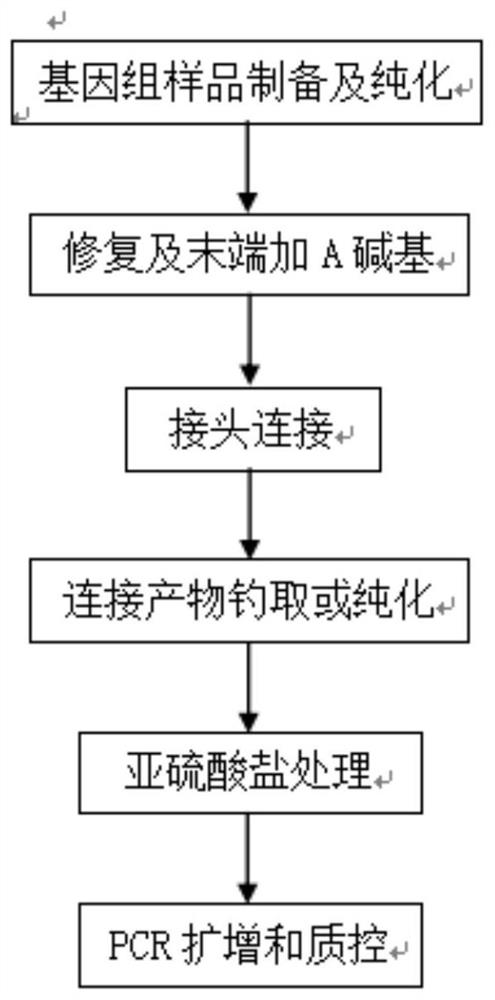
A kind of micro-degradation genomic DNA methylation library sequencing method and its kit
A genome and methylation technology, applied in the field of genomics and molecular biology, can solve problems such as undetectable, achieve the effect of low initial amount, reduce preference, and improve reaction efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Reagent configuration:
[0042] The composition of Repair Buffer includes MgCl 2 , NaCl, DTT, dATP, dTTP, dCTP, dGTP, Tris-HCl, and the concentrations are 10mM, 50mM, 5mM, 4mM, 1mM, 1mM, 1mM, 60mM respectively, wherein the pH value of Tris-HCl is 7.6;
[0043] Repair Enzymes are T4Polynucleotide Kinase, T4DNA Polymerase and Taqpolymerase, and the concentrations are 8U, 2U and 0.04U respectively;
[0044] Buffer A includes Tris-HCl, MgCl 2 , DTT, ATP, and the concentrations were 190mM, 27mM, 14mM, 7mM;
[0045] Buffer B includes PEG6000 and PEG8000, the concentration of PEG6000 is 30%, the concentration of PEG8000 is 20%
Embodiment 2
[0047] Severely degraded FFPE sample DNA methylation library construction
[0048] Genomic DNA was extracted using a commercial kit (Qiagen FFPE extraction kit). After extracting DNA from a puncture micro-tissue slide, methylation library construction was taken as an example. The DNA was eluted into 30ul of ultrapure water.
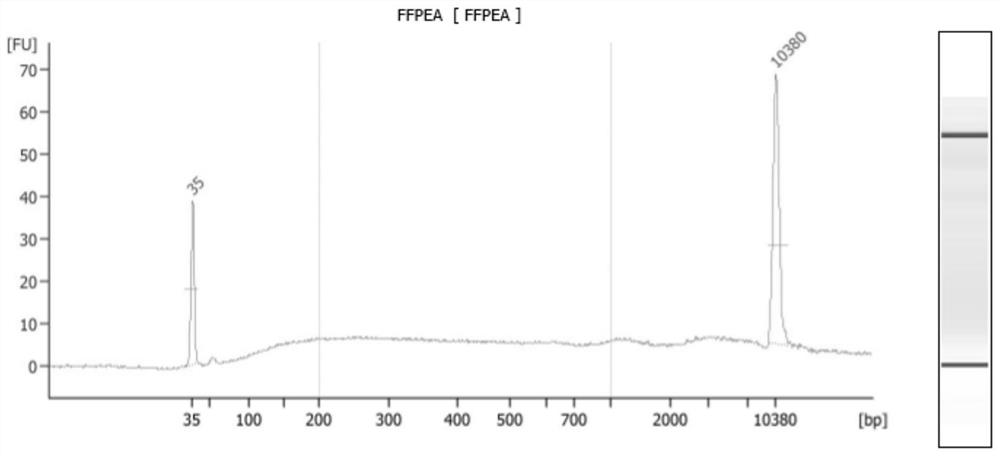
[0049] 1. Take 1uL samples respectively for Qubit HS dsDNA concentration determination and 2100 analyzer to evaluate the degree of sample degradation (such as figure 2 ), the results showed that the samples were distributed almost uniformly in the interval of 50bp-10kb, the samples were completely degraded, and the total amount of samples detected by Qubit was 23ng.
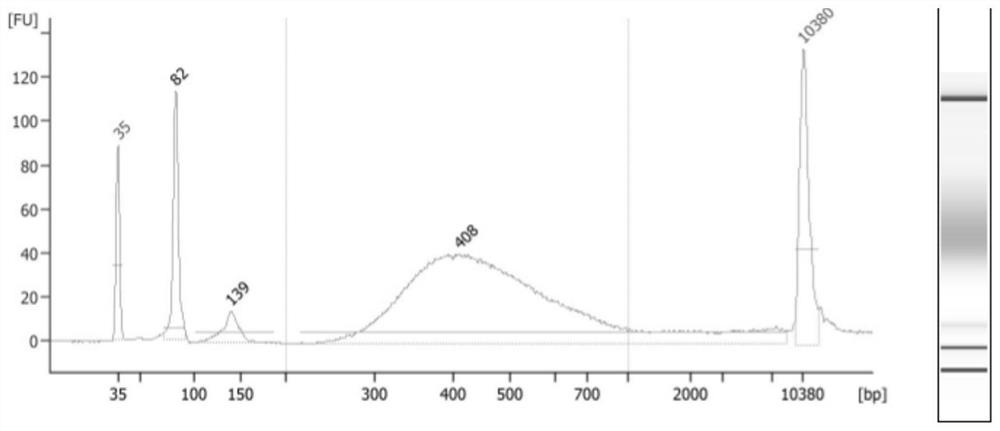
[0050] 2. DNA screening and fragmentation in different ranges: 19.5ul of Ampure XPBeads (Agencourt) was added to 30ul of DNA samples for 3 minutes, and the magnetic beads were adsorbed on the magnetic stand. After clarification, transfer the supernatant to a new EP tube, then add 30uL of A...
Embodiment 3
[0076] In this example, 5uL of DNA Repair Buffer, 2uL of Repair Enzymes and 13uL of ultrapure water were sequentially added to a 30uL DNA sample, mixed evenly and reacted on a PCR instrument according to the following process, 16°C, 40min; 75°C , 25min; 72°C, 25min; 37°C, 10min. After the reaction, add 11uL of Buffer A, 12uL of Buffer B, 1uL of the annealed joint, 3uL of water and 3uL of DNA ligase in the tube, mix well, and incubate at 24°C for 10min.
[0077] Other steps are with embodiment 2.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com