Construction method of single cell transcriptome sequencing library and applications thereof
A transcriptome sequencing and single-cell sequencing technology, applied in chemical libraries, biochemical equipment and methods, combinatorial chemistry, etc., can solve the problems of uneven transcriptome coverage, small number of detected cells, large background noise, etc. The effect of reducing library costs, reducing sample processing time, and simplifying experimental operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0033] Embodiments of the present invention are described below through specific examples, and those skilled in the art can easily understand other advantages and effects of the present invention from the content disclosed in this specification. The present invention can also be implemented or applied through other different specific implementation modes, and various modifications or changes can be made to the details in this specification based on different viewpoints and applications without departing from the spirit of the present invention.
[0034] It should be noted that the library construction in the present invention is based on the mouse mononuclear macrophage leukemia cell RAW264.7.
[0035] The main reagents, instruments and materials used in the present invention are shown in Table 1 below.
[0036] Table 1
[0037]
[0038] The specific implementation process is as follows:
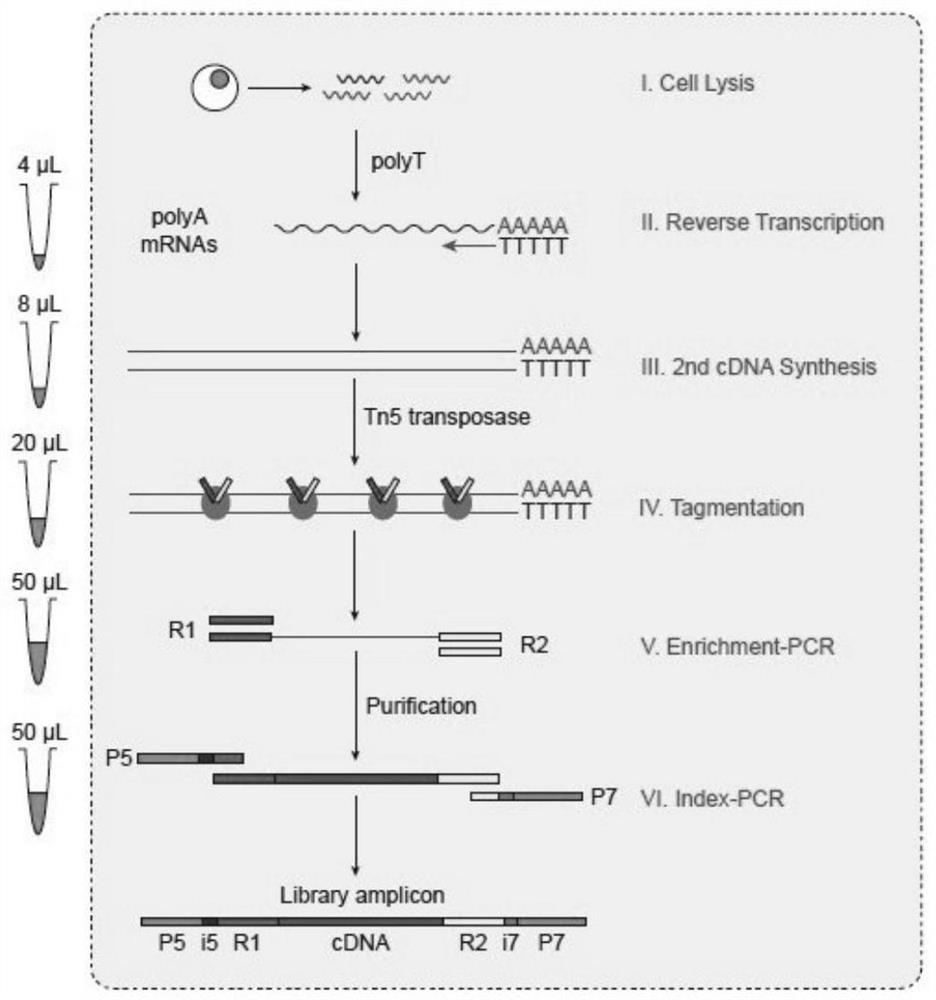
[0039] In this example, single-cell library construction in a 96-well plate is take...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com