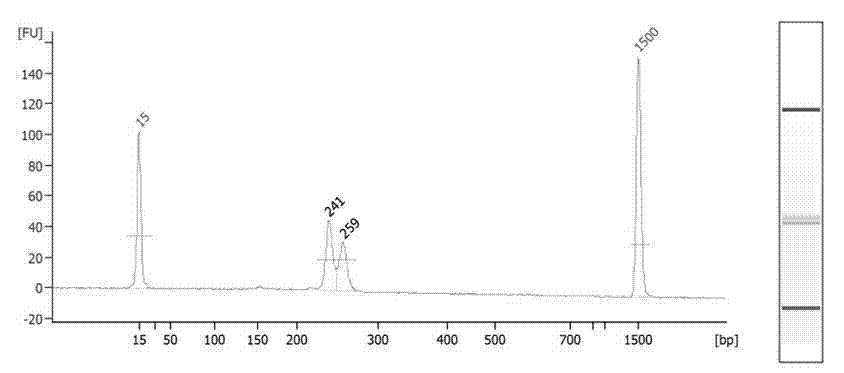
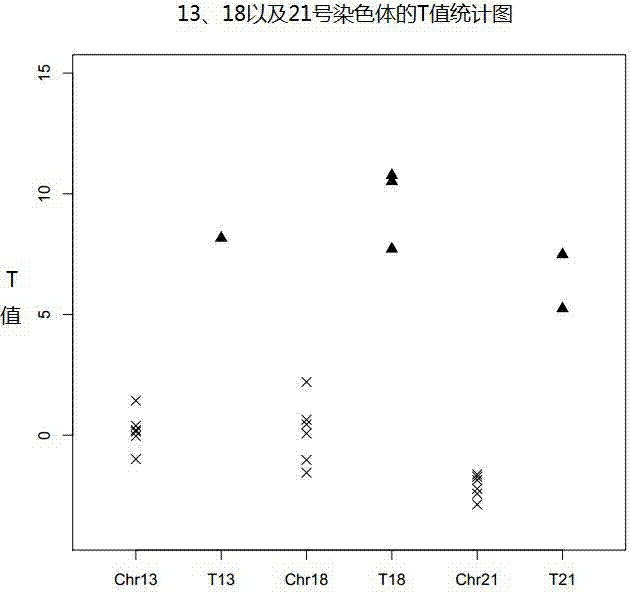
Semi-specific amplification primer group, method and kit for quickly detecting chromosome number abnormality
A technology for chromosome number and amplification primers, which is applied in the field of semi-specific amplification primer sets and kits for rapid detection of abnormal chromosome number, can solve the problems of unsatisfactory research and application, low throughput, and high cost. Achieve the effects of improved sequencing depth, short library construction time, and improved detection cycle
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0089] Example 1 The acquisition of LINEs region primers
[0090] This patent designs a set of special primers, which can bind to a series of LINEs regions on the genome during PCR annealing. At the same time, it ensures the existence of certain specific SNP polymorphisms, which can be used to locate the relative position of an amplified fragment in the original genome.
[0091] Through the Repeat Library provided by www.repeatmasker.org, extract all LINEs regions, and use the BWA algorithm in the whole genome to search for fragments with a length of 130 bp-180 bp in the extracted LINEs region, which must meet the following conditions : The number of SNP sites in the fragment region includes 10-20. After subsequent manual screening one by one, we selected one of the regions as an example. And designed a pair of primers as follows:
[0092] forward primer 1a
[0093] 5'-GGCTATAAACGCAGGCCACTNNNNNNNNNNNNNNNNNGGGAGGGGAACATCACAC-3'
[0094] or forward primer 1b
[0095] 5'-GG...
Embodiment 2
[0108] Example 2 Detection method for abnormal number of chromosomes
[0109] The detection method of the present invention can be used for, but is not limited to, abnormal chromosome numbers such as trisomy 13, trisomy 18, and trisomy 21 in adults. It can also be used to non-invasively detect abnormal chromosome numbers in fetuses by analyzing the peripheral blood of pregnant women.
[0110] 1. Extraction of sample DNA:
[0111] In this embodiment, a total of 8 maternal blood samples were drawn (sample numbers are: S_042, S_053, S_055, S_058, S_064, S_066, S_071, S_078). 5 ml of peripheral blood was extracted from each sample, and after two high-speed centrifugation at 4°C, a plasma sample from which blood cells were removed was obtained. Plasma samples were extracted using a DNA extraction kit produced by Qiagen, or other conventional DNA extraction methods.
[0112] 2. Library construction:
[0113] Target fragment amplification: select forward primer 1b and revers...
Embodiment 3
[0132] Embodiment 3 Preparation of kit
[0133] The first round of specific amplification primers obtained in Example 1:
[0134] Forward primer 1a:
[0135] 5'-GGCTATAAACGCAGGCCACTNNNNNNNNNNNNNNNNNGGGAGGGGAACATCACAC-3'
[0136] or forward primer 1b:
[0137] 5'-GGCTATAAACGCAGGCCACTCBDVHBDVHBDVHBDVHBDVHGGGAGGGGAACATCACAC-3'
[0138] Reverse Primer 1:
[0139] 5'-TCTGAGGAATCAGCAATGCAGCGATCATGGTGGTTTGCTGCA-3'
[0140] The design structure of the primer pair is as follows Figure 6 shown.
[0141] Primers designed for sequencing library preparation obtained in Example 1:
[0142] Forward primer 2:
[0143] 5'-AATGATACGGCGACCACCGAGATCTACACGGCTATAAACGCAGGCCACTC-3'
[0144] Reverse primer 2:
[0145] 5'-CAAGCAGAAGACGGCATACGAGATNNNNNNNNTCTGAGGAATCAGCAATGCAGCGAT-3'
[0146] Wherein, B, D, V, H and N in the two sets of primer pairs are degenerate bases. NNNNNNNNN in reverse primer 2 is a specific base, which is the tag sequence of the sample, and the tag sequence is diffe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com