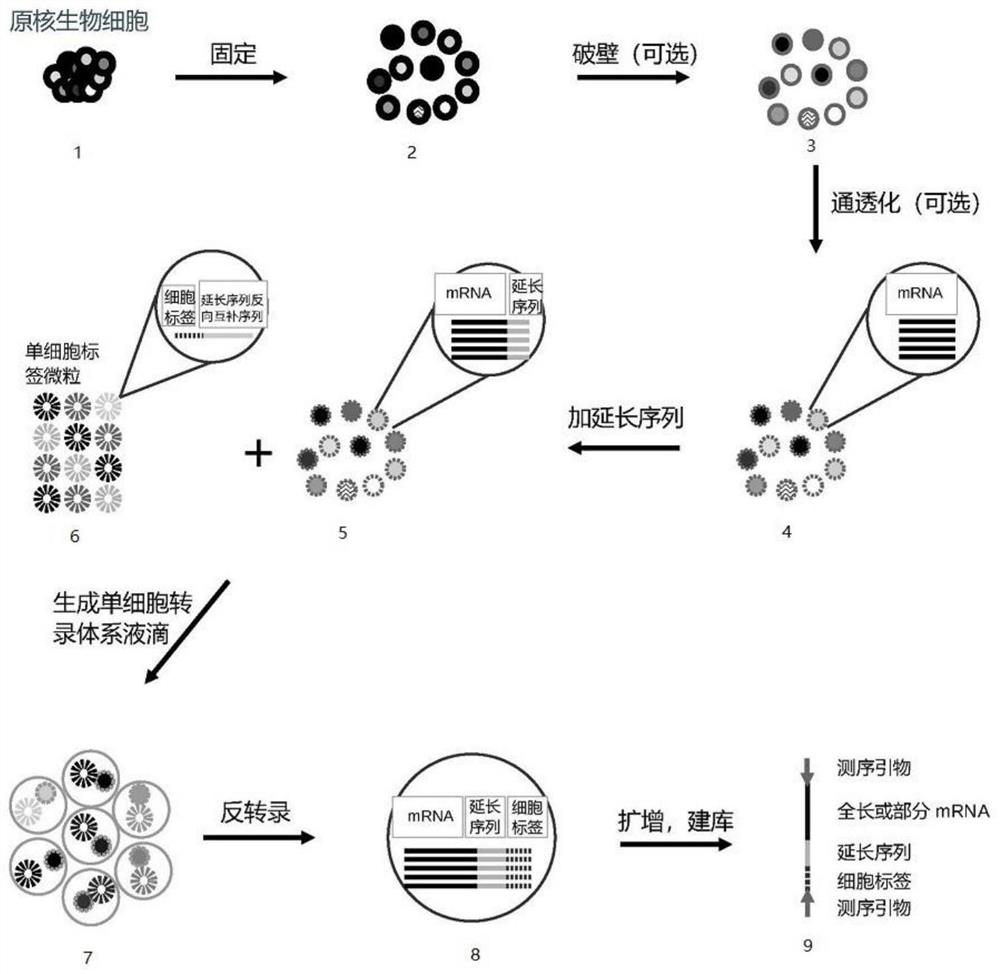
Method for constructing prokaryote single cell RNA sequencing library
A technology for prokaryotic and sequencing libraries, applied in the field of constructing prokaryotic single-cell RNA sequencing libraries, can solve the problems of complicated operation, unstable effect, complicated and time-consuming experimental operation, etc., and achieve the effect of reducing preference, high efficiency and easy operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0063] Example 1 L-type Escherichia coli single-cell RNA sequencing
[0064] 1. Cell fixation and permeabilization.
[0065] Resuspend 5000 or 20000 L-type E. coli cells in PBS containing 0.01% formaldehyde and 0.1% Tween20 and incubate at room temperature for 15 minutes. Centrifuge, then wash twice with PBS containing 1% BSA and once with PBS. Centrifuge to remove supernatant. Since L-type E. coli has no cell wall, there is no need for wall-breaking of prokaryotic cells here. Cells treated in this step still retain their single-cell environment, but the reagents and other reactants for the extended sequence reaction can pass freely.
[0066] 2. Connect the 3' end of bacterial RNA to extend the sequence.
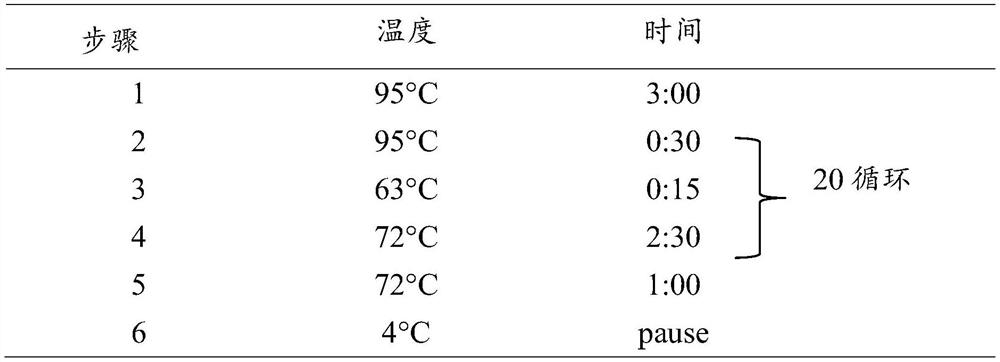
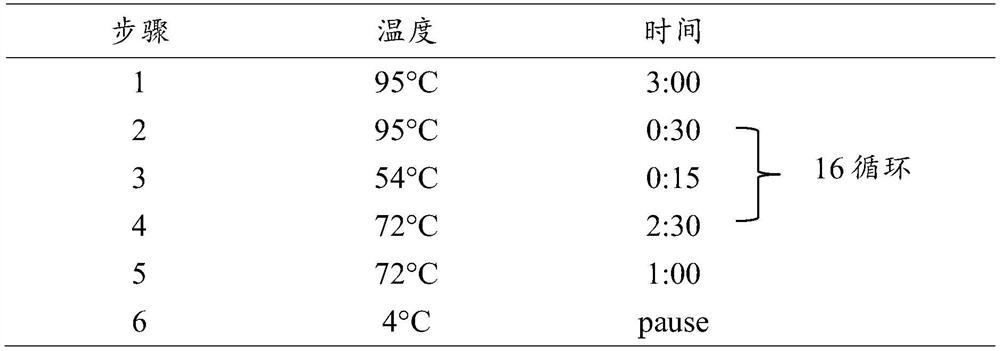
[0067] 2.1 Add 20 μL of 3'-end linker ligation solution: 1×T4 RNA Ligase Buffer, 0.5 μM 3'-end RNA linker sequence (that is, the extended sequence of this example: 5'- / 5rApp / CTGTCTCTTATACACATCTGACGCTGCCGACGA / 3ddC / -3', 5rApp represents 5 '-phosphorylated ribonucleotide b...
Embodiment 2
[0094] Example 2 Sequencing and data analysis of the files obtained in Example 1
[0095] 1. Sequencing using the Illumina NextSeq platform. The read length was set as: read1: 80bp, i5 index: 16bp, i7 index: 8bp, read2: 80bp. . Among them, the sequencing data read by read2 is the mRNA partial sequence of the transcriptome, the sequencing data read by read1 is the reverse complementary sequence of the mRNA partial sequence of the transcriptome, the sequencing data read by i5 index is the cell tag, and the sequencing data read by i7 index is the cell label. The sequencing data for the library are split markers.
[0096] 2. Split the library through the sequencing data read by the i7 index, and classify the data of the sequencing data read by the i5 index into the cell to which the sequencing fragment belongs; the reverse complementary sequence of the data sequence read by read2 and the sequencing data read by read1 Align the reference genome of E. coli to confirm the correspo...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com