CRISPER-Cas9-system-mediated sheep MSTN (myostatin) gene knock-out and exogenous gene site-specific integration method
An exogenous gene and gene knockout technology, which is applied in the fields of molecular biology and animal genetics and breeding, can solve problems such as unreported research, and achieve the effects of improving safety, reducing length, and improving integration efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] The optimization of embodiment 1 CRISPER-Cas9 carrier
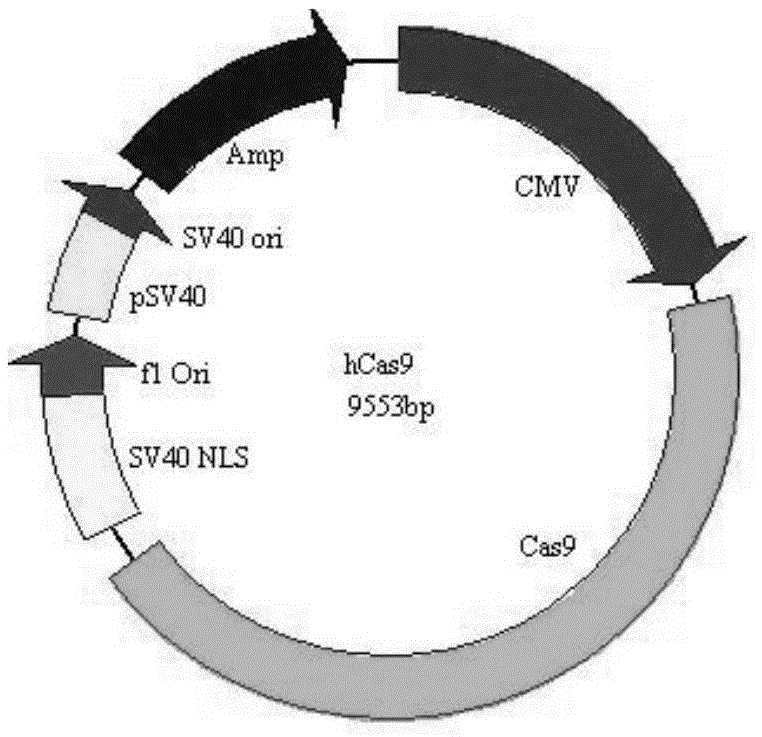
[0039] The CRISPR-Cas9 expression vector purchased from Beijing Zhongyuan Company was optimized. The nucleotide sequence of the optimized CRISPER-Cas9 vector (i.e. hCas9 plasmid) is shown in SEQ ID NO: 1, and the hCas9 plasmid map is shown in figure 1 .
Embodiment 2
[0040] The construction of embodiment 2gRNA expression vector
[0041] According to the sheep MSTN gene sequence (GeneID100860887), the gRNA sequence was designed for the exon 1 sequence of MSTN, and the gRNA expression vector based on the CRISPER-Cas9 system was constructed. The gRNA expression vector includes four parts: U6 promoter, target sequence, gRNA backbone and termination signal.
[0042] Among them, the DNA sequence of the gRNA action site is as follows:
[0043] 5′-CGATGACTACCACGTTACGA-3′ (gRNA1 target site)
[0044] 5′-CGTTACGACGGAAACGGTCA-3′ (gRNA2 target site)
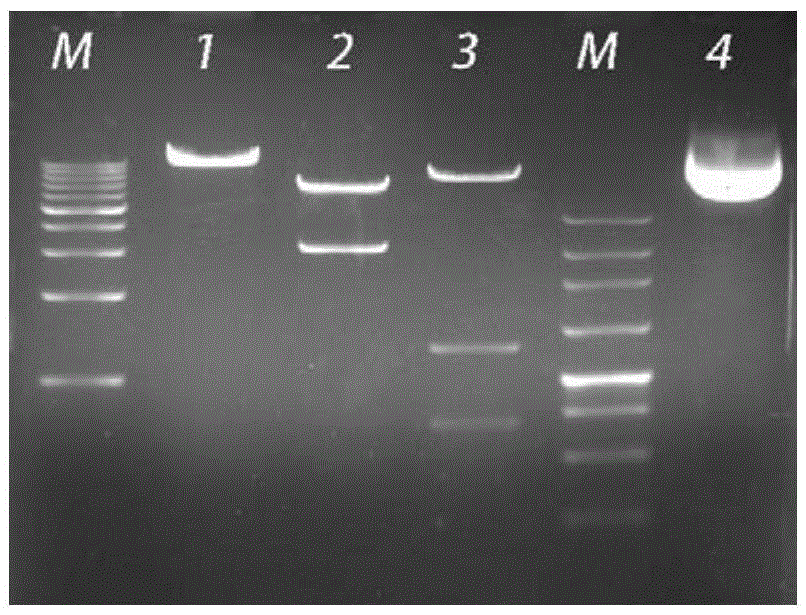
[0045] Use biological software to design gRNA sequences according to the gRNA action sites (gRNA1 target site and gRNA2 target site), clone them into the PMD-19T vector, transform Escherichia coli Trans-110, pick a single colony after plating, and perform bacterial Liquid PCR, identified by electrophoresis and sequencing, the single colony with correct sequencing was inoculated in LB medium containing...
Embodiment 3
[0046] Example 3 Efficiency Detection of CRISPER-Cas9 System
[0047] Premier5 software was used to design PCR primers spanning different target sites, and the genomes of sheep fibroblasts transfected with hCas9 plasmids and RNA1-MSTN / RNA2-MSTN48h plasmids were extracted, and the genomes were used as templates for PCR amplification. Primers are as follows:
[0048] MSTN-B-F: 5′-CTATTTATGCTGCTTGTTGC-3′
[0049] MSTN-B-R: 5′-CTATCTCCCAATCCTTCACC-3′
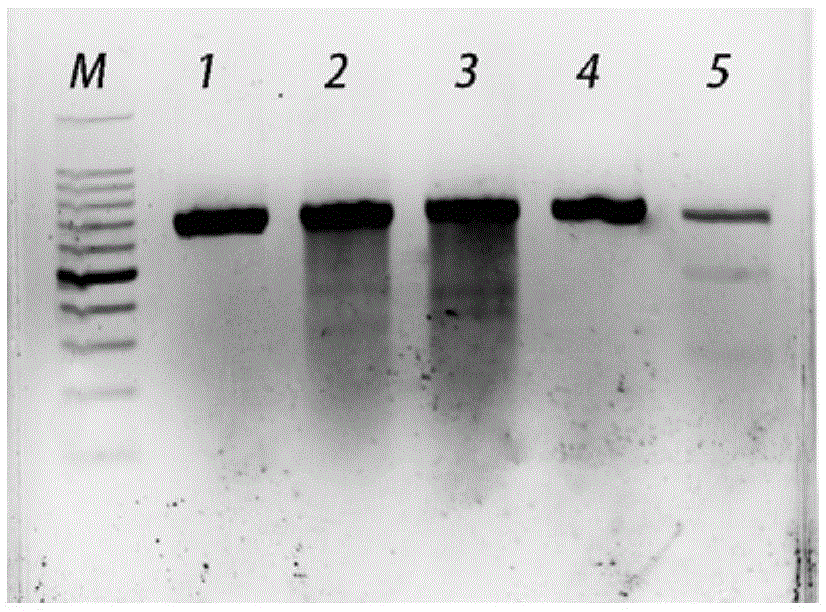
[0050] The total PCR reaction system is 50 μL, premixed ExTaq 25 μL, upstream and downstream primers (100 mmol L -1 ) each 2 μL, genomic DNA 2 μL, sterilized water 19 μL. PCR reaction conditions: pre-denaturation at 94°C for 10 min; denaturation at 94°C for 1 min, annealing at 50°C for 30 s, extension at 72°C for 45 s, 35 cycles; 10 min at 72°C, 30 min at 16°C. The size of the amplified fragment is 666bp. The fragments containing gRNA1 and gRNA2 target sites were amplified from the sheep genome, detected by 1.5% agarose gel elec...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com