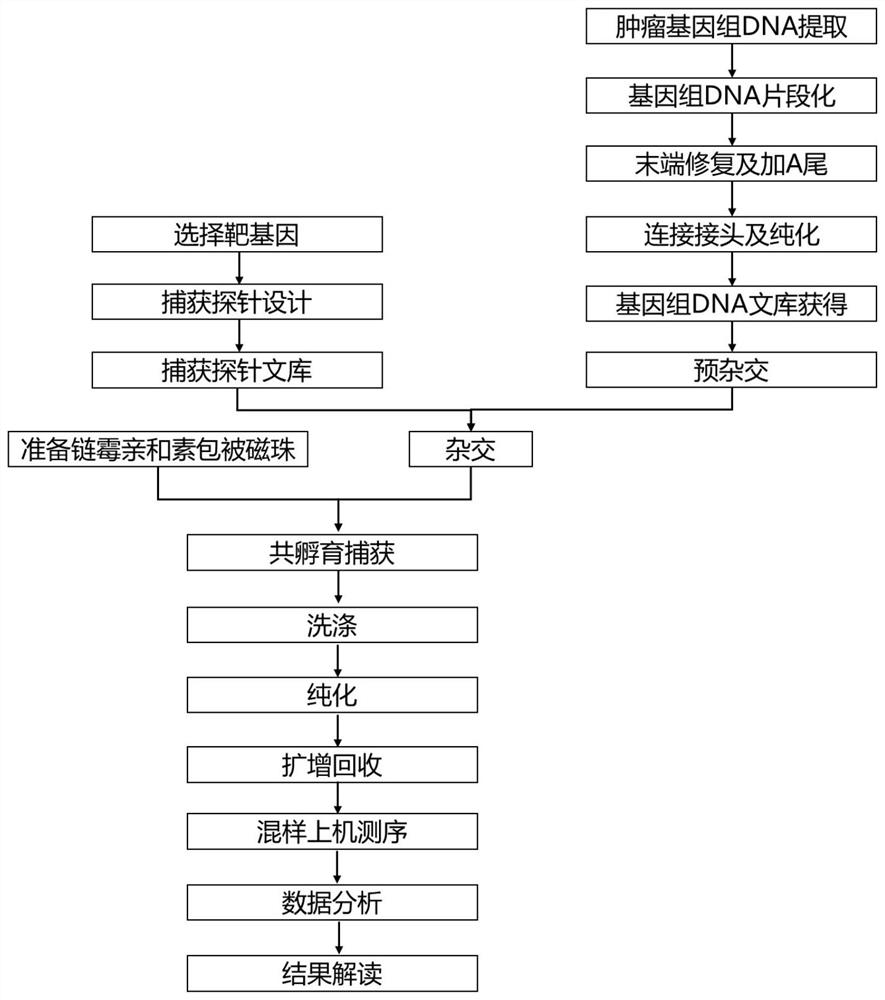
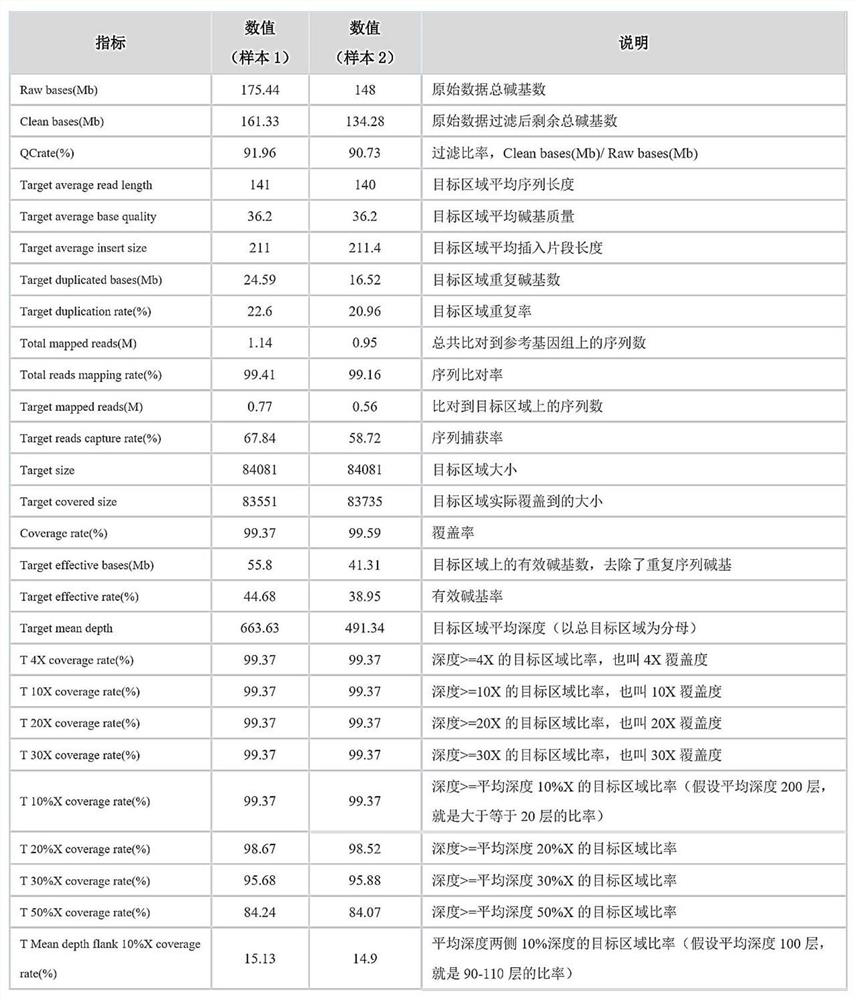
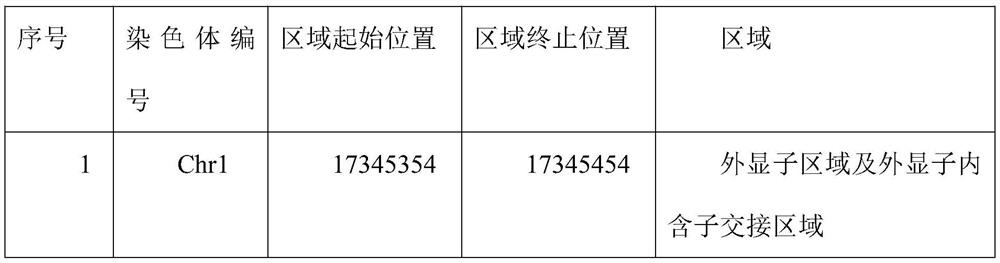
Method for capturing genetic colorectal cancer genome target sequence
A technology of colorectal cancer and genomics, applied in biochemical equipment and methods, combinatorial chemistry, microbial measurement/testing, etc., can solve the problem of not being able to screen multiple key areas of gene mutation load in a targeted manner and not well satisfying individualization problems such as specific user needs, long single-sample analysis time, etc., to achieve the effect of reducing detection cost and sequencing data volume, reducing non-specific capture, and improving sequencing depth
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Preparation of detection quality control products for high-throughput capture sequencing of hereditary colorectal cancer gene mutations.
[0050] (1) Two commonly used stable human colorectal cancer cell lines HCT-116 and HCT-15 were purchased from ATCC;
[0051] (2) Adopt fetal calf serum medium (provided by Thermo Fisher Company) to cultivate these two strains of human tumor cell lines, culture conditions: 37 ℃ constant temperature, 5% CO , humidity 50%; Cultivate until the cell density reaches the area of the culture dish 80- Passage at 90%, collect in the logarithmic phase of cell growth, centrifuge at a speed of 800-1000r / min to take the precipitate, extract the genomic DNA of each cell line separately, and purify and elute through the column;
[0052] (3) Dilute the purified genomic DNA of each cell line to 100±5ng / μL with Tris-EDTA buffer solution. The appearance is a transparent liquid without visible impurities, and the purity is 1.9>OD260 / 280>1.7 , to obtai...
Embodiment 2
[0056] capture probe design
[0057] Select the key region of hereditary colorectal cancer genome mutation, design and synthesize multiple capture probes according to the key region, and some examples of the key region are shown in Table 2; examples of the sequence design of the capture probe are as SEQ ID NO:1~ 2641; the length of the capture probe is 100bp, and the capture probes are connected end to end, covering the exon region and exon-intron junction region of each target region; all probes are labeled with biotin, and the The capture probes are 2641 probe mixtures, and the design method of the capture probes is:
[0058] (1) Determine the exon region and exon-intron junction region of each target region according to the human whole genome standard sequence HG19 provided by the public data website NCBI;
[0059] (2) Starting from the exon region in each region and the first exon in the exon-intron junction region, design the capture probe according to the standard seque...
Embodiment 3
[0066] Preparation of Genomic Mutation Capture Sequencing Kit for Hereditary Colorectal Cancer
[0067] (1) Design and synthesize the capture probes of the target regions shown in Example 2;
[0068] (2) Mix the designed capture probes with the same mass, dilute the mixture to a working concentration of 25ng / μL, and store at -20°C;
[0069] (3) Separately pack the capture probe mixture and the quality control obtained in Example 1.
[0070] (4) Prepare instructions, outer packaging, assemble and seal.
[0071] (5) The amount of capture probe mixture used is 6 μL / 3 reactions, 12 μL / 6 reactions, 24 μL / 12 reactions, 48 μL / 24 reactions; the amount of quality control products is 30ng-50ng / reaction.
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting point | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com