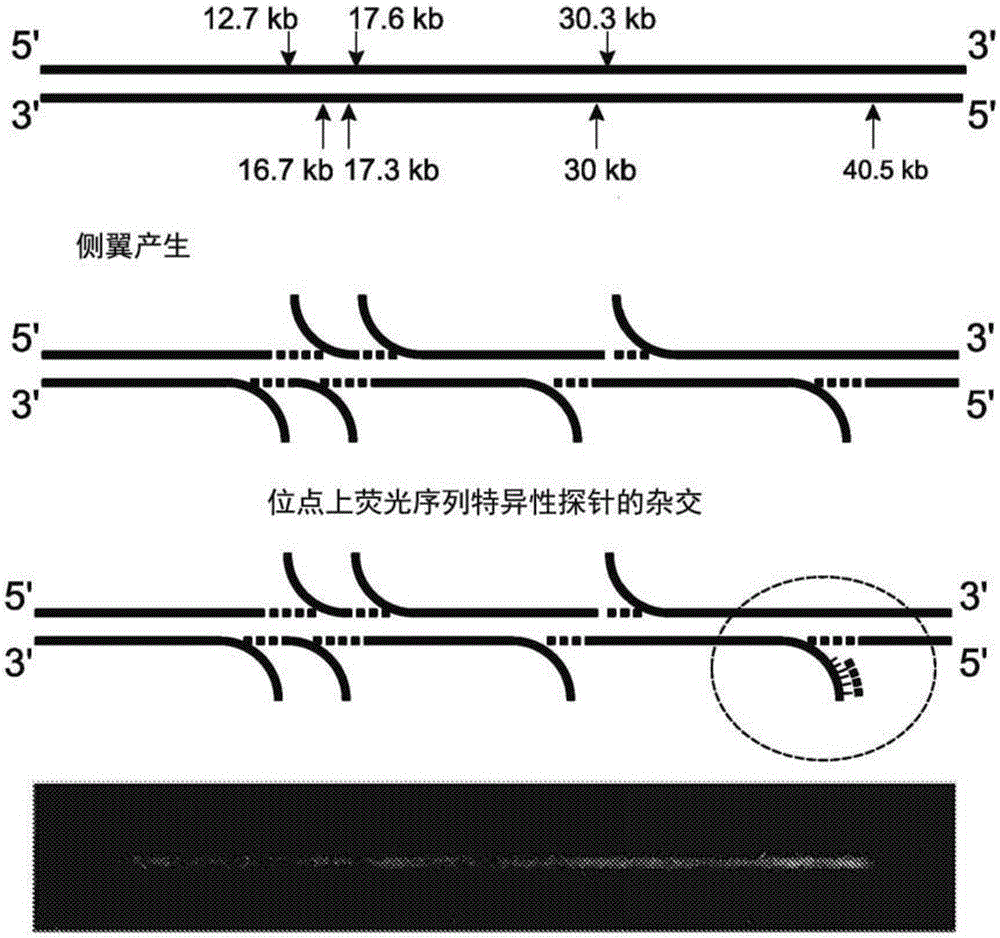
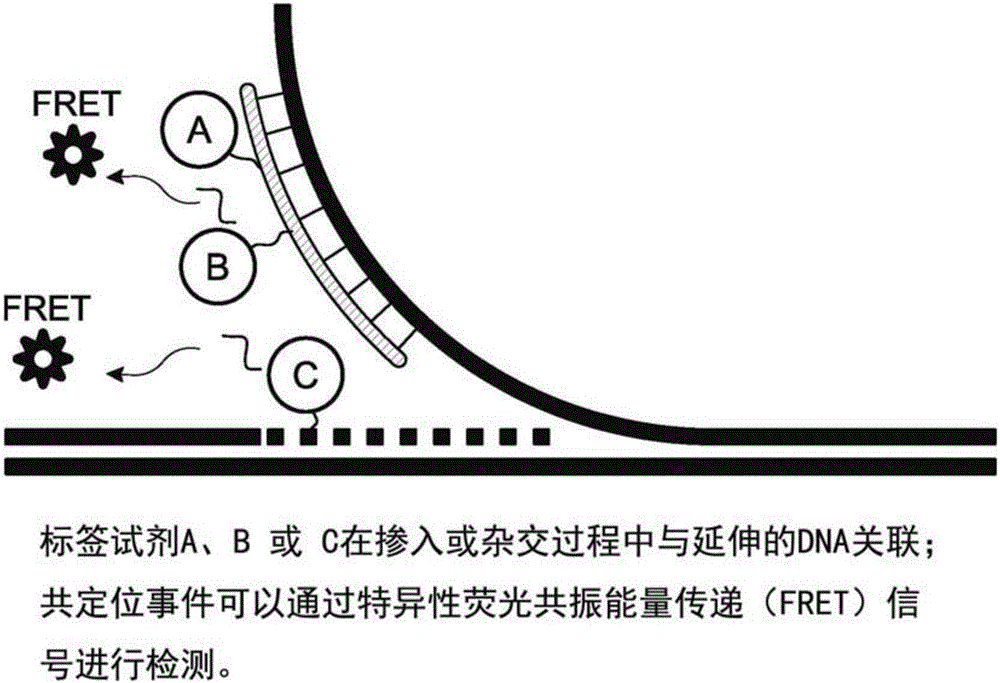
Analysis of polynucleotides
A polynucleotide and nucleic acid technology, applied in the field of nanotechnology and nucleic acid analysis, can solve the problems of inability to directly quantify the copy number and low accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0108] Example 1: De novo generation of genome maps
[0109] De novo assembly of the human genome map, using automation Nanochannel chips and an automated imaging system purchased from BionanoGenomics, San Diego, California, were used to extract genomic DNA from human cell lines and perform site-specific labeling, followed by linearization and analysis in nanochannels at 50x coverage, where the throughput Running (3-4hrs) for 5-10Gb / chip and further 50-200Gb / chip in the future. To detect site-specific markers and determine distances between markers, raw molecules (>20kb) were used in pairwise pattern matching and de novo assembly processes. image 3 The assembled human genome map is shown, indicating the ease of generating genome maps from large complex genomes. Thus, genome maps can be generated from large complex genomes using the methods described herein.
Embodiment 2
[0110] Example 2: Identification of Structural Changes Compared to a Reference Sequence
[0111] In the assembled genome of Example 1, hundreds of small and large structural variants were detected. An exemplary tandem repeat on chromosome 5 is as Figure 4 shown. Based on the site-specific marker patterns, the genomic sequence was assembled and compared to the pattern of a reference genomic sequence32. A tandem duplication of genomic region 34 comprising an additional 3 tandem copies was identified. From this, de novo genome assembly and structural variant analysis based on site-specific marker patterns can be performed.
Embodiment 3
[0112] Example 3: Preparation of labeled DNA
[0113] DNA was isolated using the plug lysis method. Those skilled in the art will recognize that suitable isolated DNA samples can be prepared using a variety of techniques well known in the art in addition to the exemplary procedures provided herein. Epstein-Barr virus (EBV) infected cells were grown to log phase and harvested. Prepare 2% agarose and water bath by immersing 2% agarose in boiling water for 10-15min until completely melted, put the agarose in 43°C water bath for at least 30min before use, warm the 50ml tube attached to THERMOMIXER to 50°C , and add 10-20ml of water to simulate a water bath. By placing 4x10 6 Cells were prepared by aspirating into a 15 ml tube and centrifuging at 2,200 rpm (1,000 rcf) for 5 minutes. Cells were washed twice in 1XPBS, suspended in 750 μl suspension buffer, and warmed to 43°C. 250 μl of melted agarose was added to the cell suspension and mixed, filling each well in a volume of ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com