Specific recognition sequence based T cell receptor high-throughput sequencing library construction and sequencing data analysis method
A technology for identifying sequences and cell receptors. It is applied in sequence analysis, chemical libraries, biochemical equipment and methods, etc. It can solve the problems of false positive test results, unrecognizable DNA damage and PCR errors, and poor accuracy.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
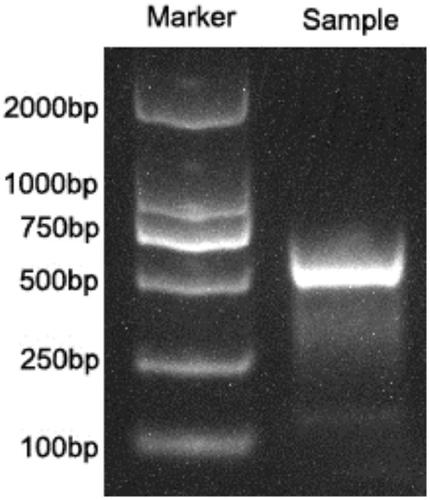
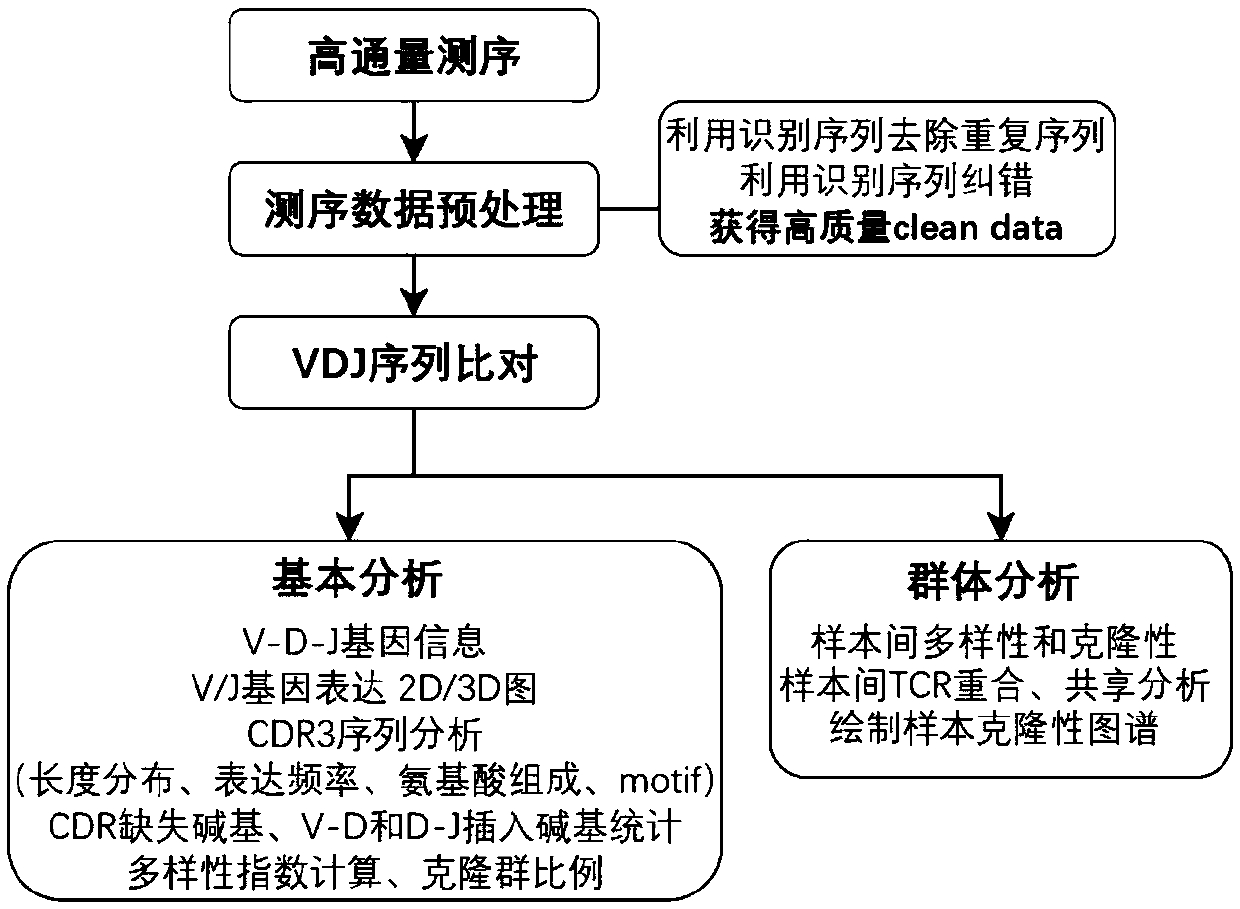
Examples
Embodiment 1
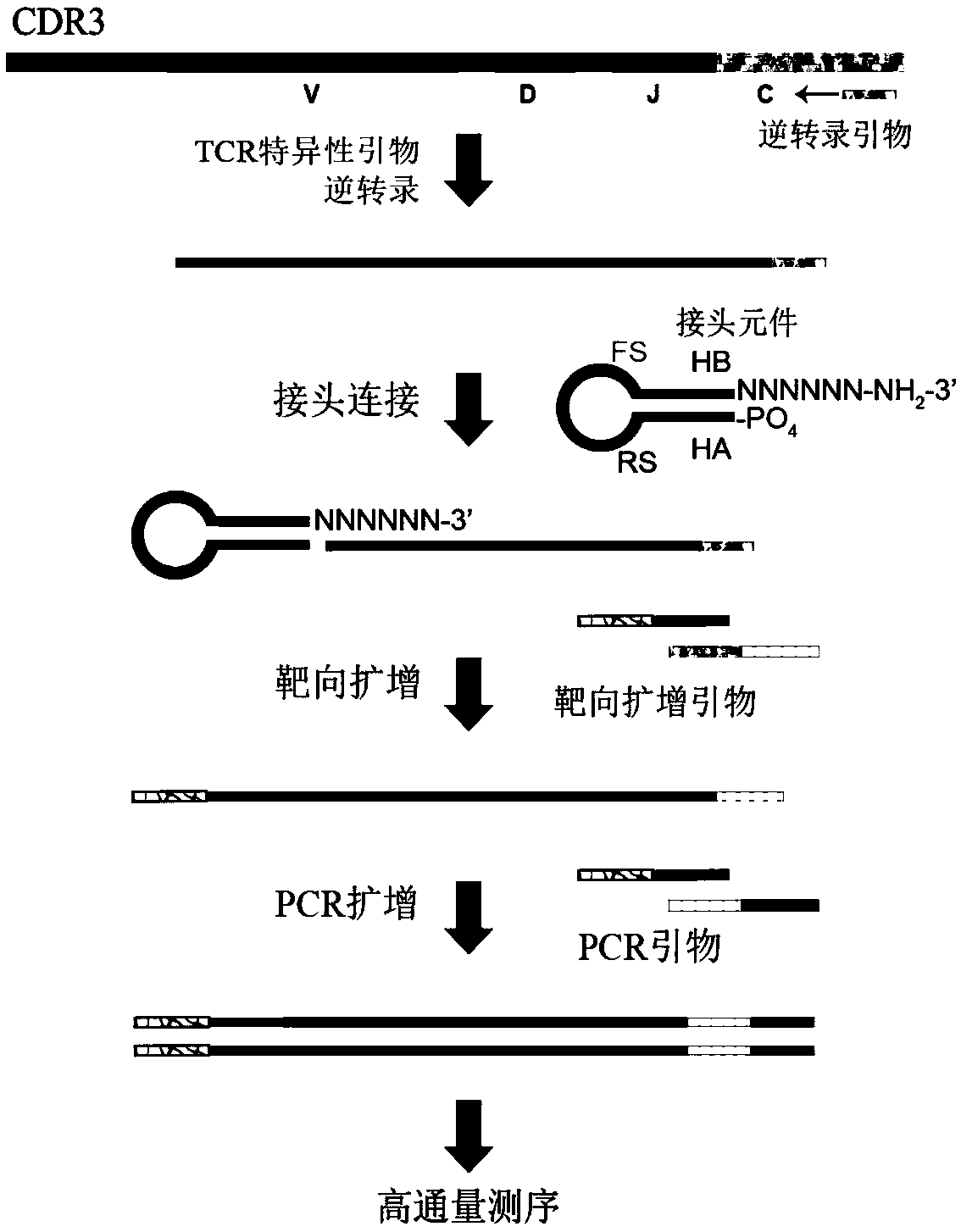
[0071] 1. Adapter elements with unique recognition sequences required for constructing TCR sequencing libraries
[0072] The sequence of the linker element is SEQ ID NO.1: (5'→3')GTGTATCCAGTGNNNNNNNGATCGTCGGACTGTAGAACTCTGAAC CACTGGATACAC NNNNNN.
[0073] Among them, GTGTATCCAGTG is the hairpin sequence A (Hairpin A, HA), NNNNNNNN is the specific recognition sequence (Recognition sequence, RS), GATCGTCGGACTGTAGAACTCTGAAC is the fixed sequence (Fixed sequence, FS), CACTGGATACAC is the hairpin sequence B (Hairpin B, HB), NNNNNN Is a random sequence (Randomsequence). The hairpin sequences A and B are complementary and annealed at high temperature to form the stem structure of the hairpin, while protruding random sequences to form sticky ends. The fixed sequence FS is the recognition sequence of the Illumina / Life library PCR primers. N represents any base in A, T, C, G; 5' has PO 4 Modification, 3' with NH 2 grooming.
[0074] The specific recognition sequence RS contained in ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com