Sebastiscus marmoratus gene screening and mining method based on simplified genome sequencing technology
A technology for genome sequencing and scorpionfish, which is applied in the field of molecular marker development to achieve the effects of increasing the rate of enzymatic cutting, accurate sequencing results, and reducing difficulty
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
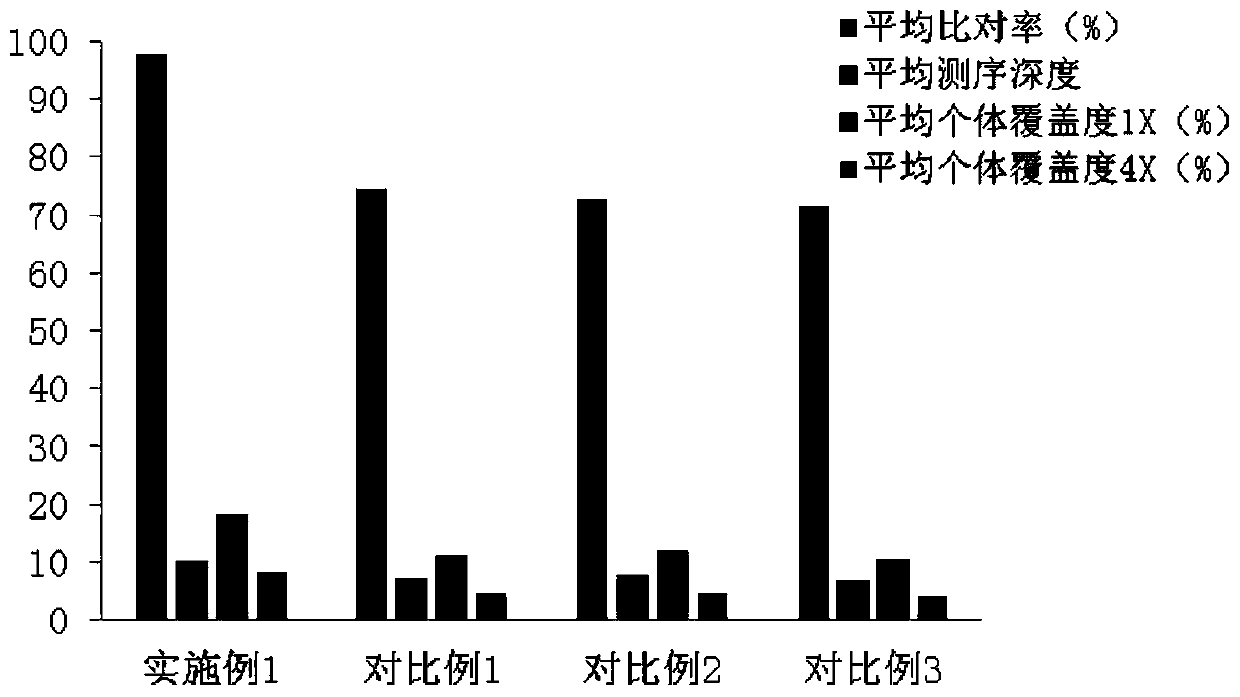
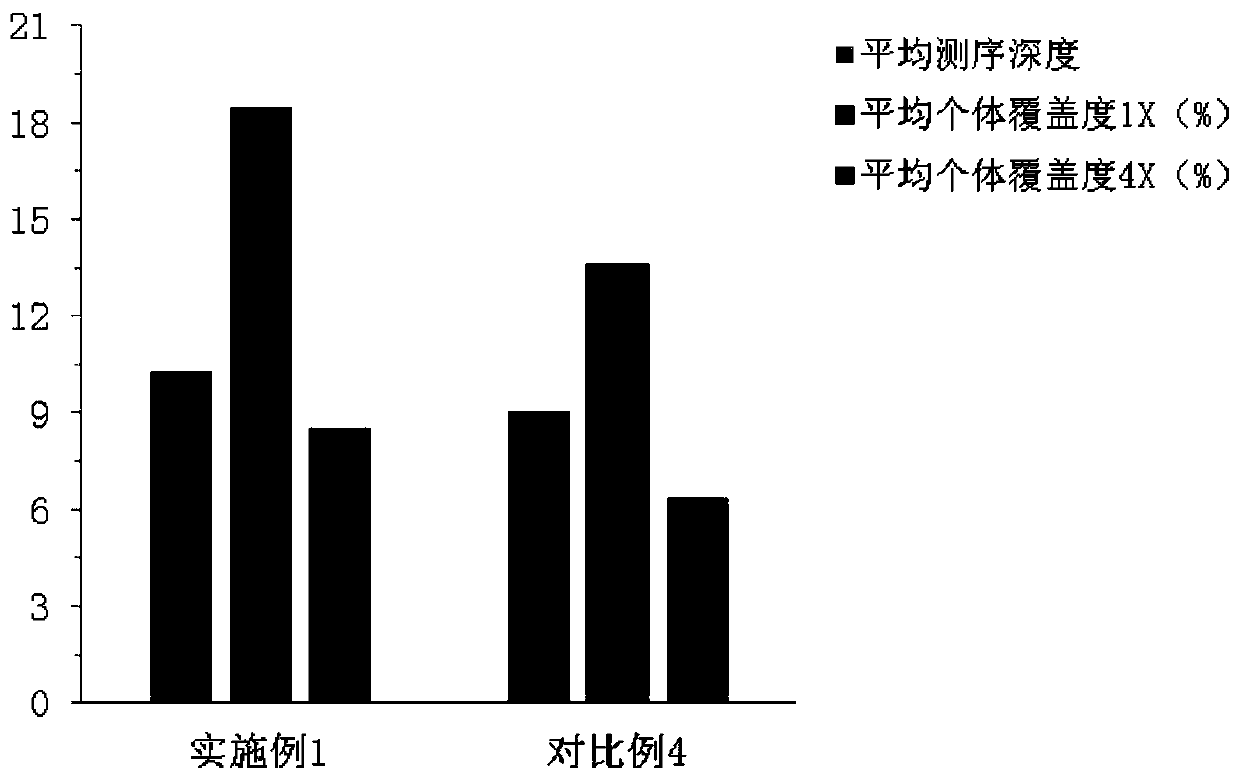
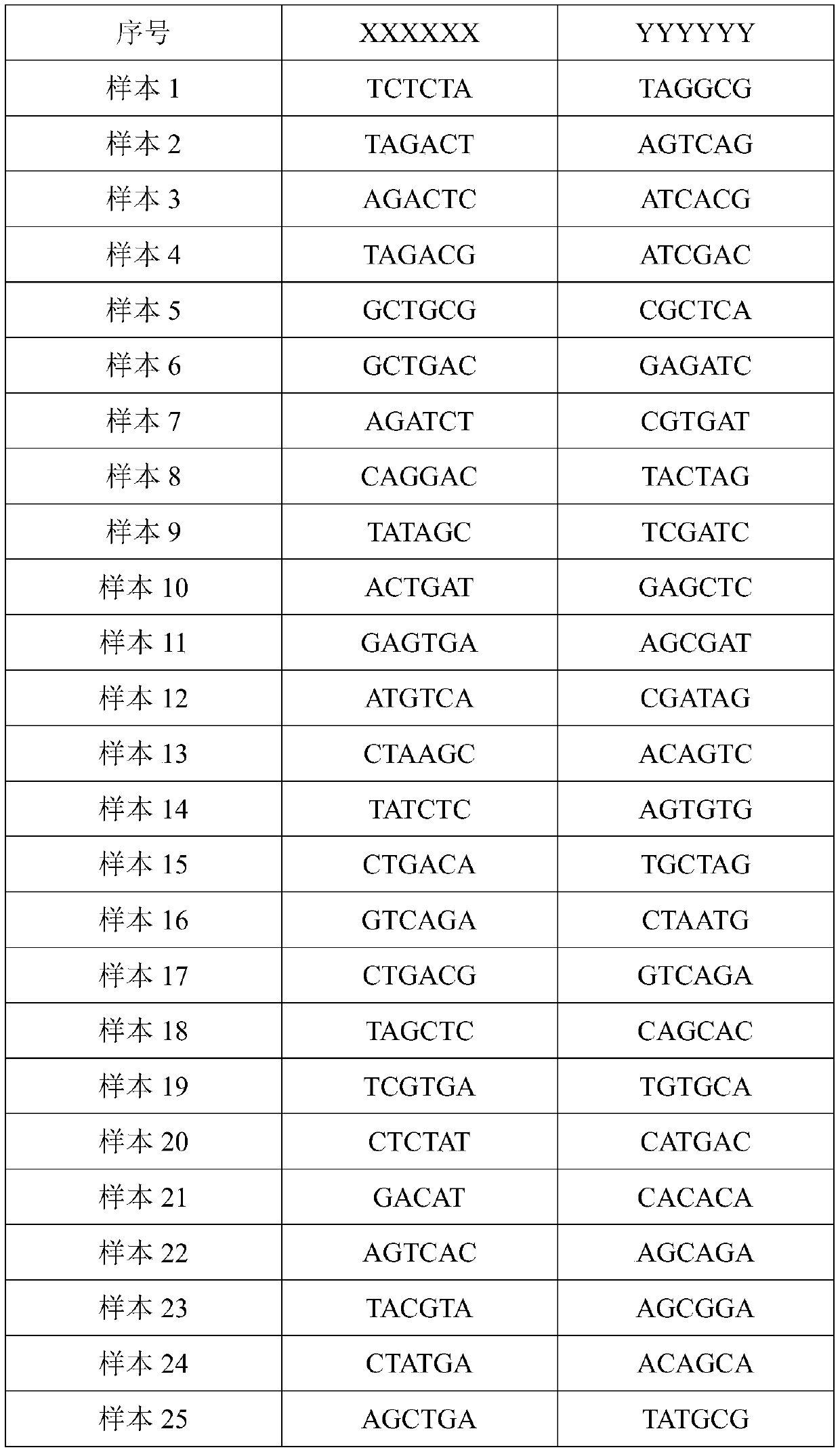
Image
Examples
Embodiment 1
[0055] The method for screening and mining the genes of the brown calamari based on the simplified genome sequencing technology includes the following steps:
[0056] The samples for this experiment were collected from the offshore waters of Qingdao, Zhoushan, and Fangchenggang in the winter of 2017. There were 10 samples of Scorpene scorpionfish in each group, a total of 30 samples, and the sample collection time was similar. Muscle samples were taken from all samples and stored in 95% ethanol solution at -20°C.
[0057] (1) Genomic DNA extraction: a. Remove the ethanol from the muscle tissue of the scorpionfish and place it in a 1.5ml centrifuge tube, add 650 μL of digestion buffer and 20 μL of proteinase K solution, shake and mix well, and place it in a 56°C oven for digestion 5h, shake once every 30min; b. After the solution is cooled to room temperature, add 650μL of equilibrated phenol into the centrifuge tube, shake and mix until the milky solution is obtained, centrifu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com