Multi-task high-order SNP upper detection method and system, storage medium and equipment
A detection method and multi-task technology, applied in the fields of proteomics, instrumentation, genomics, etc., can solve the problems of high computational complexity of detection methods, insufficient success rate of detection algorithms, and inaccurate search results, and achieve enhanced global search. ability, improve global search ability, and solve the effect of low identification accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0083] In order to make the object, technical solution and advantages of the present invention more clear, the present invention will be further described in detail below in conjunction with the examples. It should be understood that the specific embodiments described here are only used to explain the present invention, not to limit the present invention.
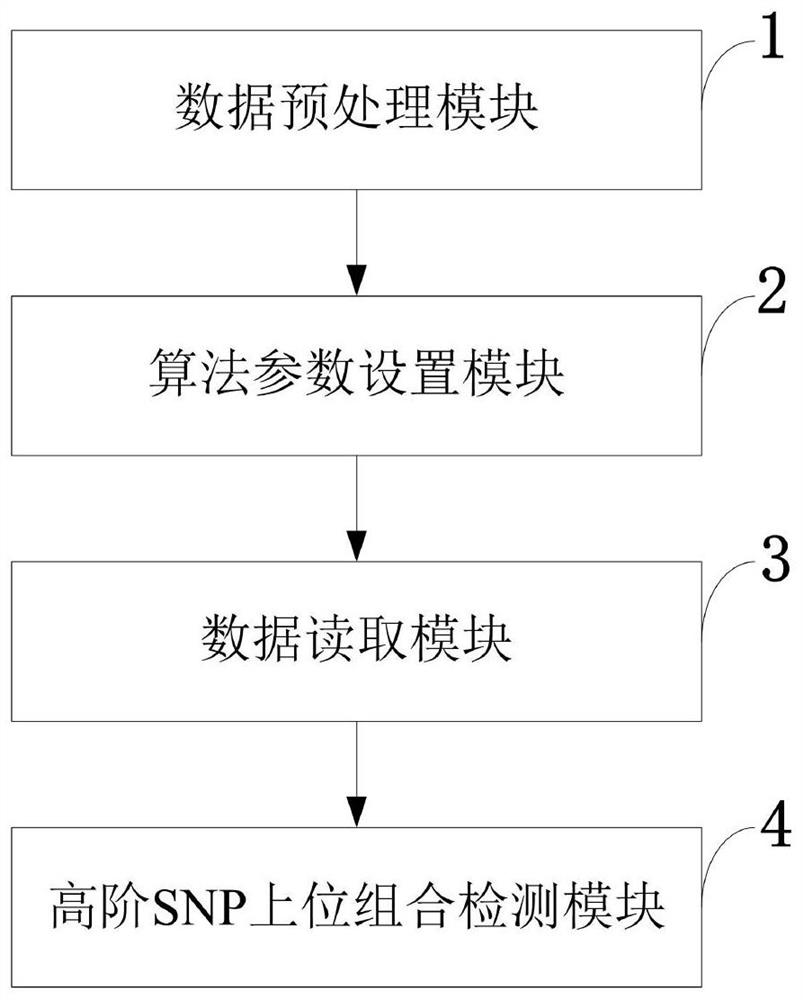
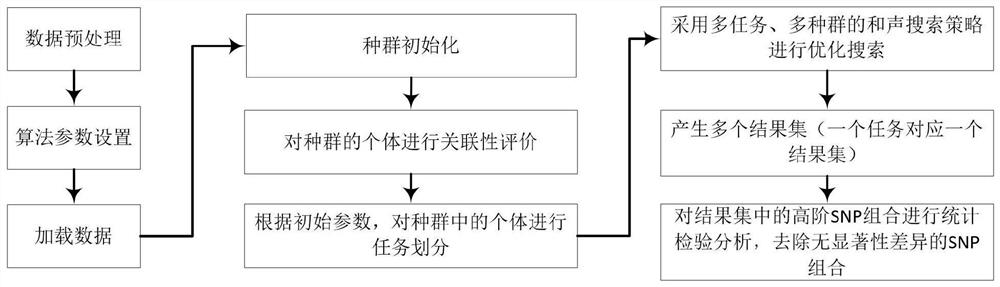
[0084] Aiming at the problems existing in the prior art, the present invention provides a multi-task high-order SNP epistasis detection method, system, storage medium, and equipment. The present invention will be described in detail below with reference to the accompanying drawings.
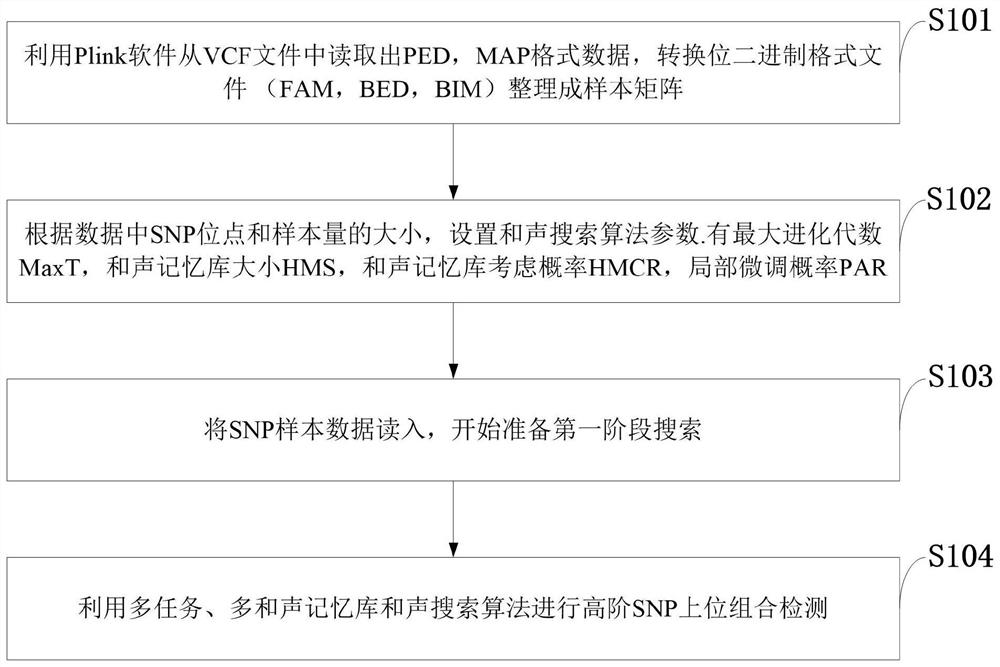
[0085] Such as figure 1 As shown, the multi-task high-order SNP epistasis detection method provided by the present invention comprises the following steps:
[0086]S101: Use Plink software to read out PED and MAP format data from the VCF file, convert bit binary format files (FAM, BED, BIM) into a sample matrix;
[0087] S102: Set the parame...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com