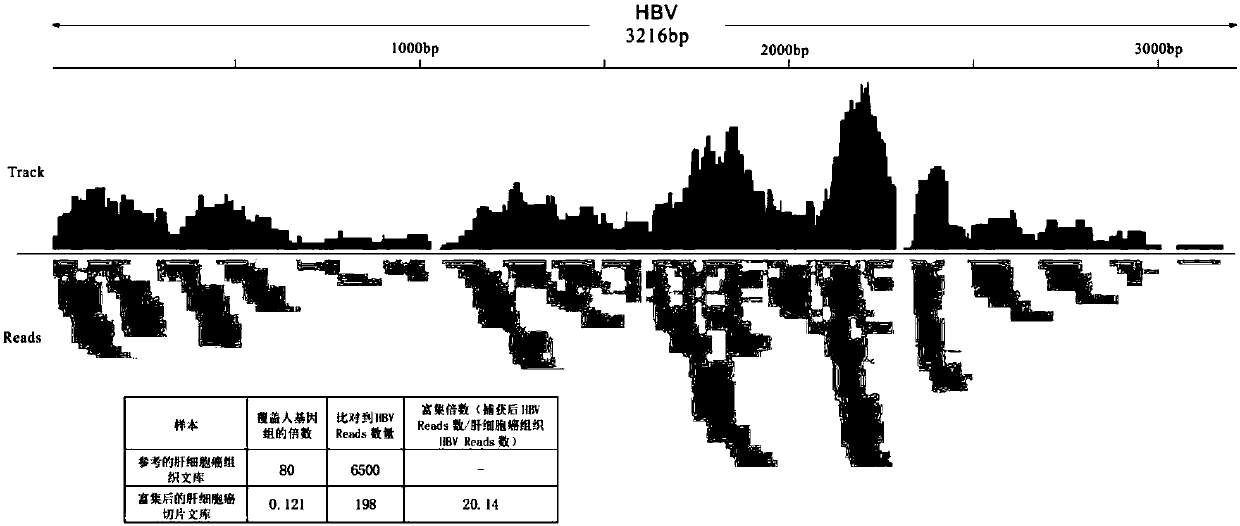
[0188] 7. The ratio of the number of sequencing reads in the enriched region to the total sequencing reads is 120 times higher than that of the unenriched sequencing reads in the total sequencing reads, and among the 91,992 reads obtained by sequencing the enriched region, There were 35,275 reads with different sequences, indicating its high complexity. Example 2: Enrichment of HBV genome in tissue sections of hepatocellular carcinoma patients
[0189] 1. Preparation of double-stranded nucleic acid library:
[0190] 1. The patent applicant owns a plasmid containing the sequence of Hepatitis B virus isolate FEN94, which contains the HBV genome sequence.
[0191] 2. shake the LB medium containing ampicillin of 10ml of DH5a transformed with Hepatitis B virus isolate FEN94 plasmid overnight, and extract the plasmid by plasmid mini kit (QIAGEN Plasmid Mini Kit, Cat.no.12125);
[0192] 3. Fragment the plasmid containing the HBV genome (40 μg / 100 μl TE solution) with a sonicator (B...
Embodiment 3
[0234] Example 3: Enrichment and sequencing of the human EGFR gene
[0235] 1. Preparation of double-stranded nucleic acid library:
[0236]1. The patent applicant owns RP11-116H11 and RP11-65D21 bacterial artificial chromosome (BAC), which covers the human genome (hg19) chromosome 7 sequence 55,034,601-55,343,001, and this region contains the EGFR gene (chr7:55,086,725-55,275,031);
[0237] 2. Take 200ml of chloramphenicol-containing LB medium transformed with RP11-116H11 and RP11-65D21 DH5a by shaking overnight, and extract the BAC plasmid by alkaline lysis;
[0238] 2. Take 20μg RP11-116H11BAC and 20μg RP11-65D21BAC and dissolve them in 100ul1xTE buffer respectively, and fragment the BAC with an ultrasonic instrument (Bioruptor).
[0239] 3. The sonicated solution was purified with QIAquick PCR Purification Kit (Qiagen), and eluted with 70 μl TE buffer;
[0240] 4. Take 5 μl of the sonicated solution, add 1 μl of 6x loading Buffer, and test the size of the sonicated fragm...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com