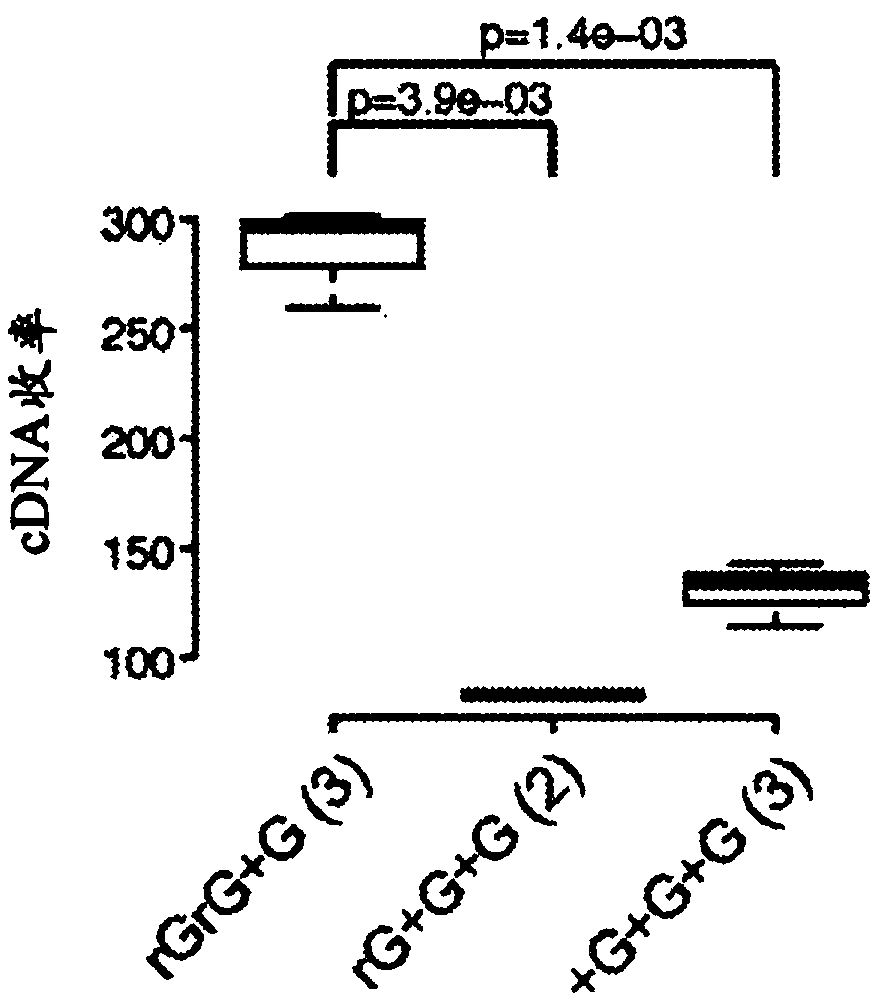
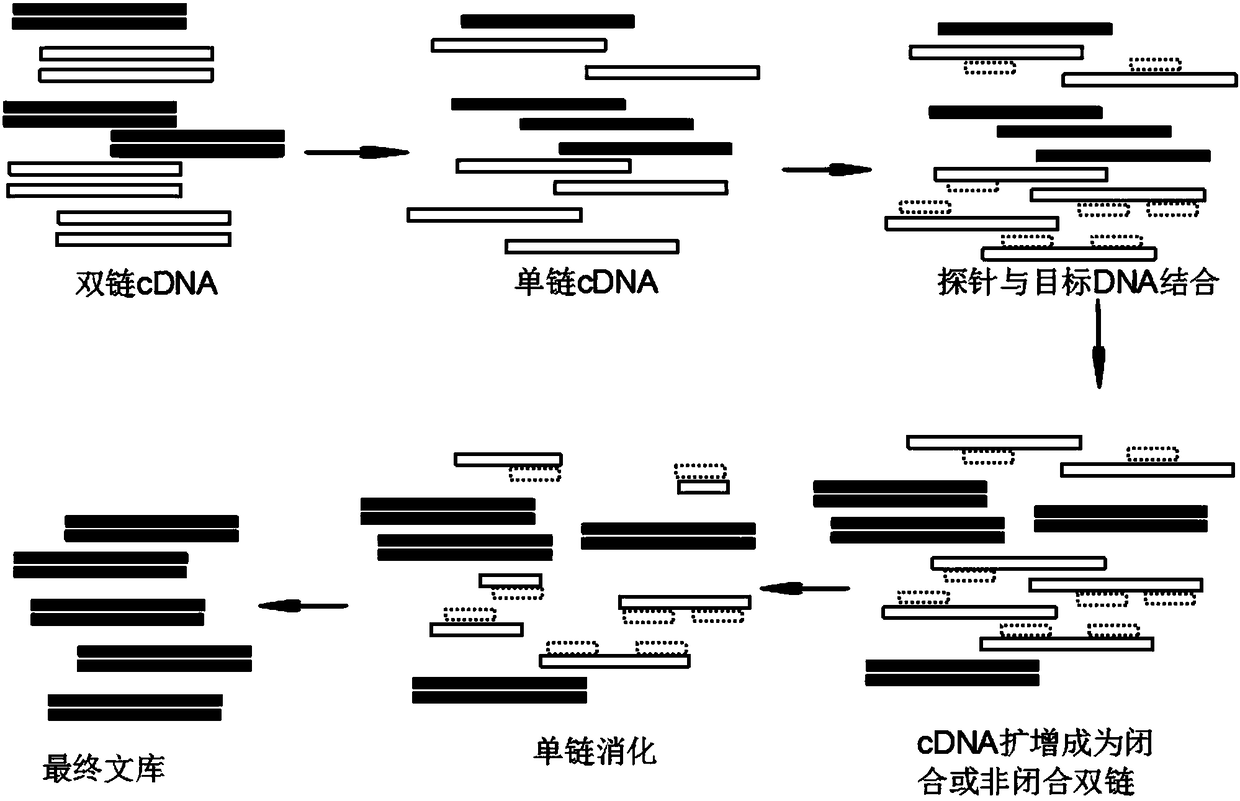
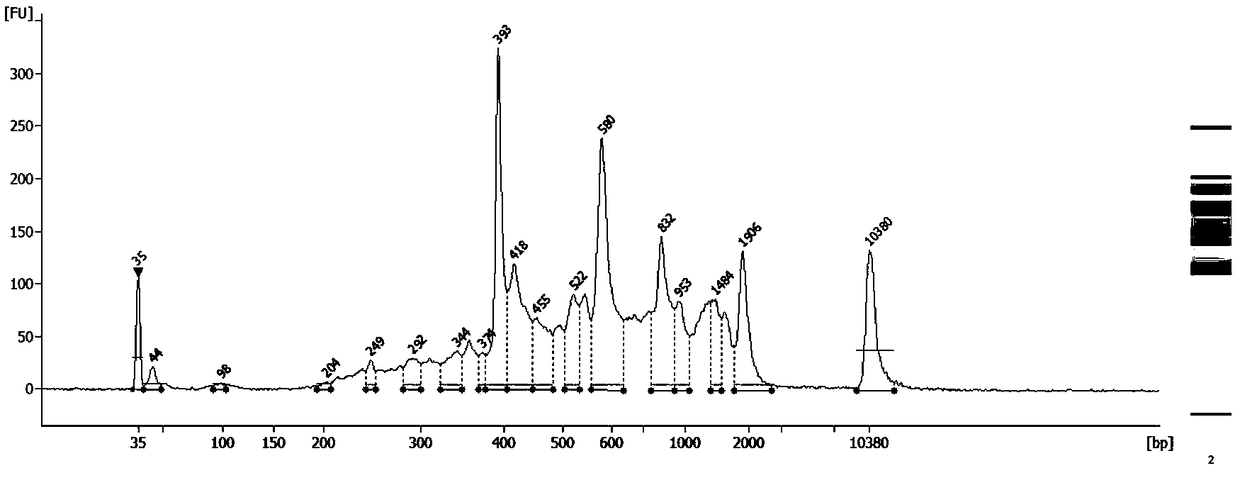
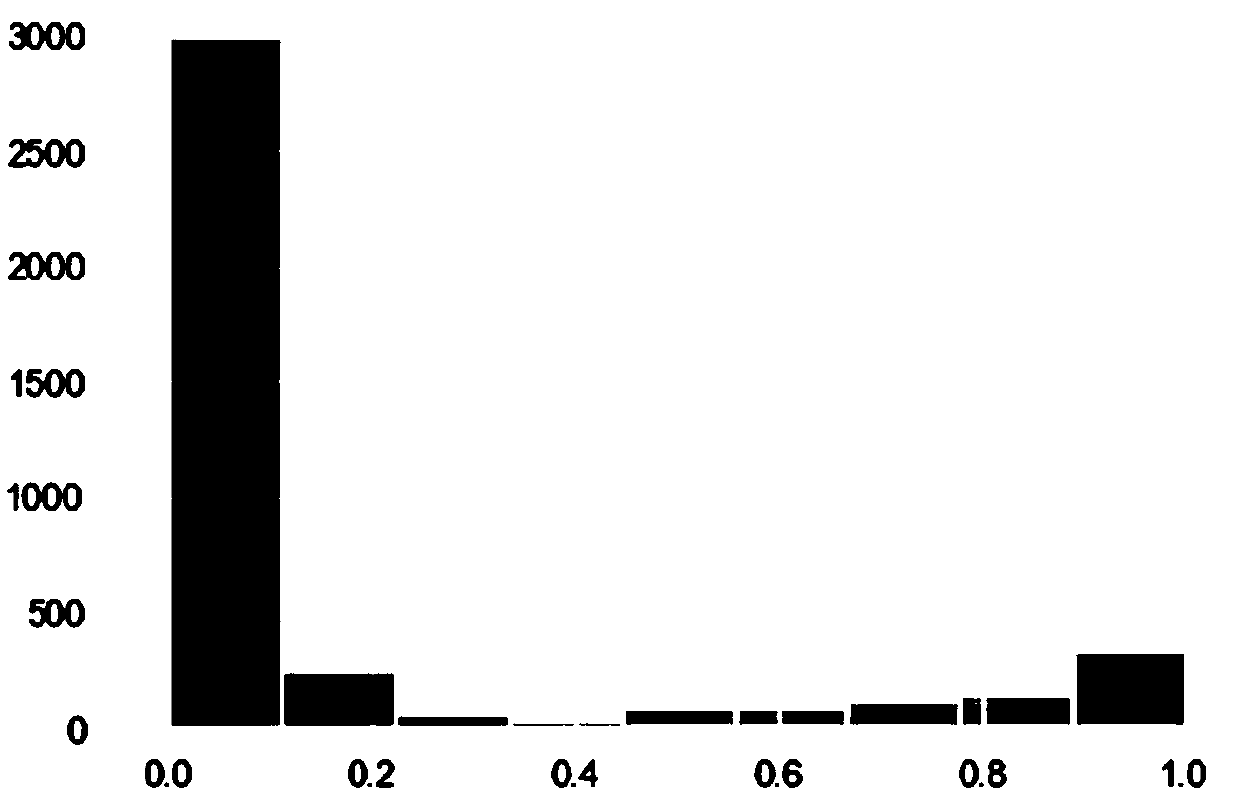
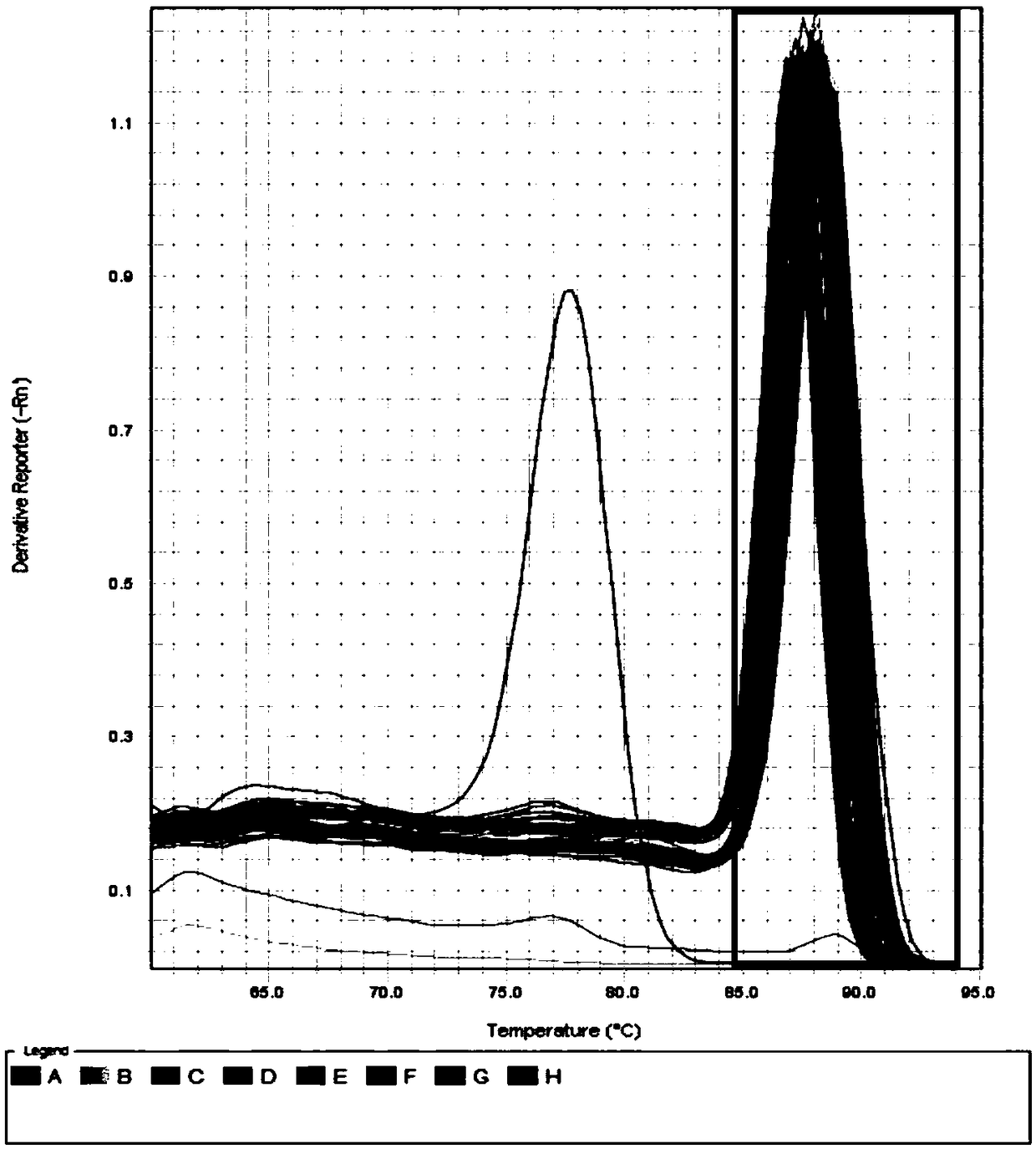
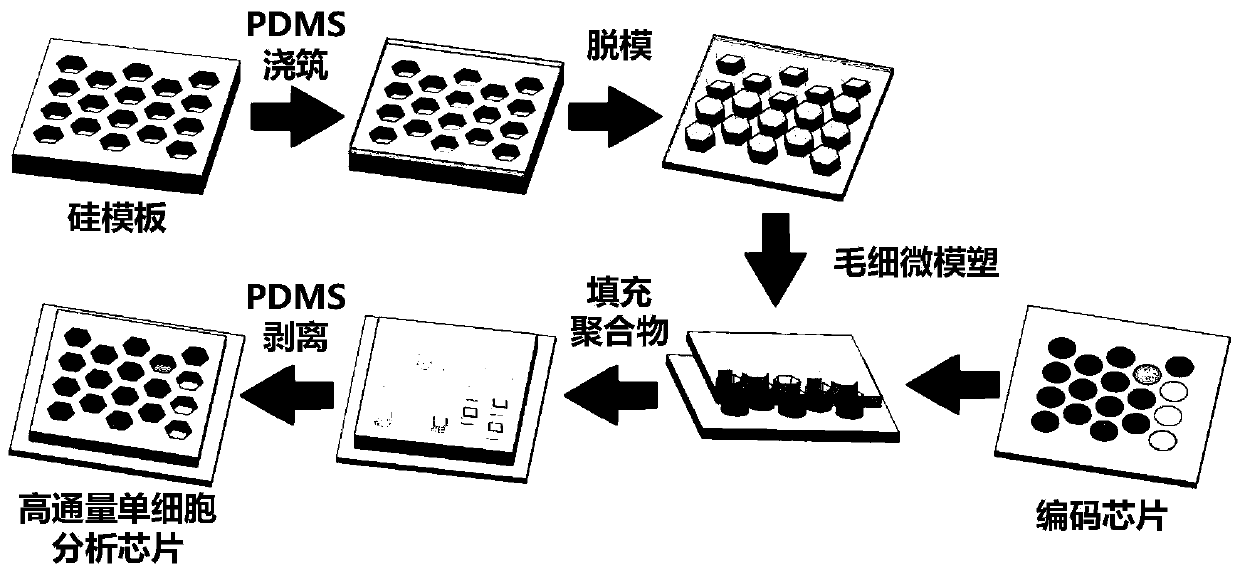
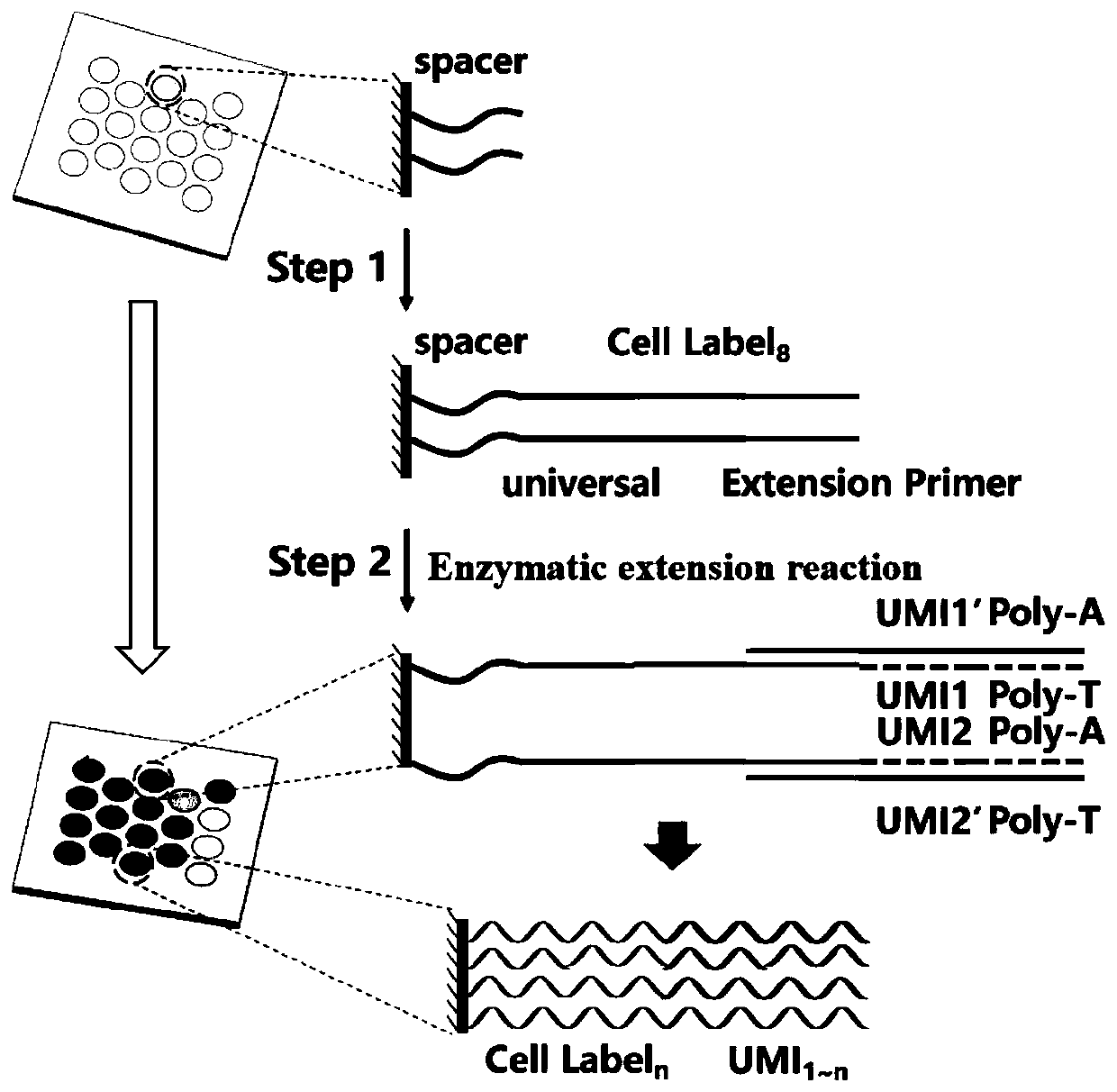
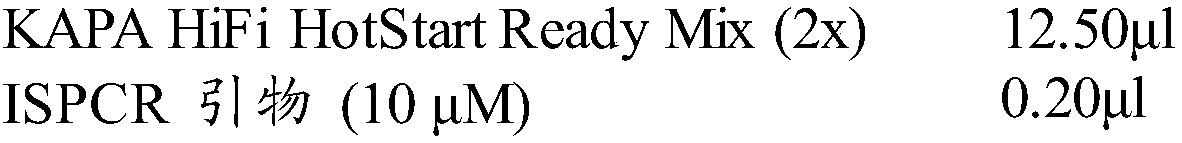Patents
Literature
112 results about "Single cell transcriptome" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Methods for determining spatial and temporal gene expression dynamics in single cells
PendingUS20190218276A1High throughput analysisHigh resolutionMicrobiological testing/measurementImmunoglobulins against virusesCell markerSingle cell transcriptome
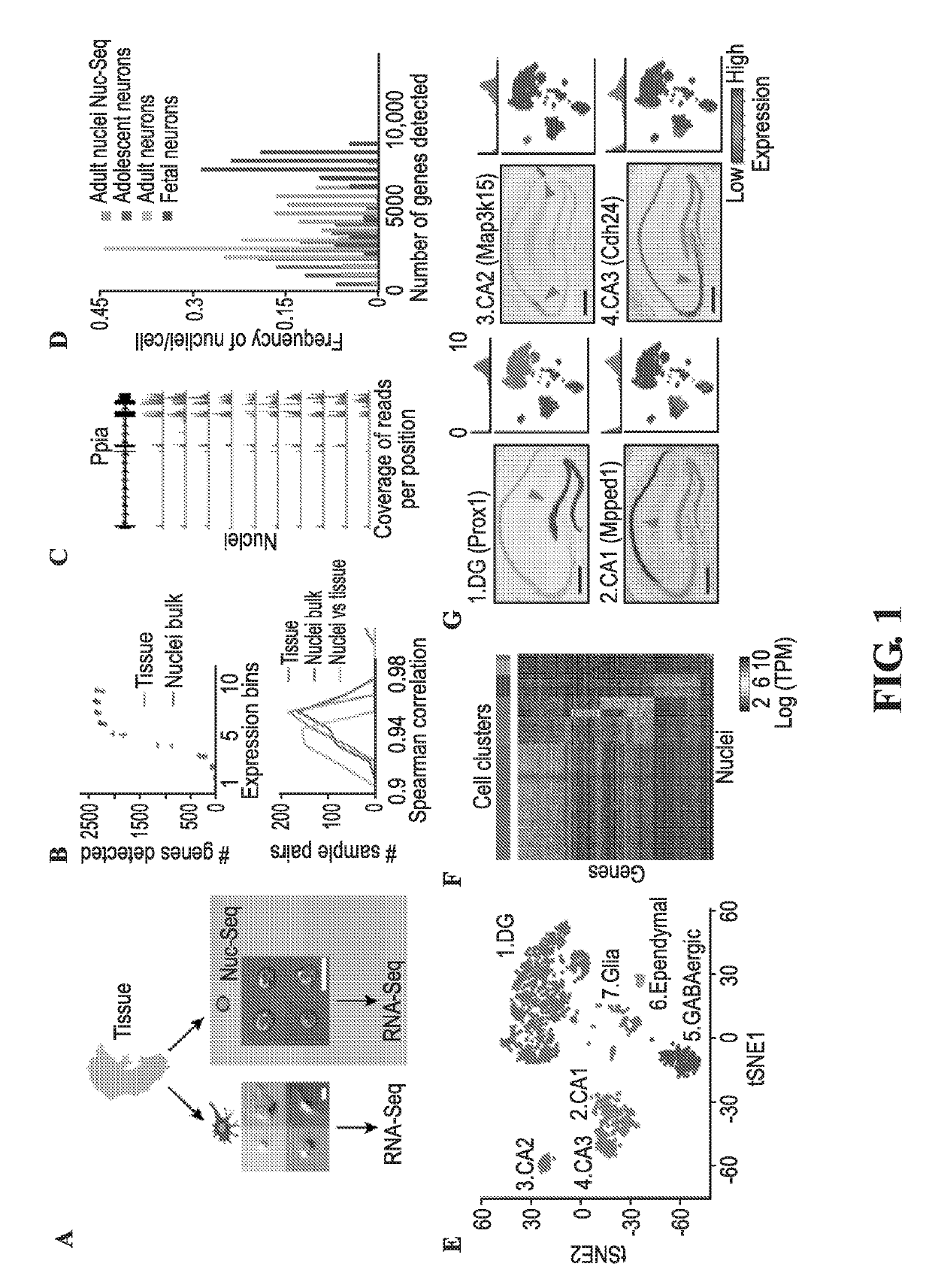
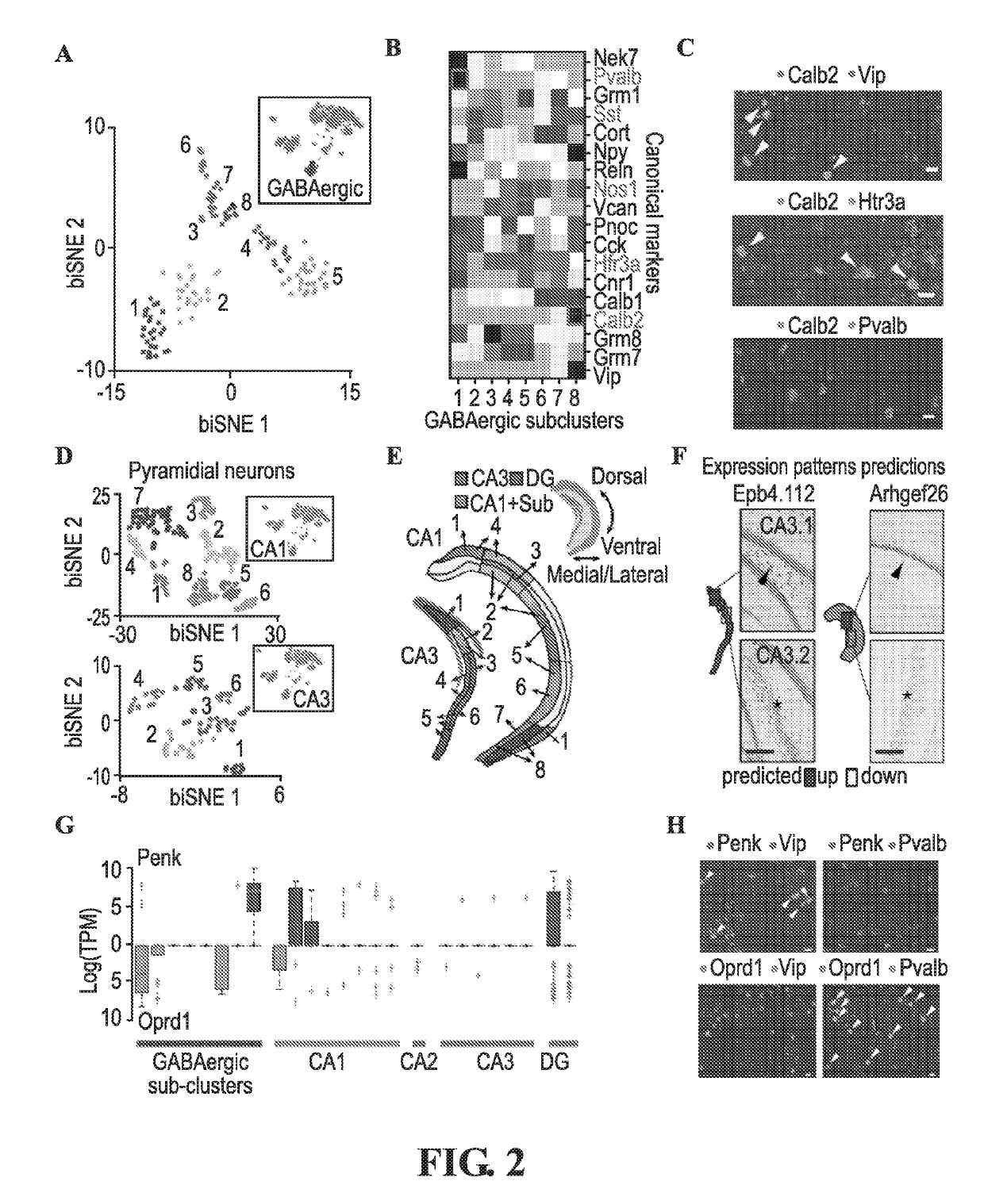
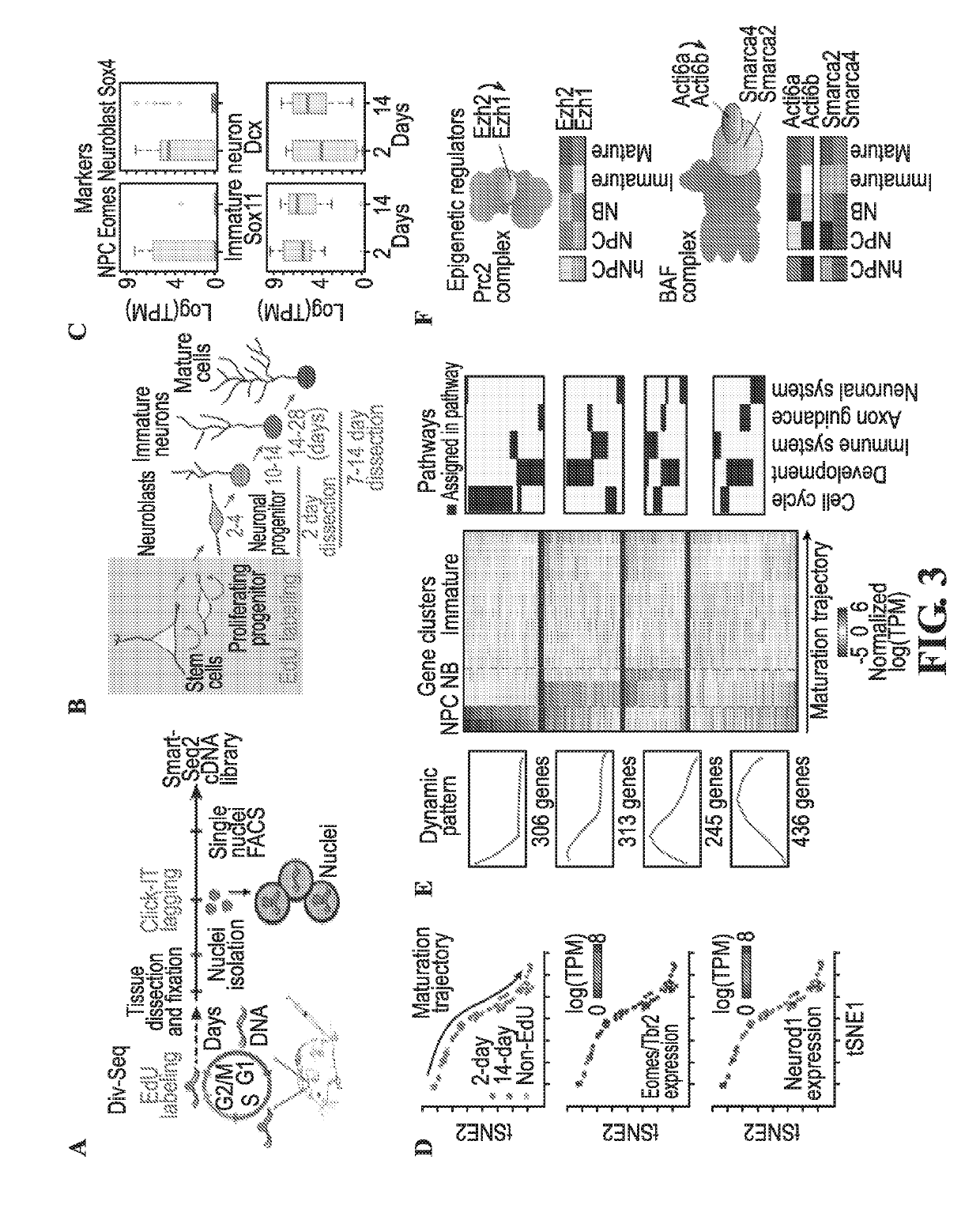
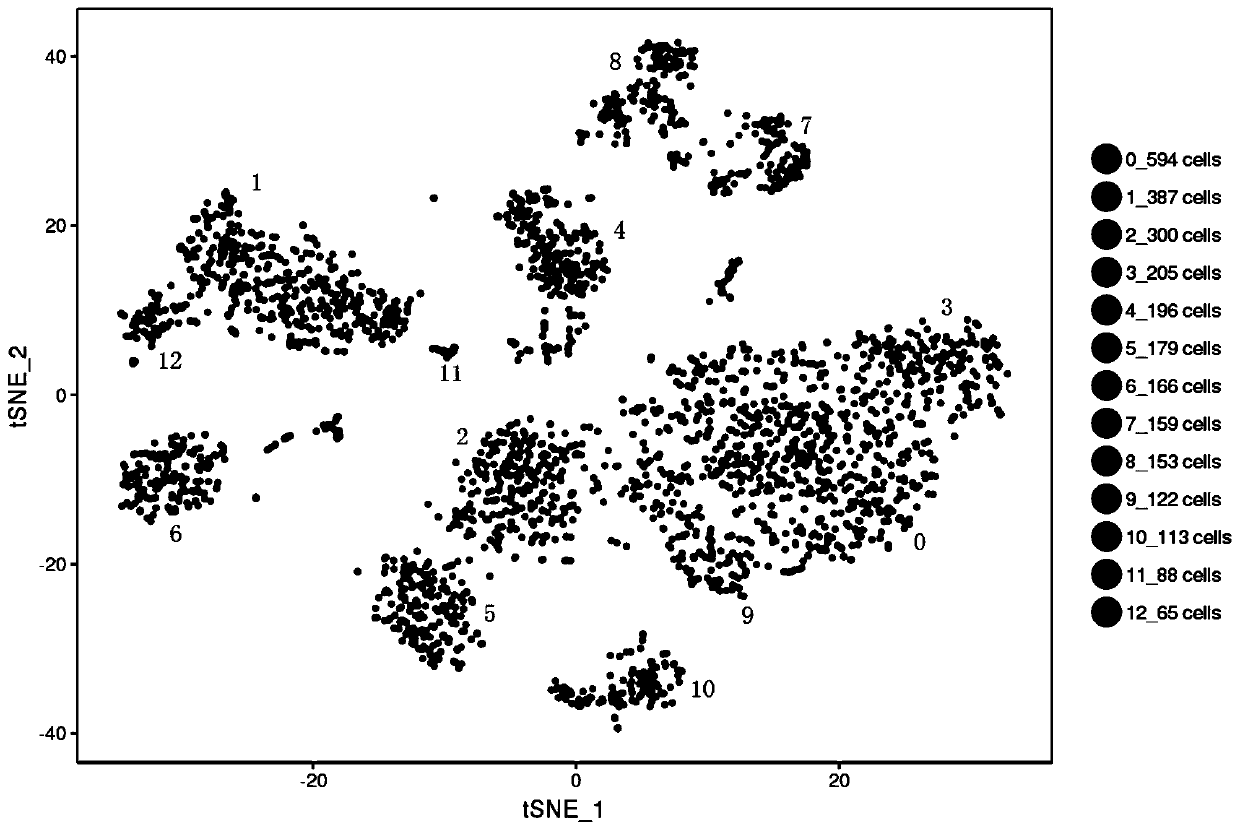
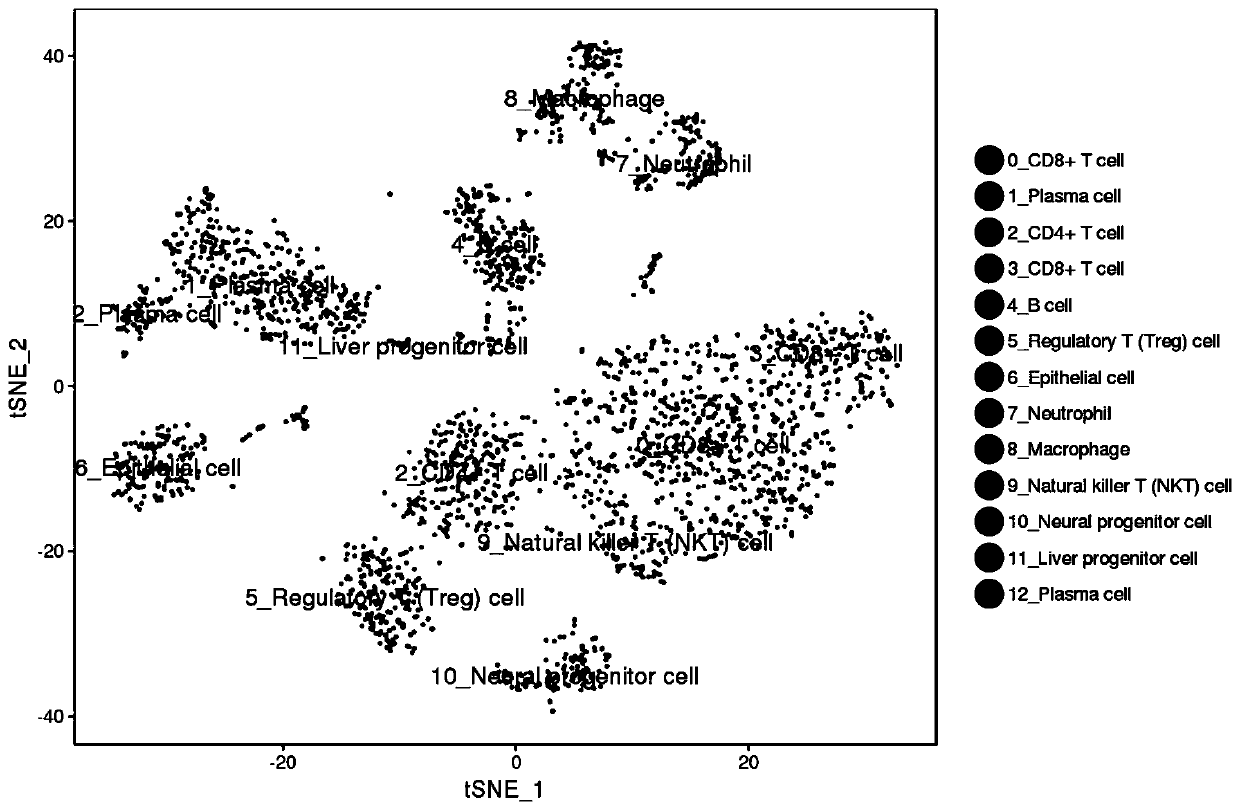
Transcriptomes of individual neurons provide rich information about cell types and dynamic states. However, it is difficult to capture rare dynamic processes, such as adult neurogenesis, because isolation from dense adult tissue is challenging, and markers for each phase are limited. Here, Applicants developed Nuc-seq, Div-Seq, and Dronc-Seq. Div-seq combines Nuc-Seq, a scalable single nucleus RNA-Seq method, with EdU-mediated labeling of proliferating cells. Nuc-Seq can sensitively identify closely related cell types within the adult hippocampus. Div-Seq can track transcriptional dynamics of newborn neurons in an adult neurogenic region in the hippocampus. Dronc-Seq uses a microfluidic device to co-encapsulate individual nuclei in reverse emulsion aqueous droplets in an oil medium together with one uniquely barcoded mRNA-capture bead. Finally, Applicants found rare adult newborn GABAergic neurons in the spinal cord, a non-canonical neurogenic region. Taken together, Nuc-Seq, Div-Seq and Dronc-Seq allow for unbiased analysis of any complex tissue.
Owner:THE BROAD INST INC +2
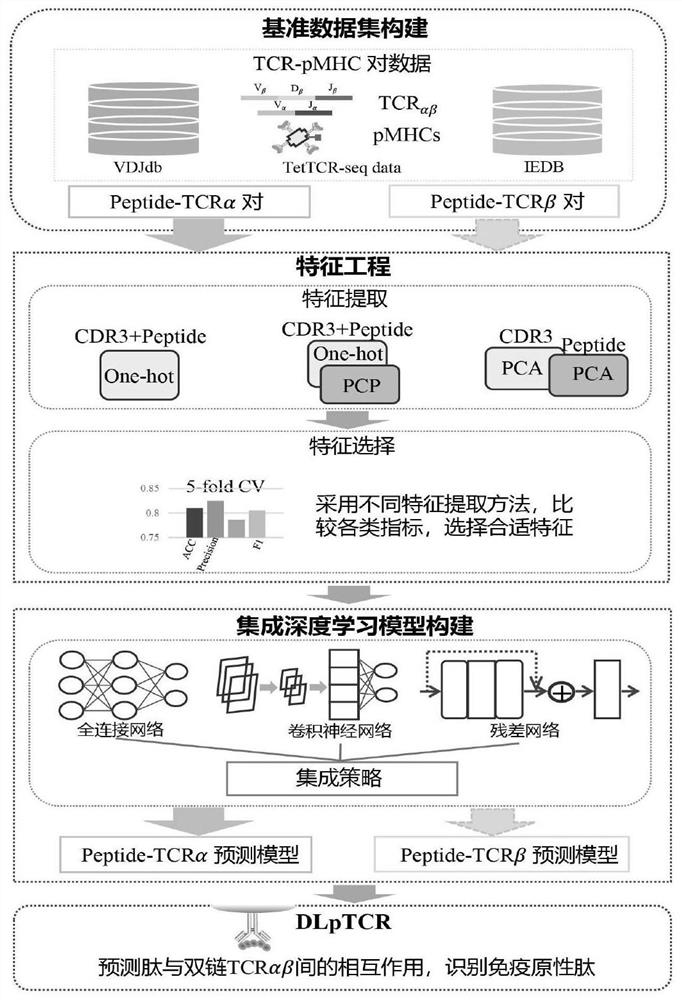
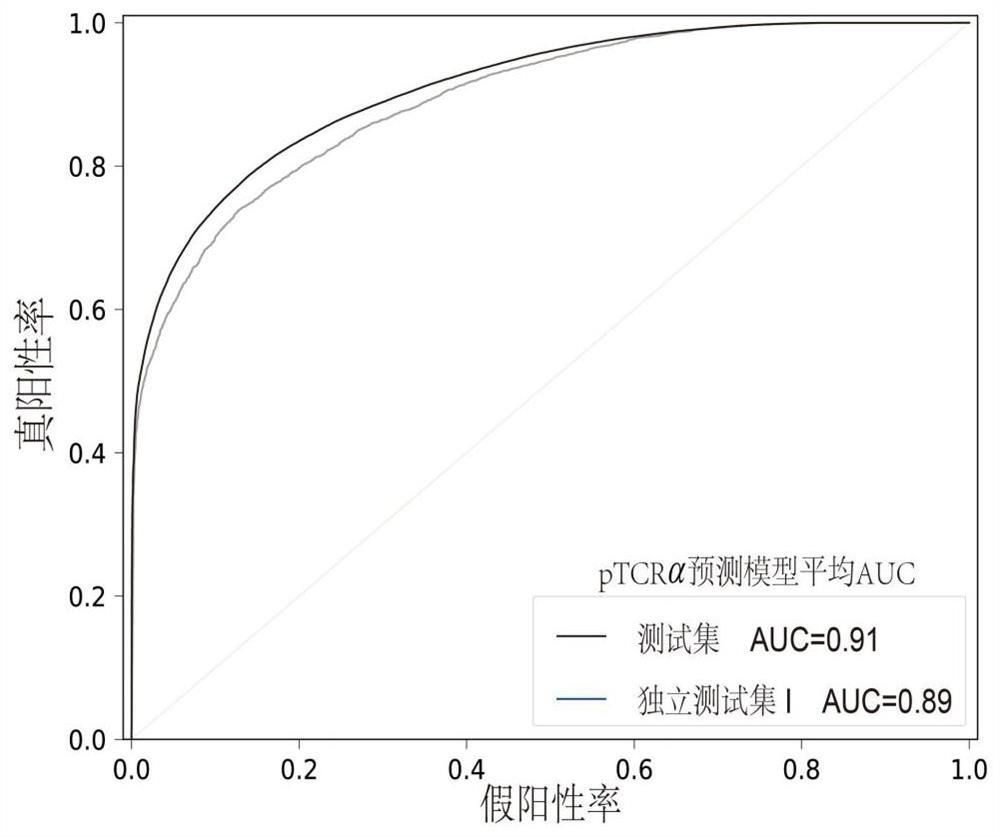
Tumor neoantigen screening method fused with single cell TCR sequencing data
ActiveCN113160887AOvercoming the high rate of misselection and omissionOvercoming problems such as insufficient immunogenicityMicrobiological testing/measurementProteomicsSingle cell transcriptomeCD8
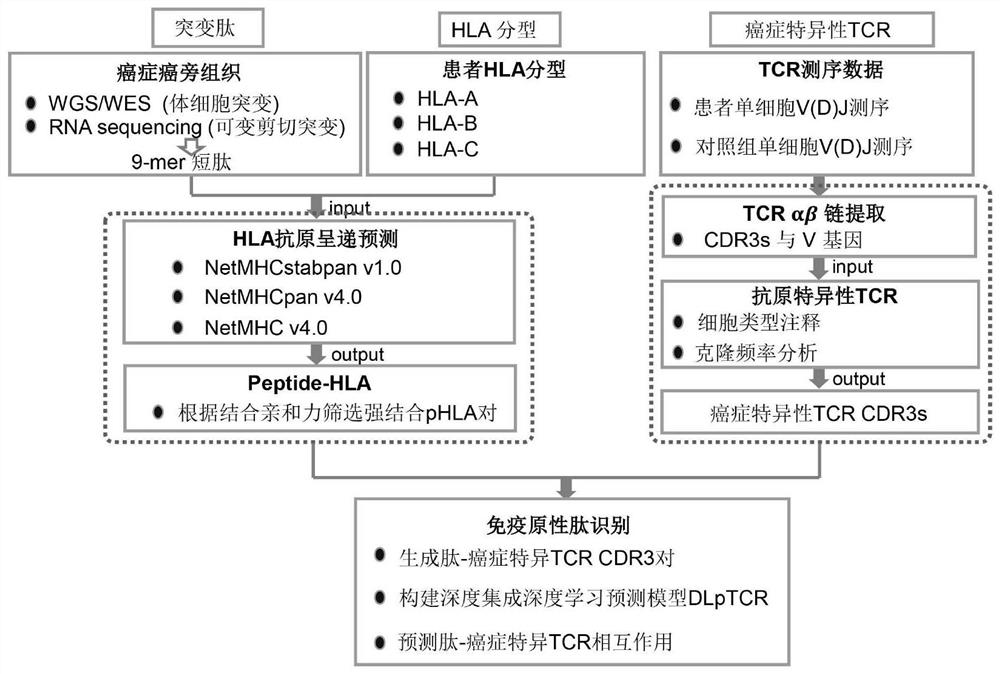
The invention discloses a tumor neoantigen screening method fused with single cell TCR sequencing data. The tumor neoantigen screening method comprises the following steps: based on whole exon sequencing data and transcriptome sequencing data, carrying out quality control, comparison and other steps through software to obtain a neomutant peptide library; predicting an HLA-I type typing by using HLA typing prediction software; in combination with single-cell TCR sequencing and single-cell transcriptome sequencing, finding cancer-specific CD8 + T cell receptors through cell type annotation and clone frequency analysis; meanwhile, based on integrated deep learning, the short peptide immunogenicity is identified through a peptide-TCR interaction prediction model, a tumor neoantigen screening method fused with single cell TCR sequencing data is provided, and the problems that a traditional tumor antigen screening method is high in neoantigen misselection and missed selection rate, insufficient in immunogenicity and the like are solved.
Owner:HARBIN INST OF TECH
Primers for immune repertoire profiling
ActiveUS20210355484A1Immunoglobulin superfamilyMicrobiological testing/measurementSingle cell transcriptomeNucleotide
Disclosed herein include systems, methods, compositions, and kits for immune repertoire profiling. There are provided, in some embodiments, primer panels enabling the determination of the nucleotide sequence of the complete variable region of nucleic acids encoding mouse B cell receptor (BCR) and T cell receptor (TCR) polypeptides. In some embodiments, the method comprises single cell transcriptomic analysis.
Owner:BECTON DICKINSON & CO
Method for annotating cell identities based on single cell transcriptome clustering results
The invention discloses a method for annotating cell identities based on single cell transcriptome clustering results. The method comprises the following steps: S1, providing survival cells, establishing a library by using 10X genetics, and sequencing to obtain transcriptome sequencing data; S2, filtering the sequencing data obtained in the step S1, then carrying out initial analysis by using thesoftware cellranger, and outputting original data; S3, analyzing the original data output in the step S2; S4, performing cell identity annotation, including the steps: S41, classifying the tag genes of the Cell Marker database according to cell types; S42, sorting the tag genes screened out by the FindAllMarkers function according to a P value; S43, taking an intersection of the tag gene of each type of cells in the Cell Marker database and a cell subset tag gene screened by the FindAllMarkers function, and scoring according to the intersection gene; and S44, sorting according to the score ofthe gene intersection, and annotating the cell type corresponding to the highest score as the cell identity of the current subgroup. By adopting the method for annotating cell identities based on single cell transcriptome clustering results disclosed by the invention, the cells can be accurately and quickly classified and subjected to identity annotation.
Owner:广州序科码医学检验实验室有限公司
METHODS AND COMPOSITIONS FOR cDNA SYNTHESIS AND SINGLE-CELL TRANSCRIPTOME PROFILING USING TEMPLATE SWITCHING REACTION
ActiveUS20160258016A1Increase the lengthHigh yieldMicrobiological testing/measurementFermentationCDNA libraryCost effectiveness
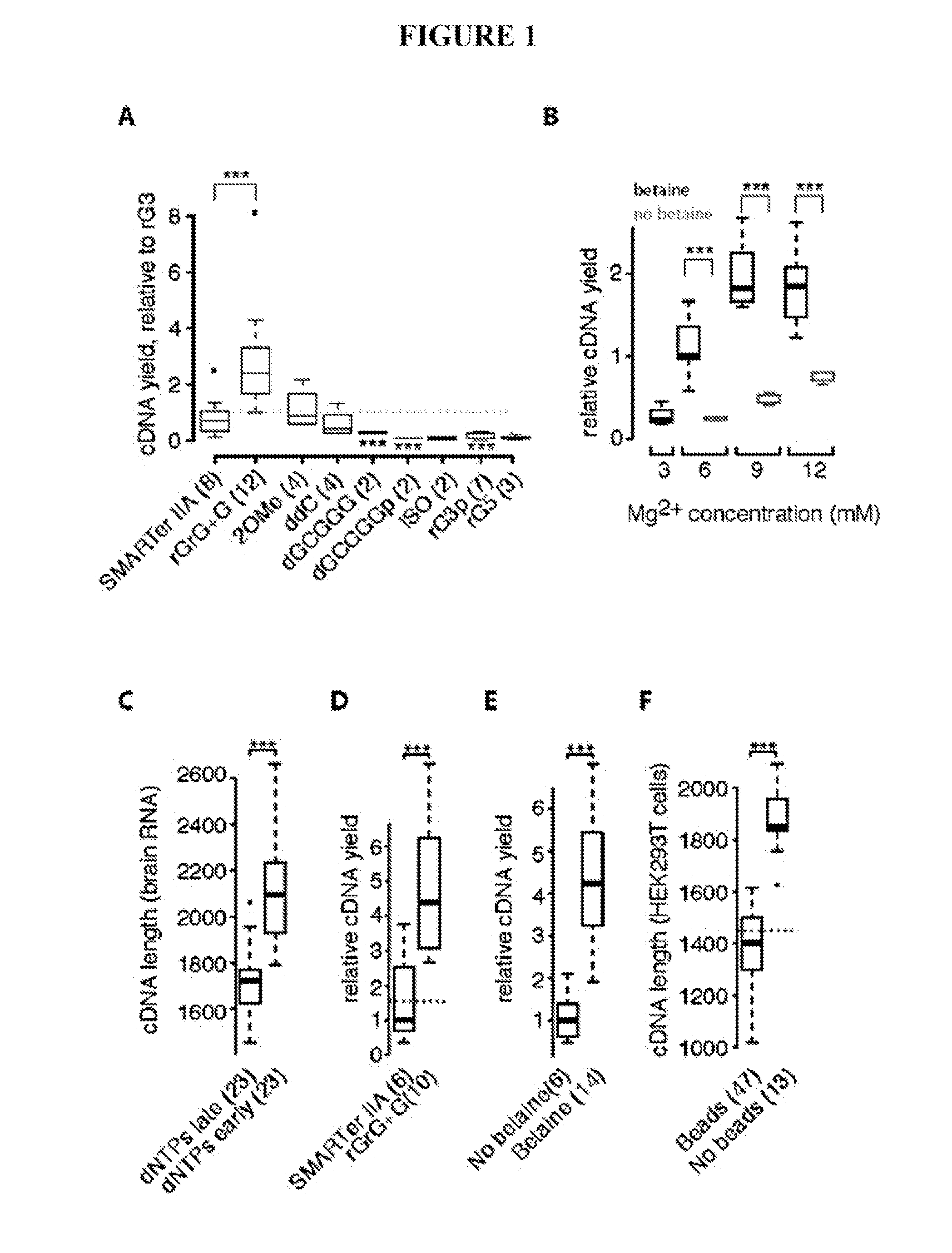
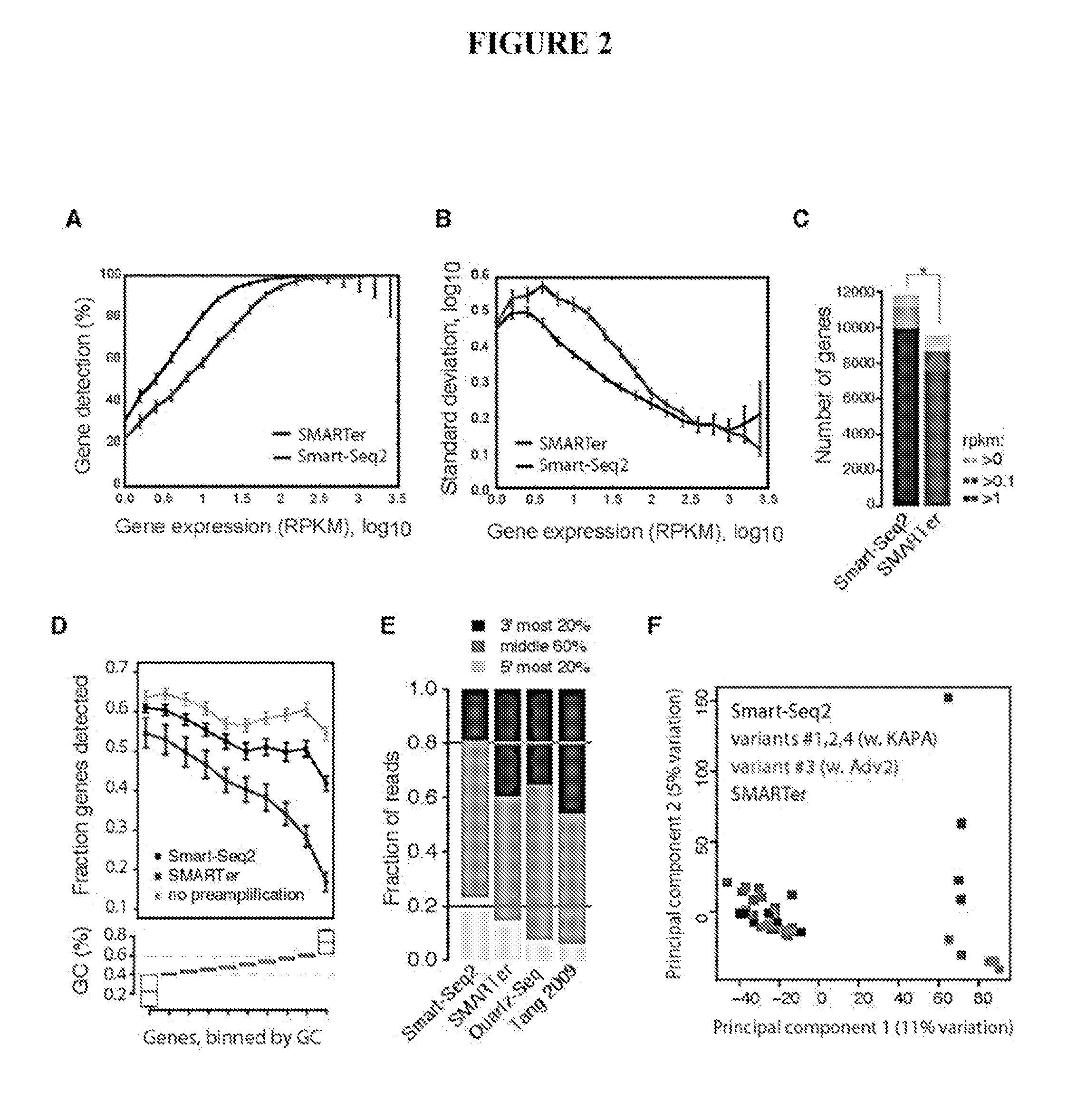
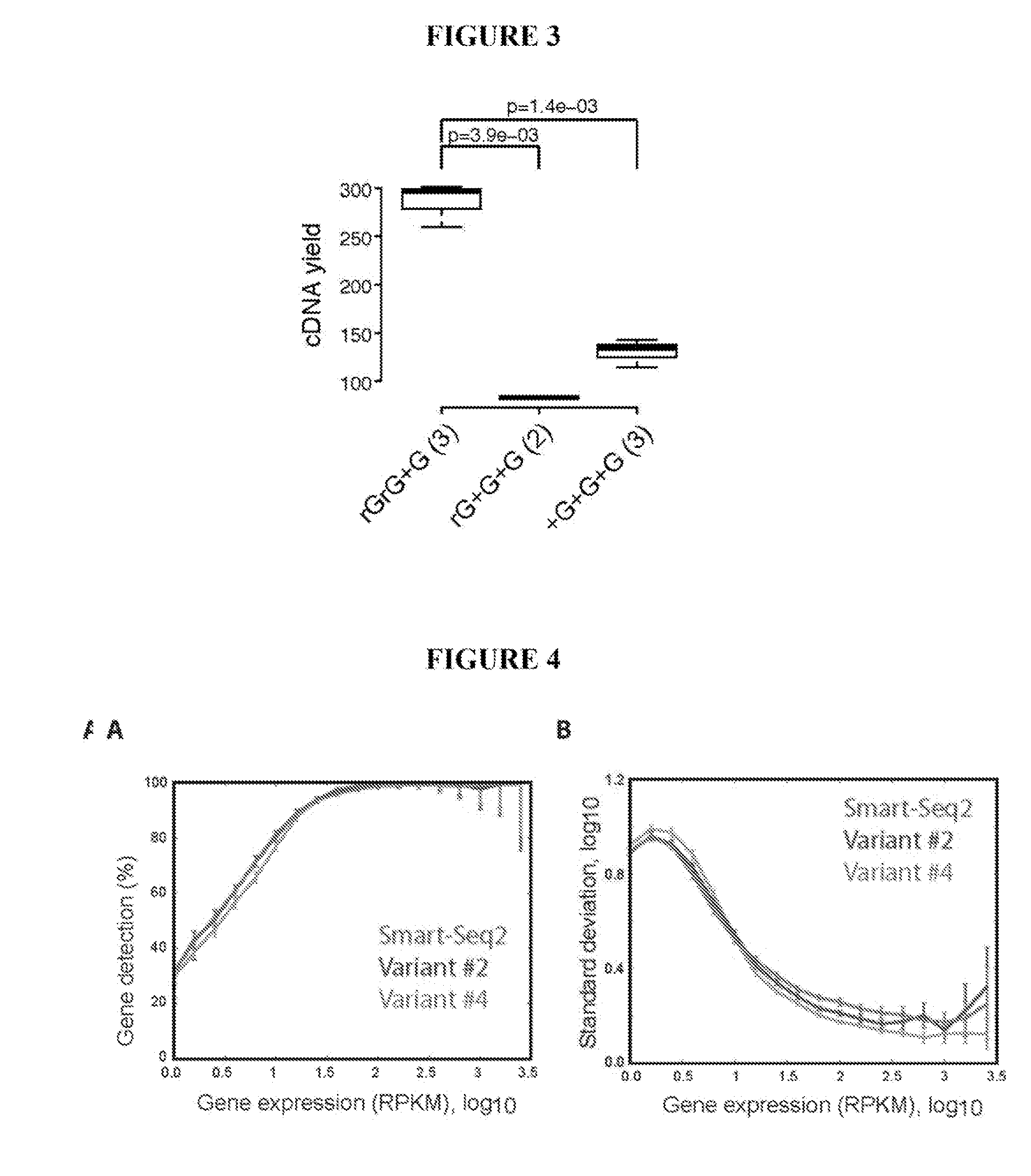
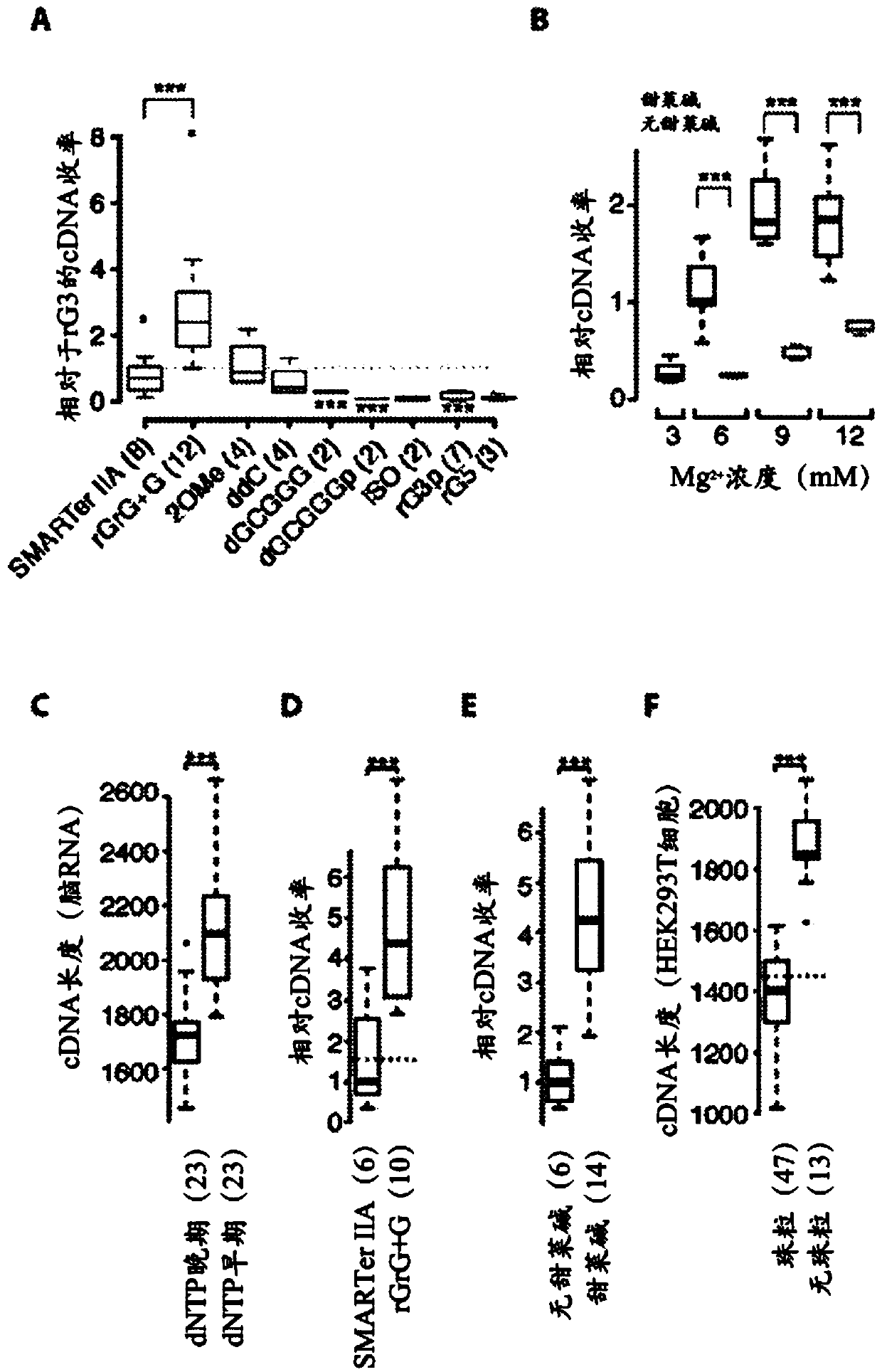
This application discloses methods for cDN′A synthesis with improved reverse transcription, template switching and preamplification to increase both yield and average length of cDNA libraries generated from individual cells. The new methods include exchanging a single nucleoside residue for a locked nucleic acid (INA) at the TSO 3′ end, using a methyl group donor, and / or a MgCb concentration higher than conventionally used. Single-cell transcriptome analyses incorporating these differences have full-length coverage, improved sensitivity and accuracy, have less bias and are more amendable to cost-effective automation. The invention also provides cDNA molecules comprising a locked nucleic acid at the 3′-end, compositions and cDNA libraries comprising these cDNA molecules, and methods for single-cell transcriptome profiling.
Owner:LUDWIG INST FOR CANCER RES
Method for establishing single cell transcriptome sequencing library and application of method
InactiveCN104389026AReduced amplificationIncrease the amount of effective dataMicrobiological testing/measurementLibrary creationSingle cell transcriptomeTranscriptome Sequencing
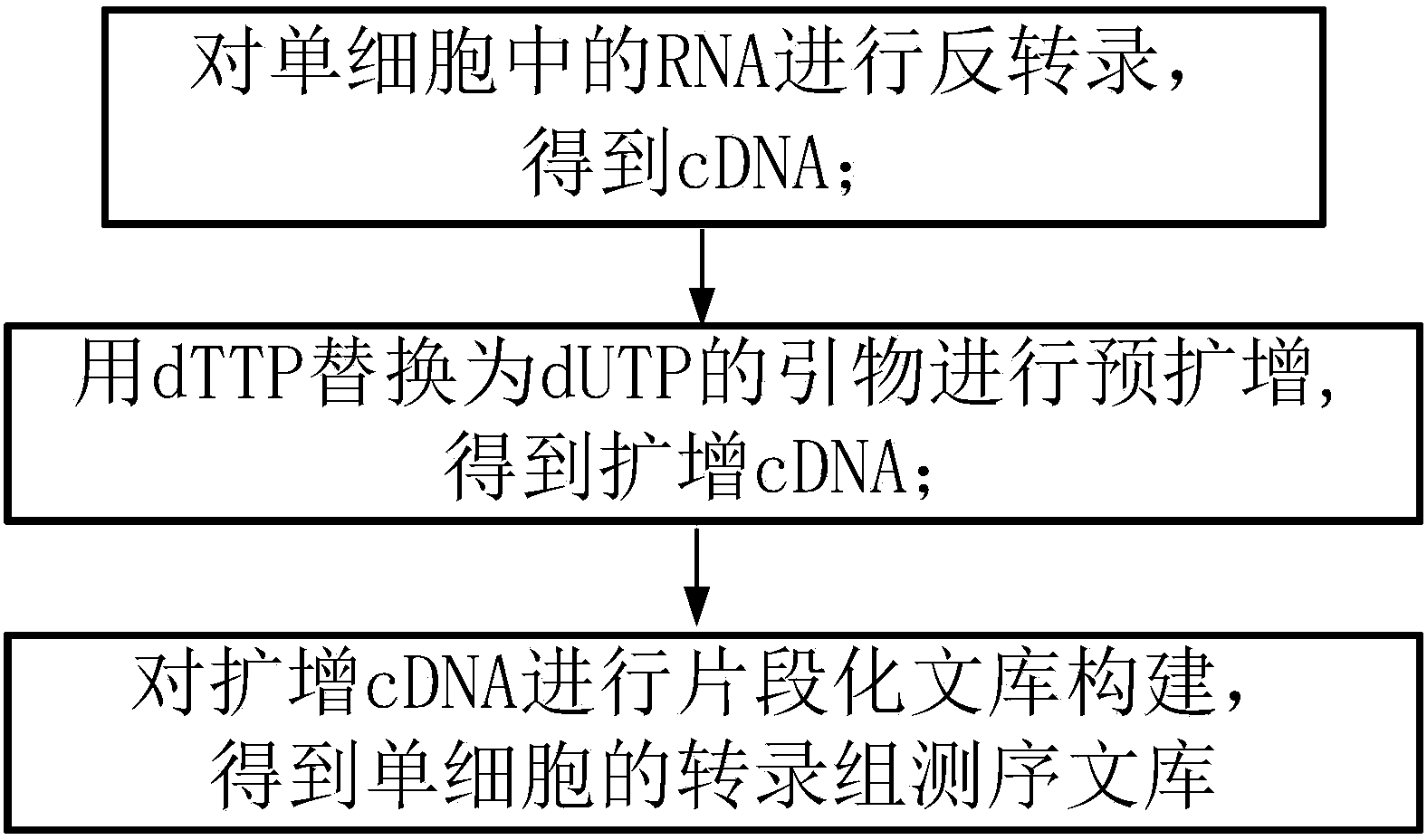
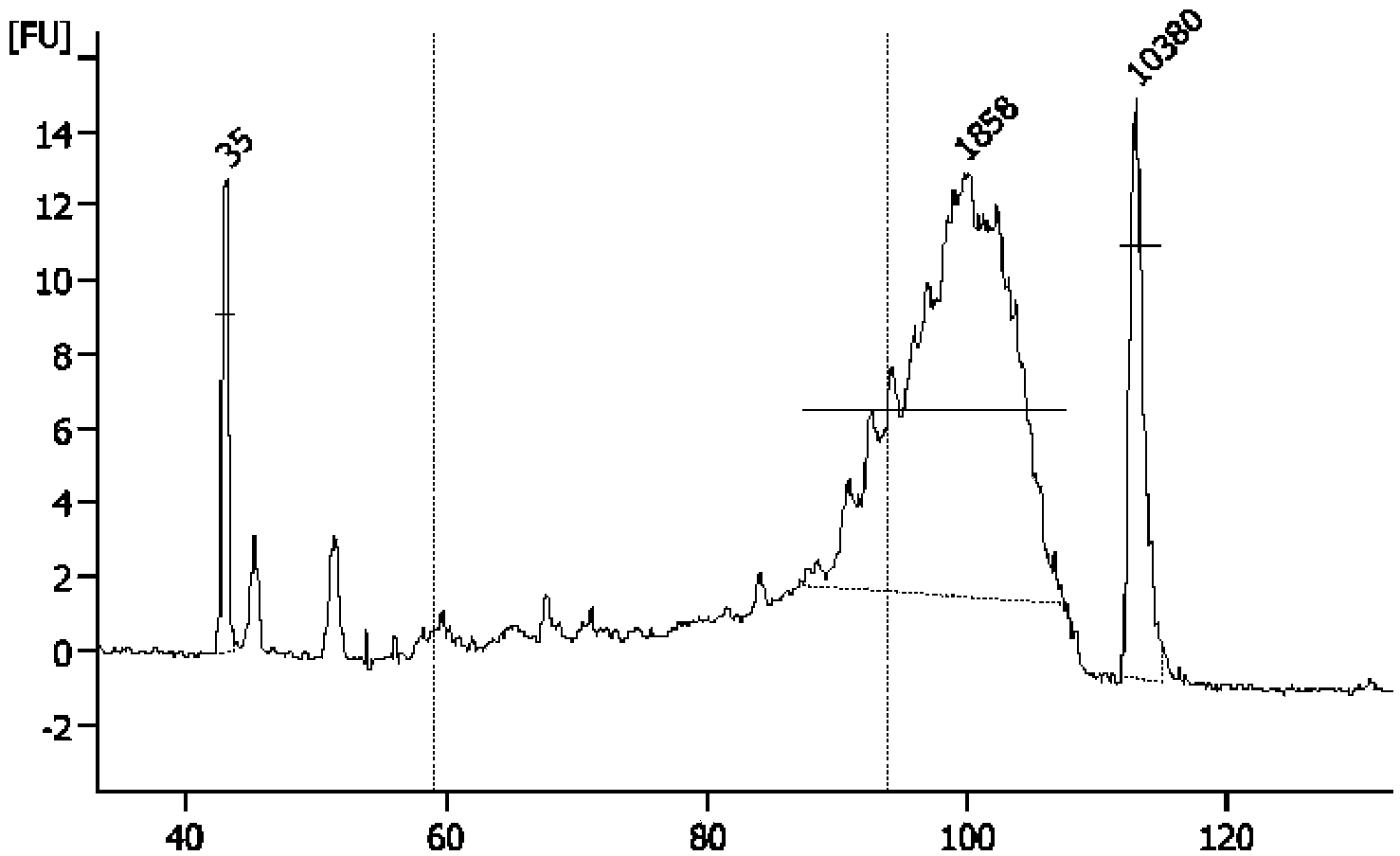
The invention discloses a method for establishing a single cell transcriptome sequencing library and application of the method. The method comprises the following steps: performing reverse transcription on RNA in a single cell, thus obtaining cDNA; performing pre-amplification on cDNA by using an amplification primer, thus obtaining amplified cDNA; and performing fragmentation library construction on the amplified cDNA, thus obtaining a transcriptome sequencing library of the single cell, wherein dTTP in the amplification primer is substituted by dUTP. According to the method disclosed by the invention, as the dTTP in the pre-amplification primer is substituted by dUTP, fragments containing the pre-amplification primer in a jointed fragment can be interrupted in an enzymic digestion step after the step of joint connection, and are removed in later high-temperature pre-denaturation and denaturation steps; furthermore, the amplification of fragments with the pre-amplification primer can be reduced, the ratio that the proportion of data with pre-amplification primer pollution in the obtained sequencing data is greatly reduced, and the effective data amount of the obtained data is greatly increased.
Owner:BEIJING NOVOGENE TECH CO LTD
Method for cell quality control through unicellular transcriptome sequencing
ActiveCN106701995AControl UniformityControl homogeneityMicrobiological testing/measurementBiostatisticsSingle cell transcriptomePrincipal component analysis
The invention provides a method for cell quality control through unicellular transcriptome sequencing. The method comprises the following steps of: performing unicellular separation so as to obtain a total unicellular number; performing transcriptome amplification on each unicell; confirming that amplification can be implemented but the unicellular number is not sufficient for analysis on sequencing results; performing sequencing and analysis, including performing main component analysis and cluster analysis on the sequencing results, comparing with a standard unicellular database, and calculating the unicellular number off from total tendency; screening, including screening based on unicellular partitioning and screening based on expression gene types, wherein the step of screening based on unicellular partitioning comprises a step of screening cells of which the activity levels are greater than 75%, and the step of screening based on the expression gene types comprises a step of classifying expression genes into a good gene group, a neutral gene group and a malignant gene group according to gene functions. By adopting the method provided by the invention, the uniformity or the homogeneity of cells can be effectively controlled.
Owner:GENEIS TECH BEIJING CO LTD
T cell subsets in lung cancer and feature genes thereof
ActiveCN109777872AMicrobiological testing/measurementBiological testingRegulatory T cellSingle cell transcriptome
The invention discloses T cell subsets in lung cancer and feature genes thereof. A single-cell transcriptome analysis technology is adopted, and by analyzing a single-cell gene expression profile of infiltrating T cells in the lung cancer tissue, the T cell subsets capable of reflecting body tumor immune states of patients with the lung cancer are separated and characterized, namely the exhaustedCD 8+ T cells or regulatory T cells capable of expressing genes including TNFRSF9, TNFRSF18 and LAYN are separated and characterized. Through research, relations between lung cancer prognosis and thenew feature genes which include TNFRSF9, TNFRSF18 and LAYN and are expressed by the cell subsets are further determined. Therefore, the T cell subsets in lung cancer and the feature genes thereof canbe used for diagnosis and monitoring of lung cancer prognosis and serve as new targets for lung cancer immunotherapy.
Owner:PEKING UNIV
Cell subset annotation method based on single cell transcriptome sequencing
ActiveCN112700820AComprehensive identificationFix the annotation problemBiostatisticsSequence analysisSingle cell transcriptomeCell subpopulations
The invention provides a cell subset annotation method based on single cell transcriptome sequencing. The cell subset annotation method comprises the following steps: 1) 10x barcode UMI identification; 2) genome comparison; 3) gene expression profile comparison; 4) low-quality cell filtering and data homogenization; 5) cell population clustering; 6) marker gene extraction; 7) cell subset annotation. The invention belongs to the technical field of biological information analysis, and provides a cell subset annotation method based on single-cell transcriptome sequencing, which solves the problem of single-cell subset annotation, so that single-cell sequencing data can support cell annotation according to a gene expression profile and / or a cell Marker gene after conventional analysis. Therefore, organic combination of different annotation methods is realized, and the distribution condition and related information of cell types are obtained.
Owner:广州华银医学检验中心有限公司 +1
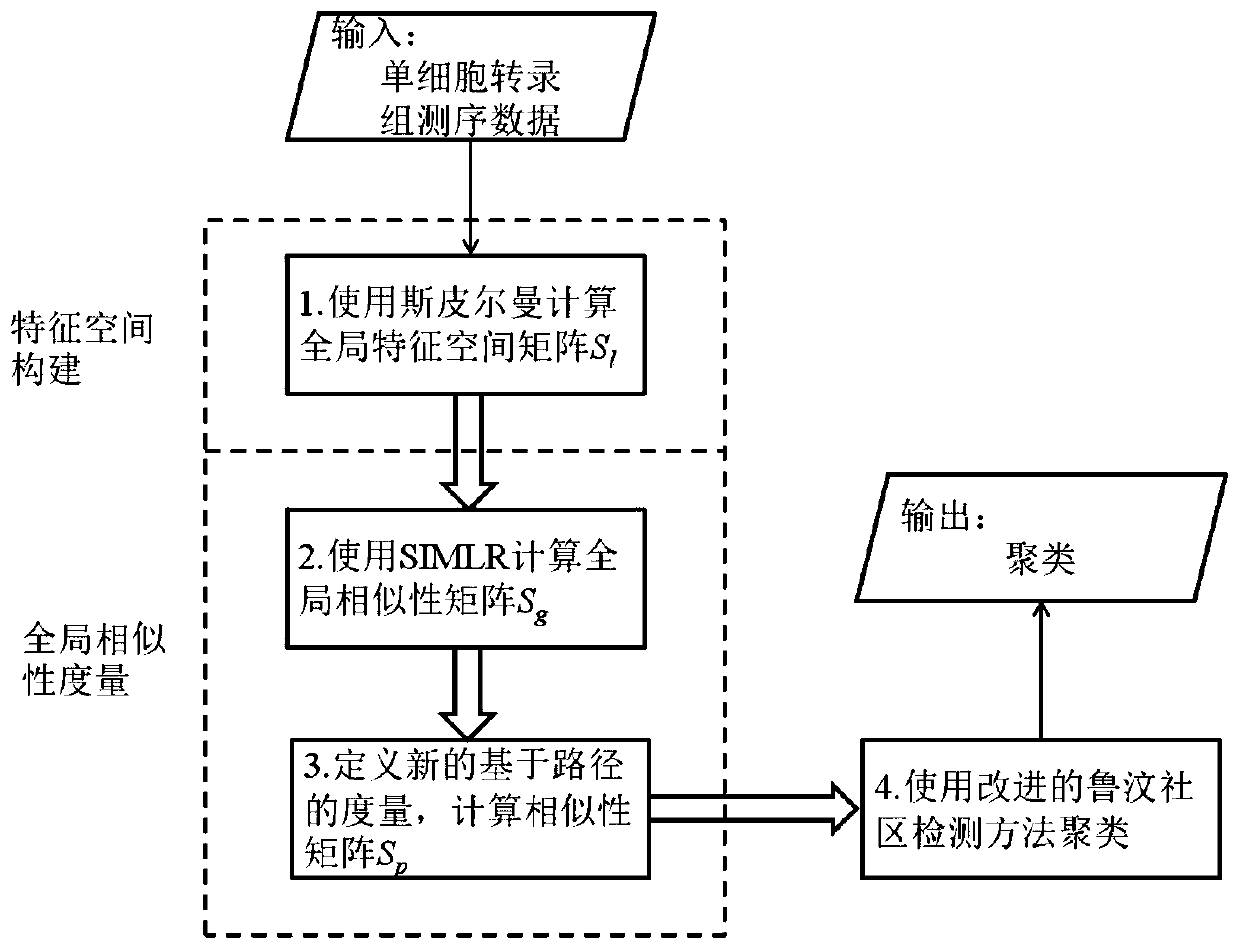
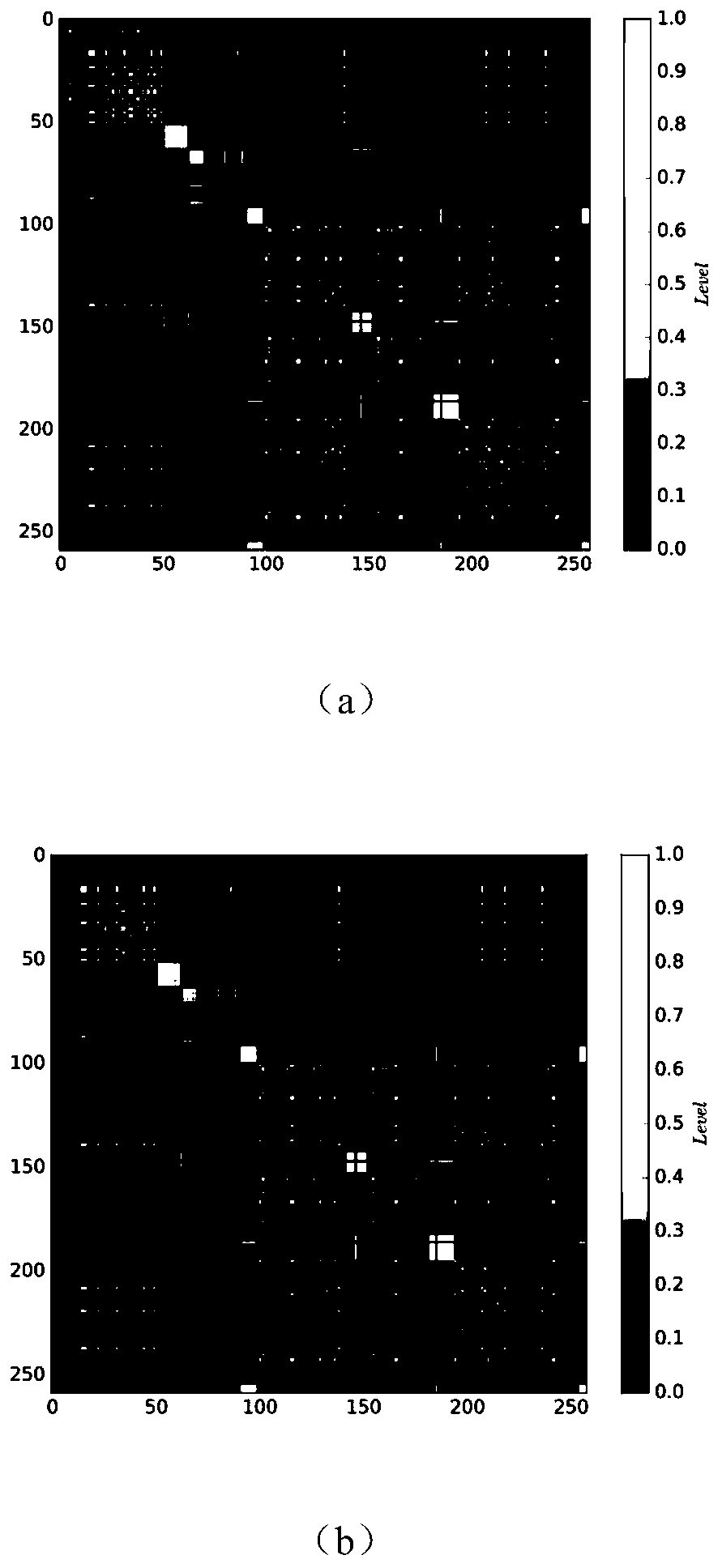
Single cell clustering method and device, electronic equipment and storage medium
The invention discloses a single cell clustering method and device, electronic equipment and a storage medium. On the basis of calculating local similarity between node pairs based on distance information, a global feature space is constructed. Based on the global feature space, the global similarity between the node pairs is calculated by using a multi-core learning method. Then, nodes on all second-order paths of the node pairs are expanded and considered, more relevant node information is added, and a more effective global similarity calculation method is constructed. Finally, by sorting the nodes according to the node degrees, and determining an initial association joining sequence of the nodes, a Louvain association detection method is improved, and clustering is carried out by usingthe method. The method is simple and effective, and compared with other methods, tests on a public single cell transcriptome sequencing data set show that the method has good prediction performance inthe aspect of single cell transcriptome sequencing data clustering.
Owner:YULIN NORMAL UNIVERSITY +1
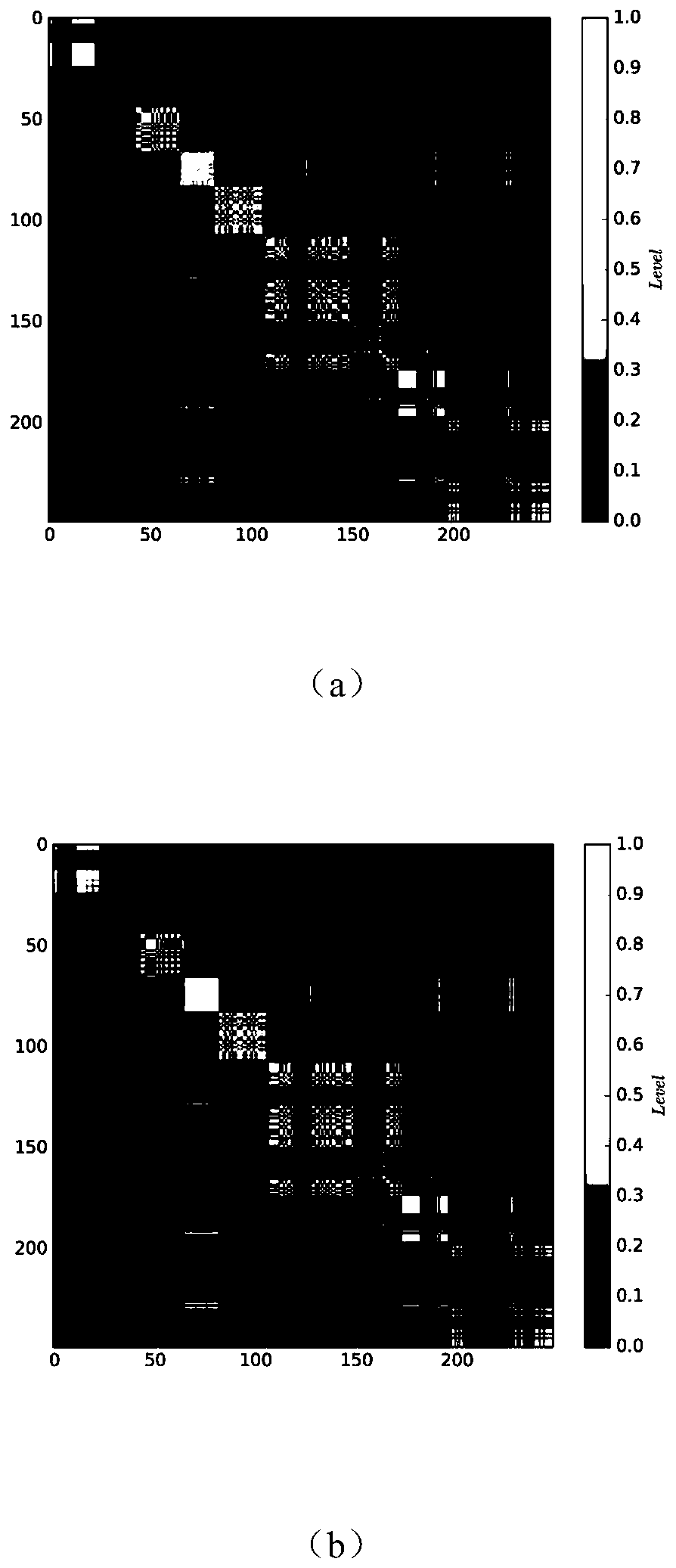
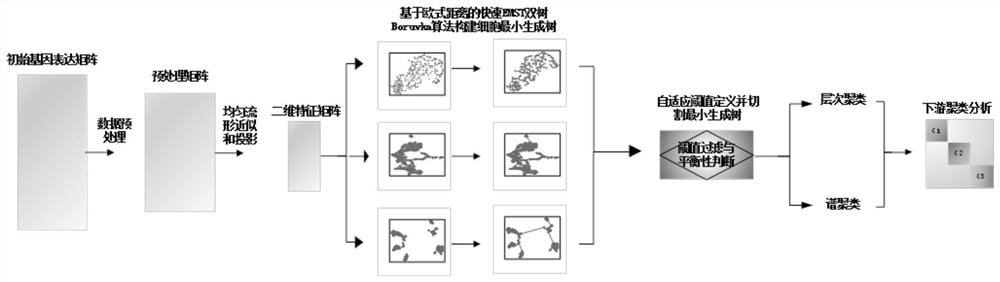
Single-cell transcriptome sequencing data clustering recommendation method based on two-dimensional distribution structure judgment
ActiveCN112750502AImprove clustering accuracyBiostatisticsCharacter and pattern recognitionSingle cell transcriptomeCell
The invention discloses a single-cell transcriptome sequencing data clustering recommendation method based on two-dimensional distribution structure judgment, which comprises the following steps of: acquiring a gene expression matrix obtained by single-cell transcriptome sequencing data of a plurality of cells, and after filtering and standardization processing, constructing a two-dimensional feature matrix and carrying out linear normalization; calculating an Euclidean distance between cells according to the normalized two-dimensional feature matrix, thereby establishing a cell minimum spanning tree; cutting the cell minimum spanning tree through a self-adaptive threshold, and determining a two-dimensional distribution structure of data according to the balance of clusters formed after cutting; and recommending and applying a hierarchical clustering algorithm for data with fuzzy inter-cluster boundaries and a continuous two-dimensional distribution structure, and recommending and applying a spectral clustering algorithm for data with obvious inter-cluster boundaries and a block two-dimensional distribution structure. According to the method, a method which is more suitable for a two-dimensional distribution structure of single cell transcriptome sequencing data in hierarchical clustering and spectral clustering can be recommended to serve as a downstream clustering analysis method, and the clustering accuracy is improved.
Owner:CENT SOUTH UNIV
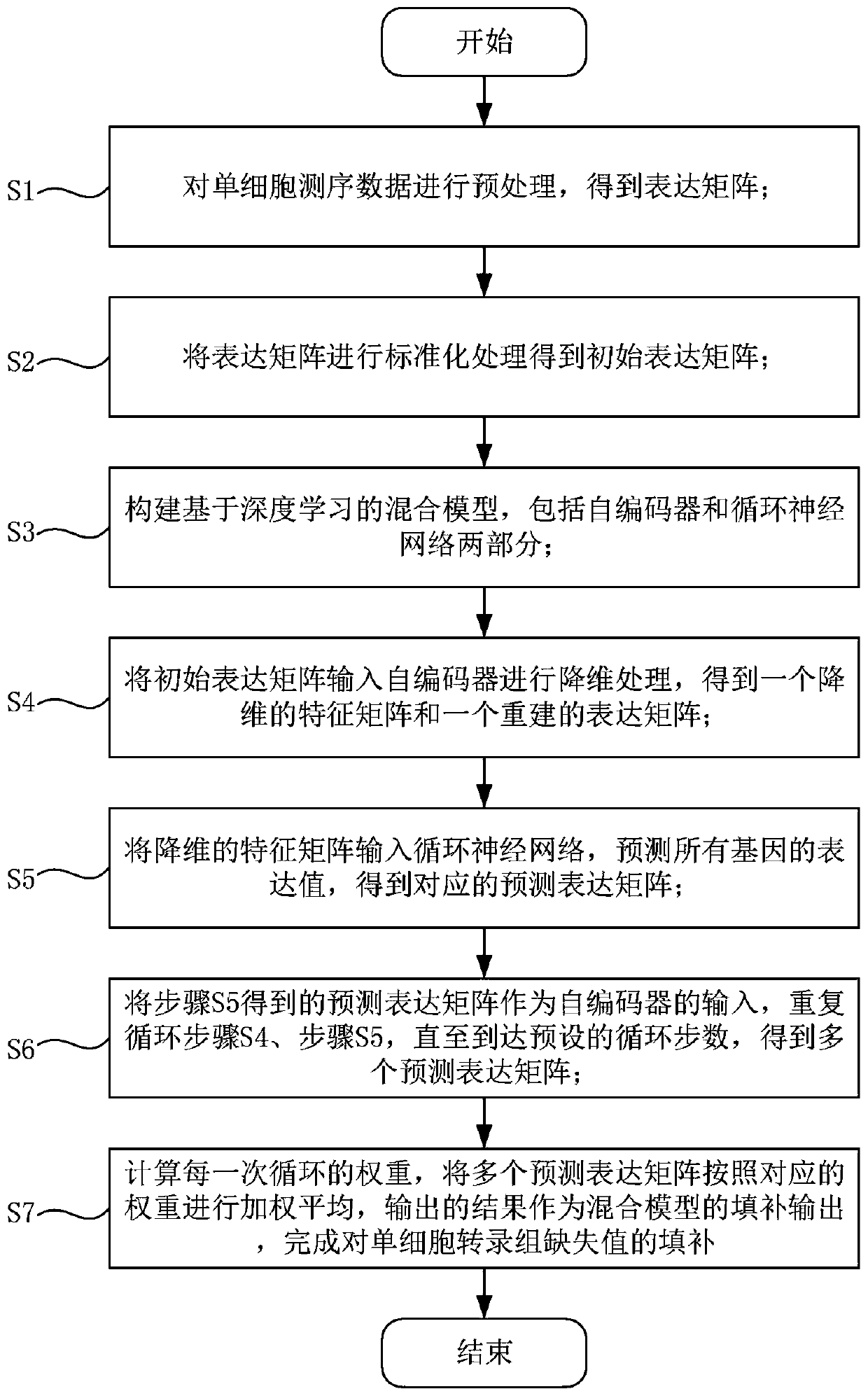
Single cell transcriptome missing value filling method based on deep hybrid network
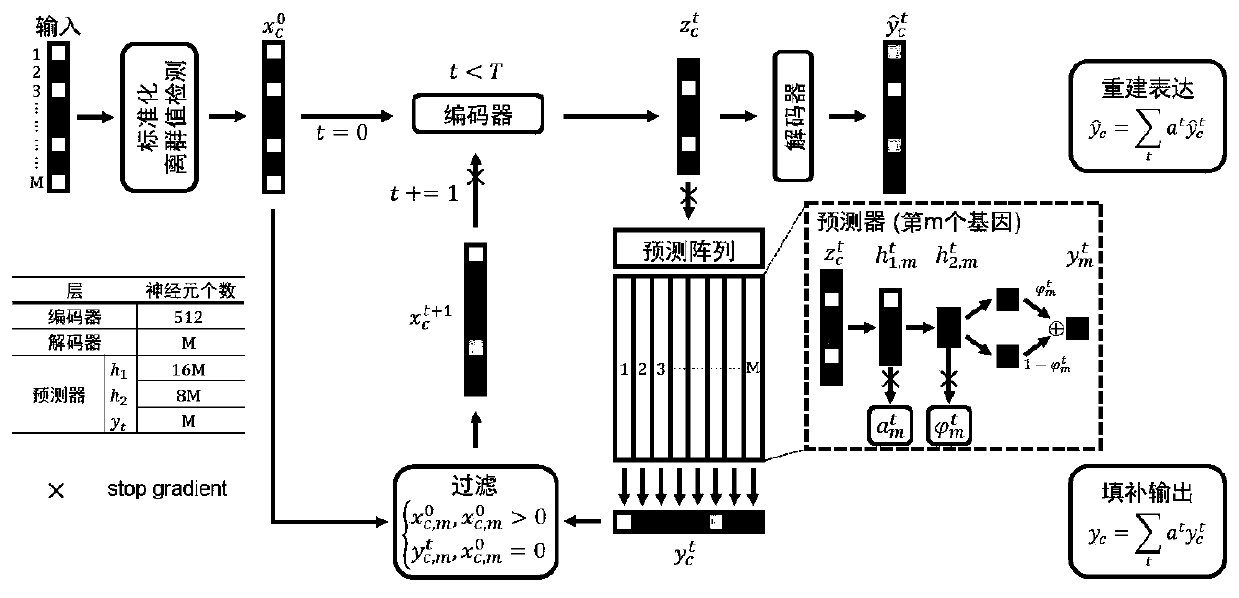
ActiveCN110957009AImprove reliabilityGuaranteed versatilityBiostatisticsInstrumentsData setSingle cell transcriptome
The invention provides a single cell transcriptome missing value filling method based on a deep hybrid network. The method comprises the steps of: carrying out sequencing and preprocessing of a singlecell, obtaining an expression matrix, and carrying out standardization processing; constructing a hybrid model based on deep learning, and inputting the standardized expression matrix into the hybridmodel for cyclic calculation to obtain a plurality of prediction expression matrixes; calculating the weight of each cycle, performing weighted average on the multiple prediction expression matrixesaccording to the corresponding weights, wherein the obtained result is filling output of the hybrid model, and filling of missing values is completed. According to the filling method provided by the invention, the fitting capability of the deep neural network to a complex function is adapted to the expression distribution of the single cells, so that the universality of the filling method to various single cell transcriptome data is ensured; and moreover, the expansibility of deep learning on a data set with an ultra-large cell number is reserved, filling of the single cell transcriptome missing value is completed, and the reliability of single cell data interpretation is remarkably improved.
Owner:ZHONGSHAN OPHTHALMIC CENT SUN YAT SEN UNIV
High-throughput single-cell transcriptome sequencing method and kit
PendingCN110684829AReduce mutual contaminationIncrease the proportionMicrobiological testing/measurementGenomicsSingle cell transcriptome
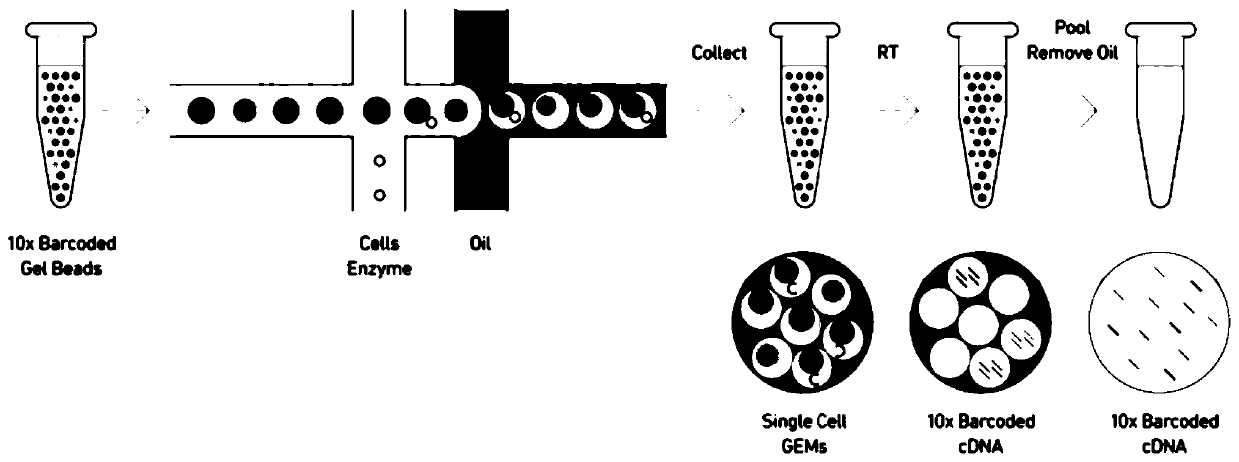
The invention discloses a high-throughput single-cell transcriptome sequencing method and a kit. The high-throughput single-cell transcriptome sequencing method includes the steps that a droplet generation system is adopted to encapsulate a single cell and labeled microbeads in a droplet, and reverse transcription is performed in the droplet. According to the high-throughput single-cell transcriptome sequencing method, a throughput of 9,000 cells which is equivalent to 10 x genomics can be achieved, after the droplet is demulsified, the microbeads are less contaminated with each other, and theproportion of valid data is increased. According to the high-throughput single-cell transcriptome sequencing method, reverse transcription is performed in the droplet, fewer reagents are required, and the cost is low. In the preferred scheme of the high-throughput single-cell transcriptome sequencing method, a SMART template conversion technology is adopted in reverse transcription, and productscan be directly used for subsequent Tn5 library construction and BGISeq-500 platform sequencing without library conversion; and multiple amplification and introduction of deviations are avoided, a droplet-based microfluidics platform is used in conjunction with a BGISeq-500 sequencing platform, and large-scale single-cell sequencing is simplified and facilitated.
Owner:MGI TECH CO LTD
Methods and compositions for cDNA synthesis and single-cell transcriptome profiling using template switching reaction
InactiveCN105579587ASugar derivativesMicrobiological testing/measurementCDNA librarySingle cell transcriptome
This application discloses methods for cDN'A synthesis with improved reverse transcription, template switching and preamplification to increase both yield and average length of cDNA libraries generated from individual cells. The new methods include exchanging a single nucleoside residue for a locked nucleic acid (INA) at the TSO 3' end, using a methyl group donor, and / or a MgCb concentration higher than conventionally used. Single-cell transcriptome analyses incorporating these differences have full-length coverage, improved sensitivity and accuracy, have less bias and are more amendable to cost-effective automation. The invention also provides cDNA molecules comprising a locked nucleic acid at the 3'-end, compositions and cDNA libraries comprising these cDNA molecules, and methods for single-cell transcriptome profiling.
Owner:LUDWIG INST FOR CANCER RES LTD
Single-cell transcriptome library establishment method and application thereof
InactiveCN108193283AIncrease profitIncrease coverageMicrobiological testing/measurementLibrary creationLysisSingle cell transcriptome
The invention provides an optimized single-cell transcriptome library establishment method. The method comprises the following steps: (1) obtaining a sample RNA through cell lysis or extraction; (2) performing reverse transcription operation on the sample to make the RNA form cDNA through reverse transcription; and (3) constructing a transcriptome library by using a separated cDNA, wherein the step (3) of constructing the transcriptome library by using the separated cDNA comprises the following steps: (3-1) performing a tail adding reaction or adopting a single-chain connection manner to add an adaptor; (3-2) performing two-chain synthesis on the cDNA and performing an amplification reaction; (3-3) performing exponential amplification; and (3-4) after transcriptome library construction isperformed on the cDNA amplification product subjected to step (3-3) treatment, removing ''useless DNA''; or after the ''useless DNA'' in the cDNA amplification product subjected to step (3-3) treatment, constructing a transcriptome library. Compared with the prior art, the method provided by the invention is more high-efficiency, simpler, more practical and more accurate, has less pollution, lessloss, low costs and good method repeatability, expands the application range of the sample and improves the precision degree of detection.
Owner:SHANGHAI MAJORBIO BIO PHARM TECH
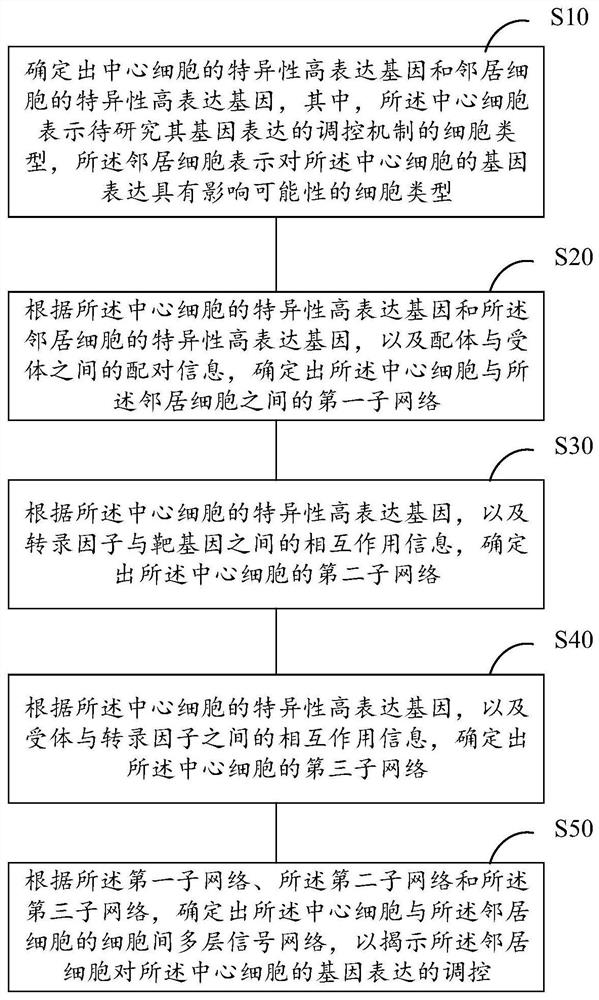
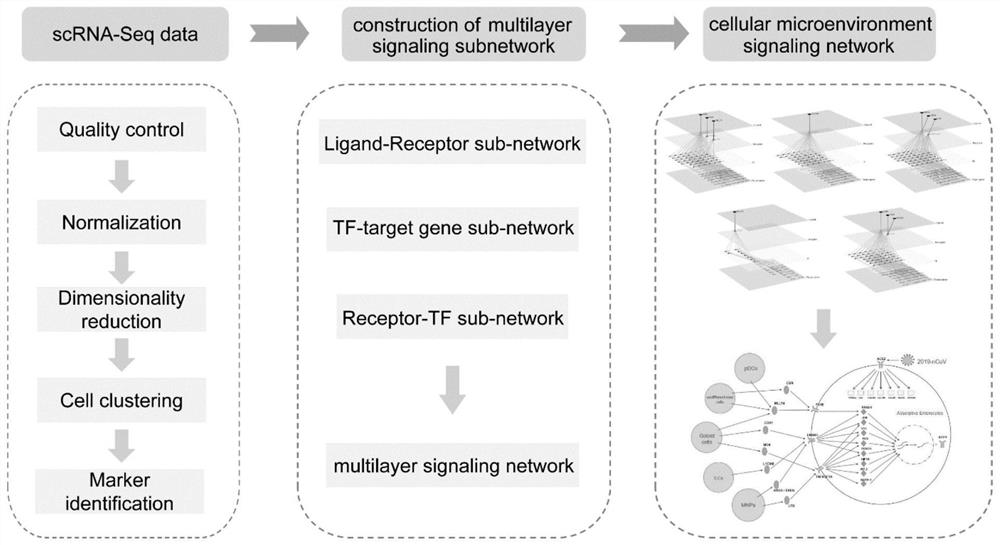
Method for determining gene expression regulation mechanism based on unicellular transcriptome data
ActiveCN111613268AComprehensive system displayAccurately reflect the impactBiostatisticsProteomicsSingle cell transcriptomeReceptor
The invention provides a method for determining a gene expression regulation mechanism based on unicellular transcriptome data. The method comprises the following steps: determining a specific high-expression gene of a central cell and a specific high-expression gene of a neighbor cell according to single cell transcriptome data; determining a first sub-network between the central cell and the neighbor cell according to the specific high-expression genes of the central cell and the neighbor cell and the pairing information between the ligand and the receptor; determining a second sub-network of the central cell according to the specific high-expression gene of the central cell and the interaction information between the transcription factor and the target gene; determining a third sub-network of the central cell according to the specific high-expression gene of the central cell and the interaction information between the receptor and the transcription factor; and according to the firstsub-network, the second sub-network and the third sub-network, determining an intercellular multilayer signal network of the central cell and the neighbor cells so as to reveal regulation and controlof gene expression of the central cell by the neighbor cells.
Owner:SUN YAT SEN UNIV
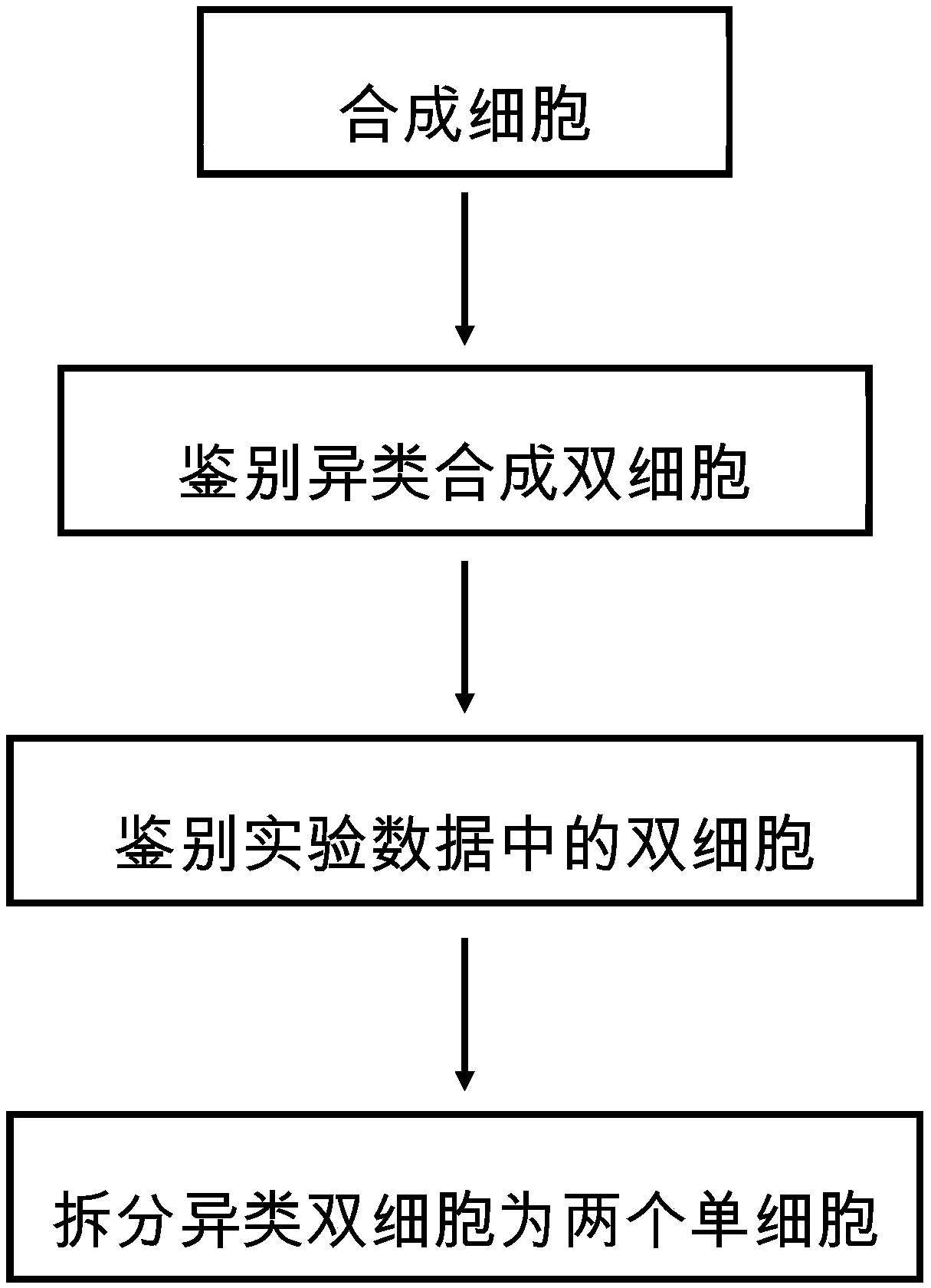
Method for analyzing double cells in single-cell transcriptome data
ActiveCN111292807AImprove the efficiency of detecting the number of cellsBiostatisticsSequence analysisSingle cell transcriptomeCell type
The invention discloses a method for analyzing double cells in single cell transcriptome data, and belongs to the technical field of single cell transcriptome sequencing. Through computer simulation,a cell expression profile is synthesized on the basis of experimental data, and double cells in the experimental data are identified by identifying and synthesizing heterogeneous double cells. The method does not depend on the existing knowledge of cell types and does not depend on the hypothesis of the total expression quantity of the cells, and the full-automatic double-cell identification can be realized on the basis of computer simulation.
Owner:SINGLERON NANJING BIOTECHNOLOGIES LTD
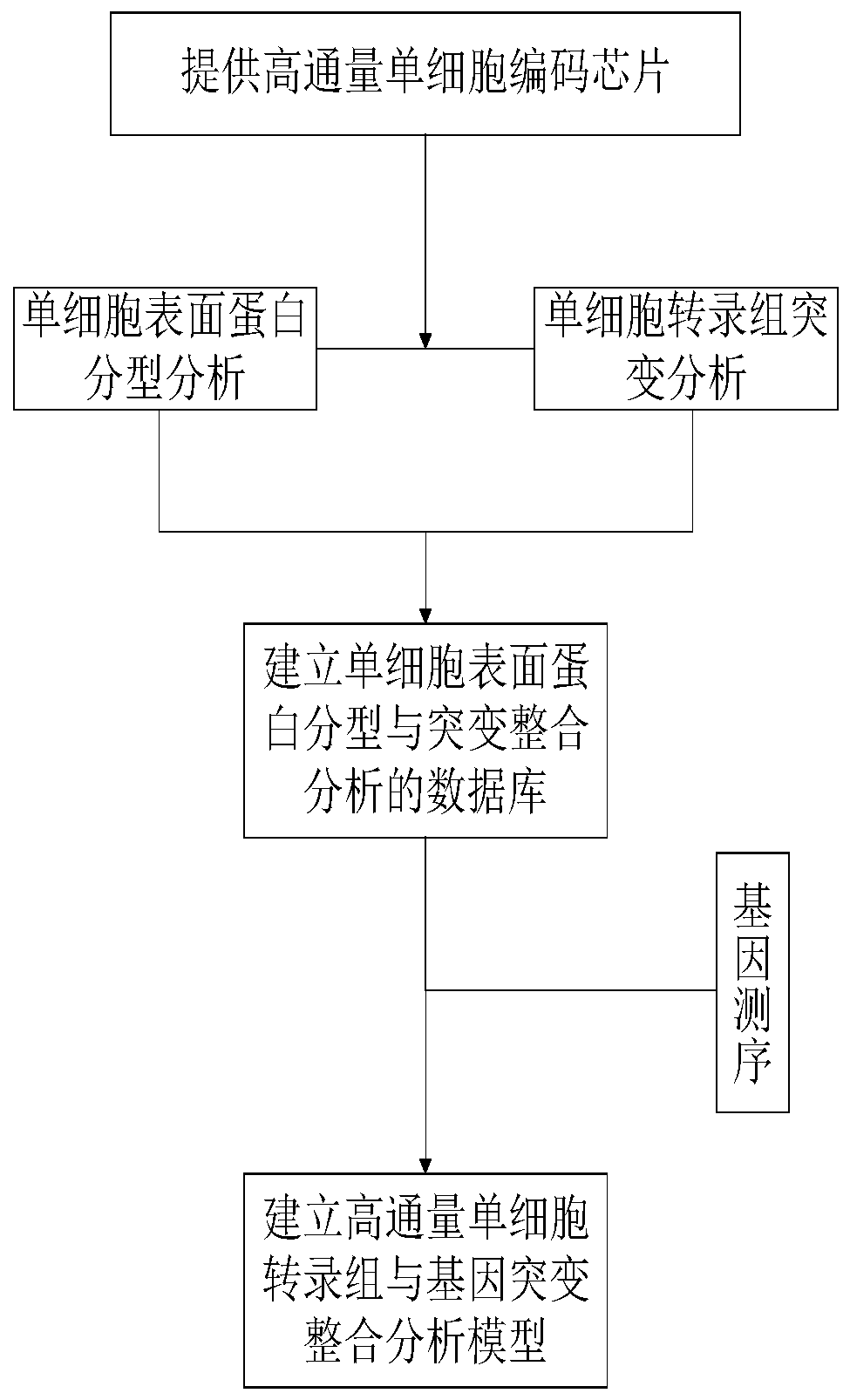
High-throughput single-cell transcriptome and gene mutation integration analysis method
InactiveCN110577983AAchieve a comprehensive understandingAchieve comprehensive characterizationMicrobiological testing/measurementSingle cell transcriptomeCell Surface Proteins
The invention discloses a high-throughput single-cell transcriptome and gene mutation integration analysis method. The high-throughput single-cell transcriptome and gene mutation integration analysismethod comprises the following steps that (1) a high-throughput single-cell encoding chip is provided; (2) single-cell surface protein parting analysis is conducted; (3) single-cell transcriptome mutation analysis is conducted; (4) a database for single-cell surface protein parting and mutation integration analysis is established; and (5) a high-throughput single-cell transcriptome and gene mutation integration analysis model is established. According to the high-throughput single-cell transcriptome and gene mutation integration analysis method, by designing the single-cell encoding chip withtriple encoding technologies of microporous spatial coordinates, cell nucleic acid labels and molecular nucleic acid labels and by combining the modes of single-cell surface protein parting, single-cell transcriptome mutation analysis and gene sequencing, gene mutation information, transcriptome information and protein expression information of single cells can correspond one by one, the completedatabase for high-throughput single-cell transcriptome and gene mutation integration analysis is formed, and the multi-omics integration analysis model is obtained.
Owner:SUZHOU INST OF BIOMEDICAL ENG & TECH CHINESE ACADEMY OF SCI
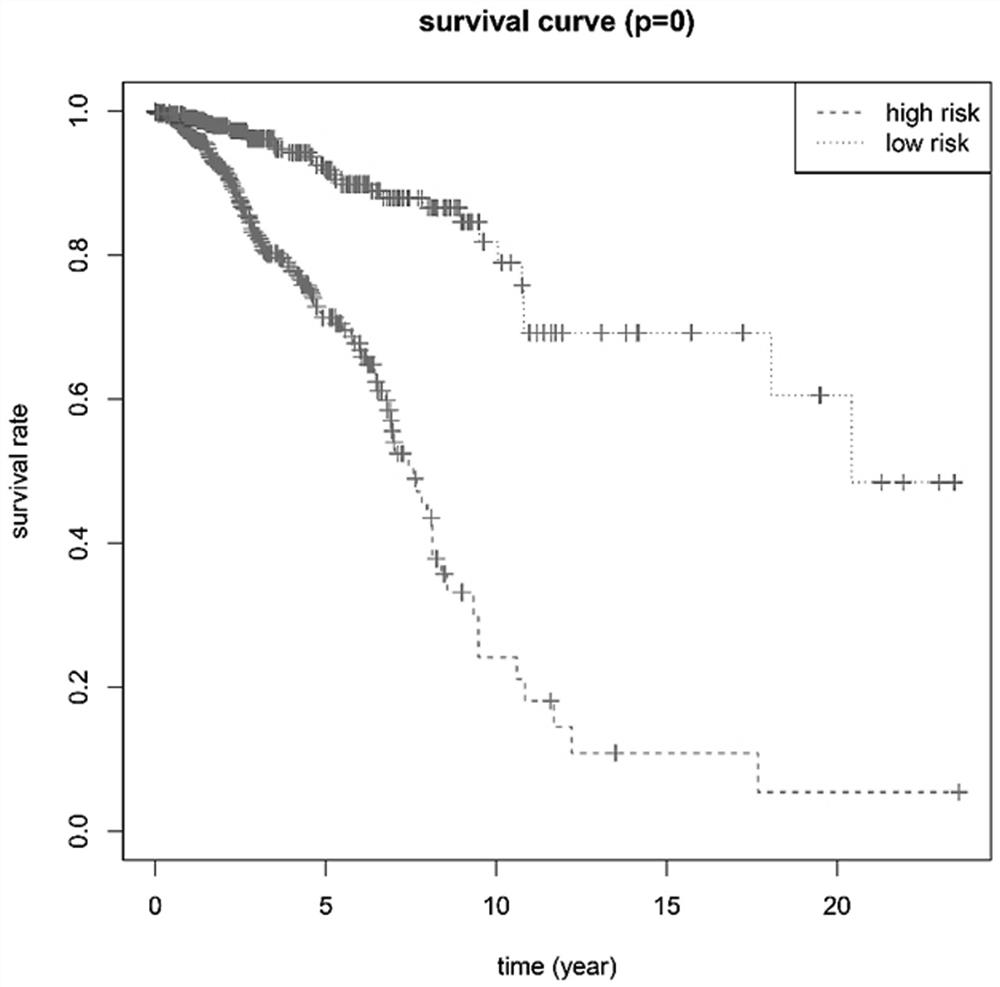
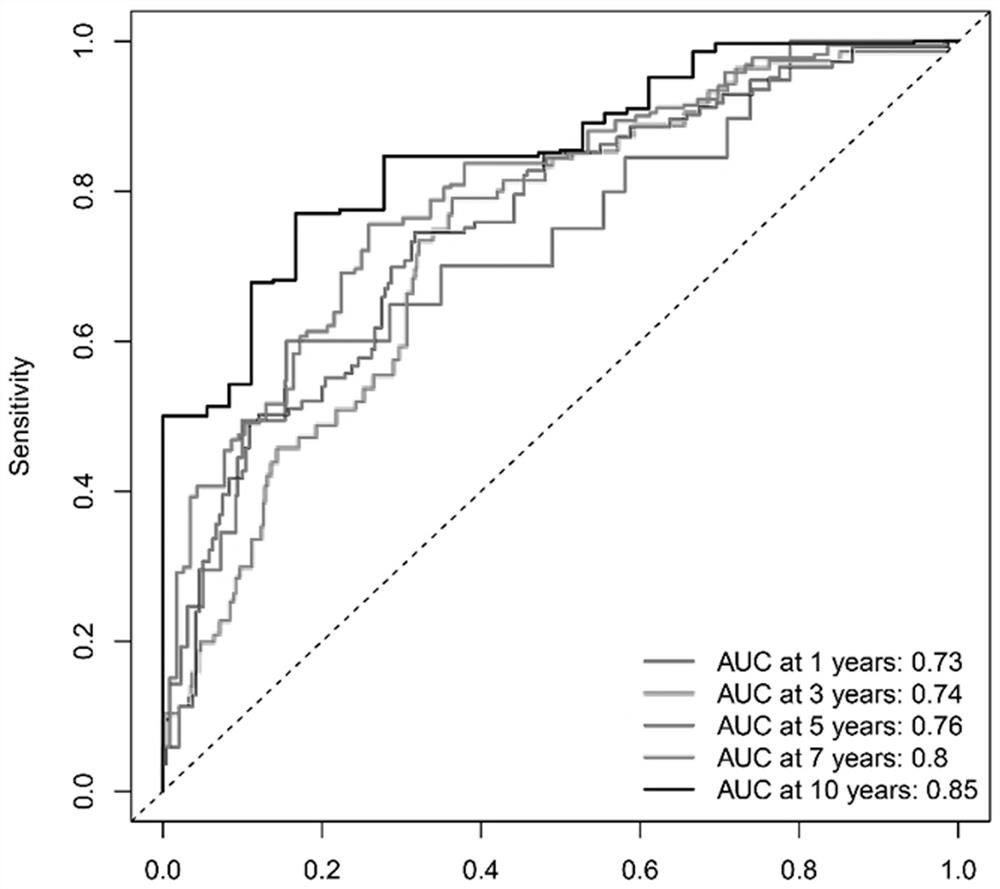
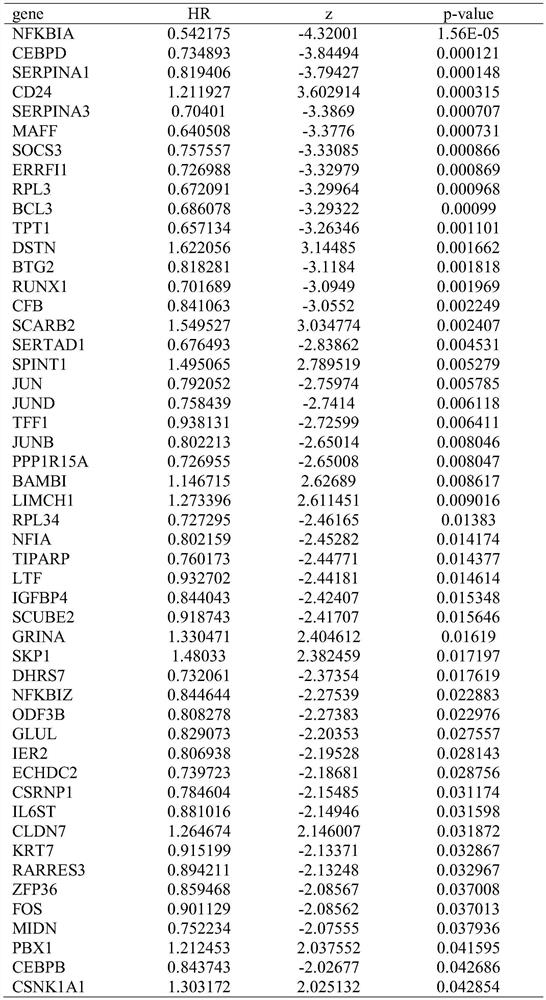
Breast cancer patient recurrence risk 20 gene prediction model based on breast cancer single cell transcriptome sequencing analysis
PendingCN112481378AAccurate judgmentPrecision therapyMicrobiological testing/measurementSequence analysisSingle cell transcriptomeAdjuvant therapy
The invention relates to the field of gene detection technologies and biomedicine, in particular to a breast cancer patient recurrence risk 20 gene prediction model based on breast cancer single-celltranscriptome sequencing analysis and an establishment method and application thereof. The model is composed of 20 genes of CEBPD, SERPINA1, CD24, ERRFI1, BCL3, DSTN, BTG2, SERTAD1, SPINT1, BAMBI, LIMCH1, NFIA, SKP1, DHRS7, ODF3B, KRT7, ZFP36, CEBPB, BHLHE40 and UGDH. The breast cancer patient recurrence risk 20 gene prediction model provided by the invention provides more accurate judgment for long-term prognosis of a breast cancer patient, and provides a basis for selection of a postoperative adjuvant therapy scheme of the patient, so that individualized accurate treatment is realized, and meanwhile, a new research perspective is provided for research of breast cancer tumor stem cells.
Owner:SHENGJING HOSPITAL OF CHINA MEDICAL UNIV
T-cell subset for cancers and characteristic genes
ActiveCN109081866AMicrobiological testing/measurementBlood/immune system cellsRegulatory T cellSingle cell transcriptome
The invention isolates and characterizes a T-cell subset capable of reflecting body tumor immunity state, namely exhaustible CD8+T-cells to express genes WARS and ACP5 as well as regulatory T-cells toexpress genes STAM and BATF, by analyzing single cell gene expression profiles of T-cells infiltrated in cancer tissues through single cell transcriptome analysis. Further determined by researches are new characteristic genes WARS and ACP5 expressed by the cell subset, and a relationship between the STAM and BATF and prognosis of tumor, as well as application of the genes in tumor prognosis diagnosis and monitoring, and application as new tumor immunotherapy targets.
Owner:PEKING UNIV
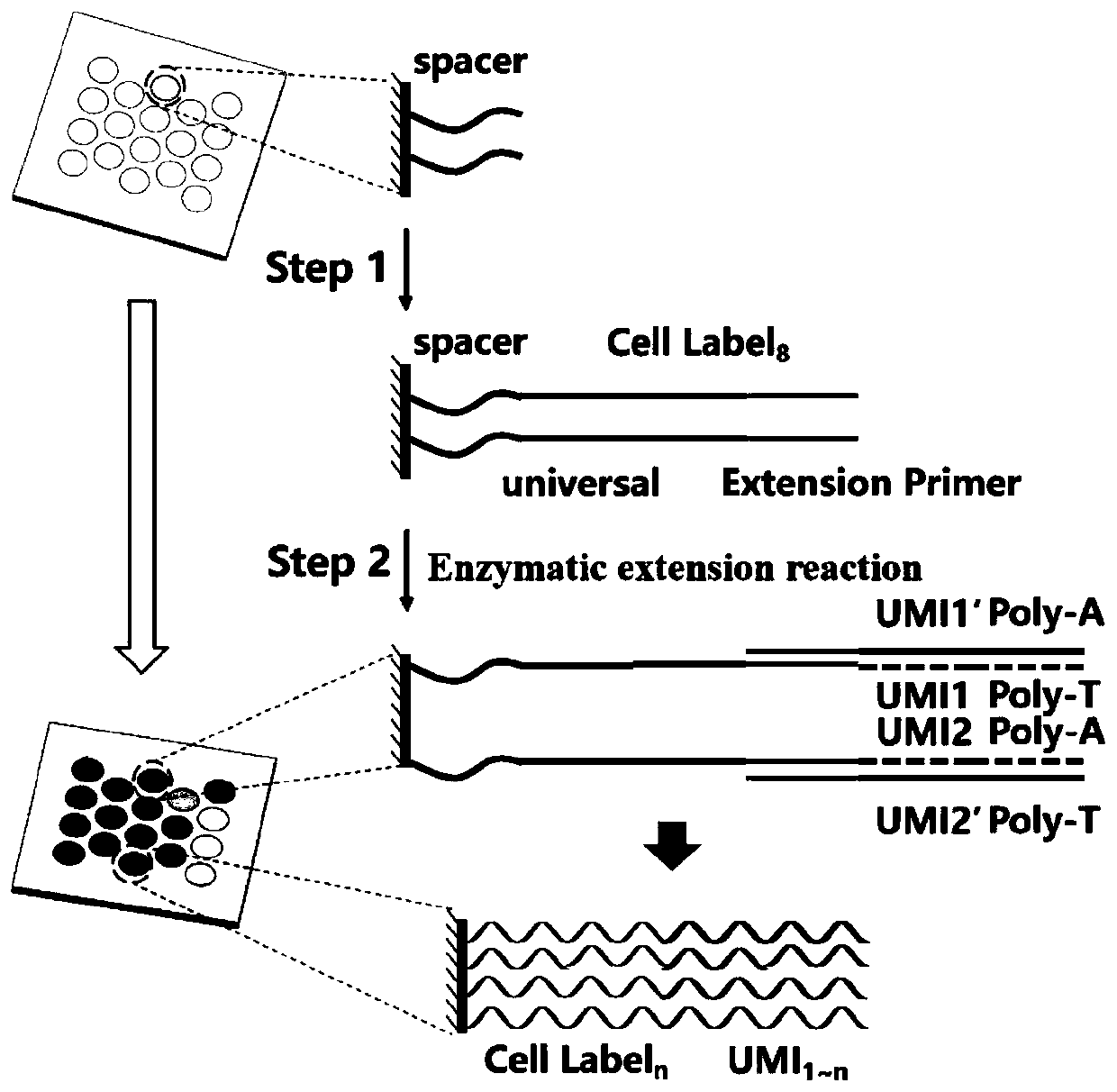
High-throughput single-cell transcriptome and gene mutation integration analysis coding chip
InactiveCN110577982AMicrobiological testing/measurementSingle cell transcriptomeNucleic acid sequencing
The invention discloses a high-throughput single-cell transcriptome and gene mutation integration analysis coding chip. According to the chip, a substrate is provided with a plurality of microholes, the micropores have a size and a shape which can only accommodate a single cell in a micropore, each micropore has a unique spatial coordinate encoding, the micropores are internally modified with a plurality of known nucleic acid sequences, the nucleic acid sequences successively include Spacer sequences, universal primer sequences, cell labels, molecular labels, and Ploy T, wherein the universalprimer sequences are used as primer-binding regions when a PCR is augmented, the cell labels are used for labeling cells from which the RNA originated, and the molecular labels are used for labeling combinative RNA. The invention provides the chip capable of being used for high-throughput single-cell transcriptomes and gene mutation integration analysis. By adopting a triple coding technique of microporous space coordinates, cell nucleic acid labels and molecular nucleic acid labels, gene mutation, the transcriptomes and protein expression information of a single cell can be matched one by one.
Owner:SUZHOU INST OF BIOMEDICAL ENG & TECH CHINESE ACADEMY OF SCI
Application of senescence markers and caloric restrictions of multiple tissue organs and cell types in delaying senescence of organism
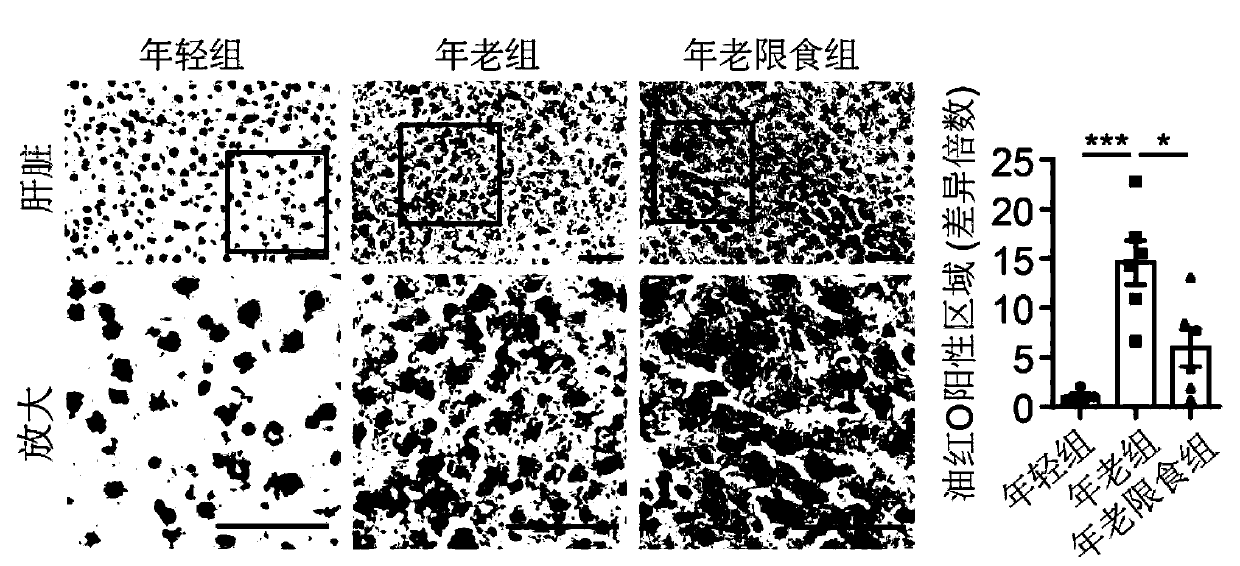
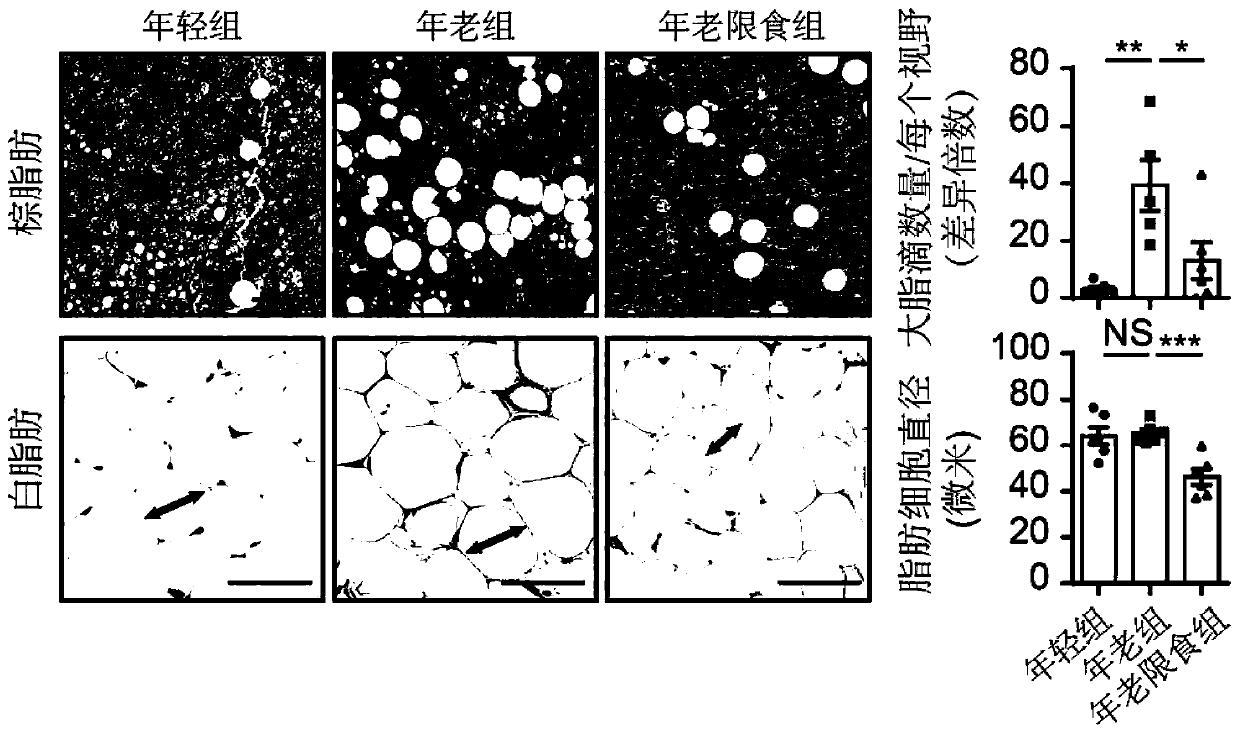
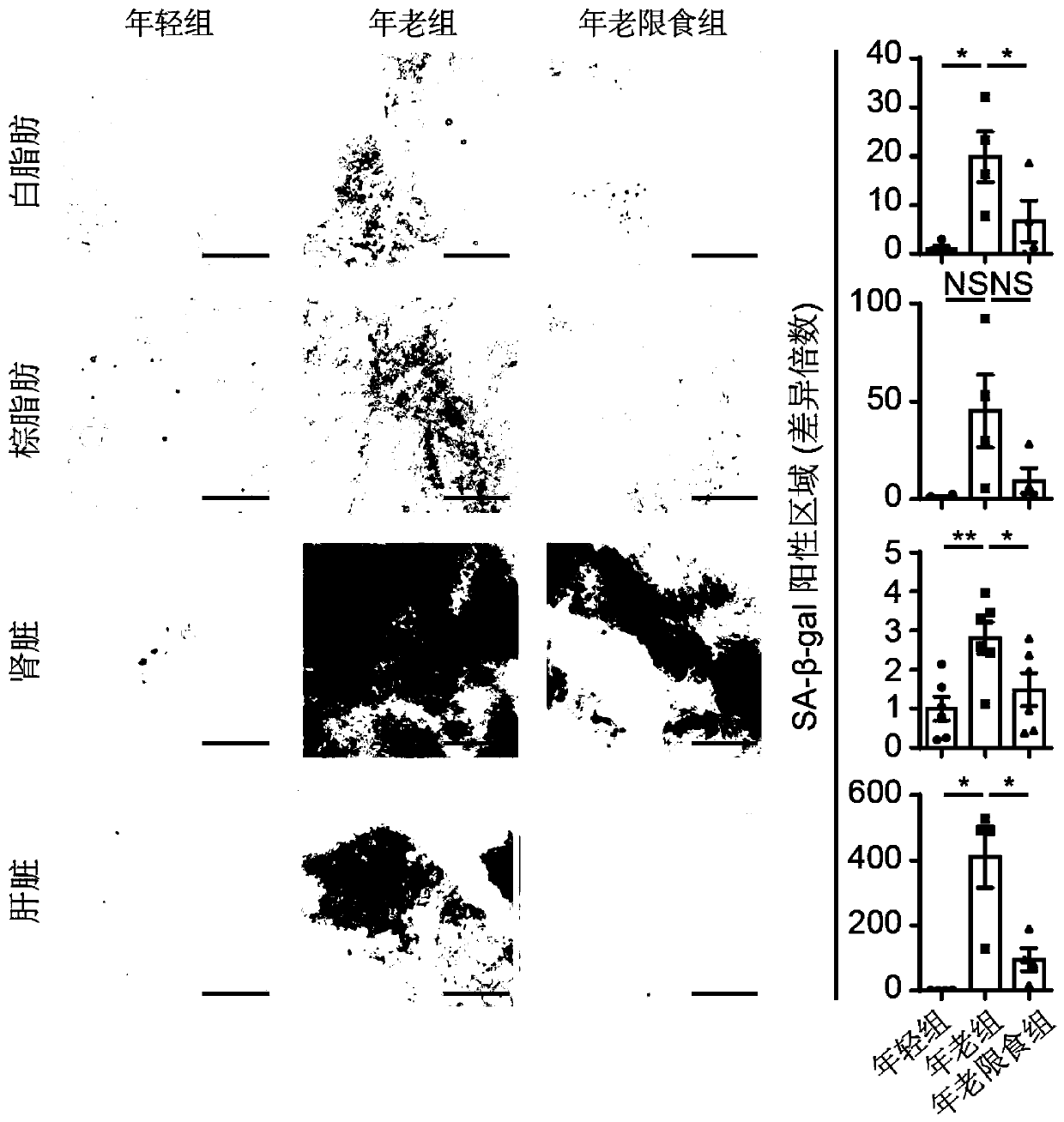
ActiveCN111218515AMicrobiological testing/measurementBiological material analysisBiotechnologySingle cell transcriptome
The invention discloses an application of senescence markers and caloric restrictions of multiple tissue organs and cell types in delaying senescence of organism. According to the invention, integration analysis of a multi-tissue single-cell transcriptome is carried out by taking a rat as a model so as to research the regulation and control effects of senescence and calorie limitation on a mammalsingle-cell level, and 160000 single cells in seven tissues of a young diet group, an old group and an old diet limitation group are systematically analyzed; based on a high-throughput single-cell transcriptome sequencing technology, a first mammal aging and CR multi-tissue organ high-throughput single-cell transcriptome map is established, the effects of senescence and CR on different tissues andcell types are comprehensively and systematically evaluated in the aspects of cell type composition, cell and tissue specificity differential expression gene, core regulation and control transcription factors, cell-cell communication networks and the like, the complex processes of senescence and CR are revealed, and a basis is provided for systematically researching the molecular regulation and control mechanisms of senescence and CR in the future.
Owner:INST OF ZOOLOGY CHINESE ACAD OF SCI
Enhanced MMLV reverse transcriptase mutant and application thereof
ActiveCN113174381AStrong reverse transcription abilityReverse transcription is efficientMicrobiological testing/measurementTransferasesReverse transcriptaseSingle cell transcriptome
The invention provides an enhanced MMLV reverse transcriptase mutant, which is characterized in that 16 amino acid sites are mutated on the amino acid sequence (as shown in SEQ ID No.1) of MMLV reverse transcriptase in a wild type, and the amino acid mutation sites are as follows: A70K, S142P, E201N, D206R, G248D, L300K, A307K, G331P, I434R, S453K, Y522G, E531G, E574G, E596G, E610G, and E645G. The MMLV enhanced MMLV reverse transcriptase mutant disclosed by the invention has stronger reverse transcription capability, can be used for effective reverse transcription of micro RNA, and can be applied to the fields of single cell transcriptase library building, micro RNA virus detection and the like.
Owner:YEASEN BIOTECHNOLOGY (SHANGHAI) CO LTD
Screening method and application of direct mechanical response cell subpopulation
ActiveCN112029710AAchieving the goal of identifying direct mechano-responsive cellsMicrobiological testing/measurementCulture processDiseaseSingle cell transcriptome
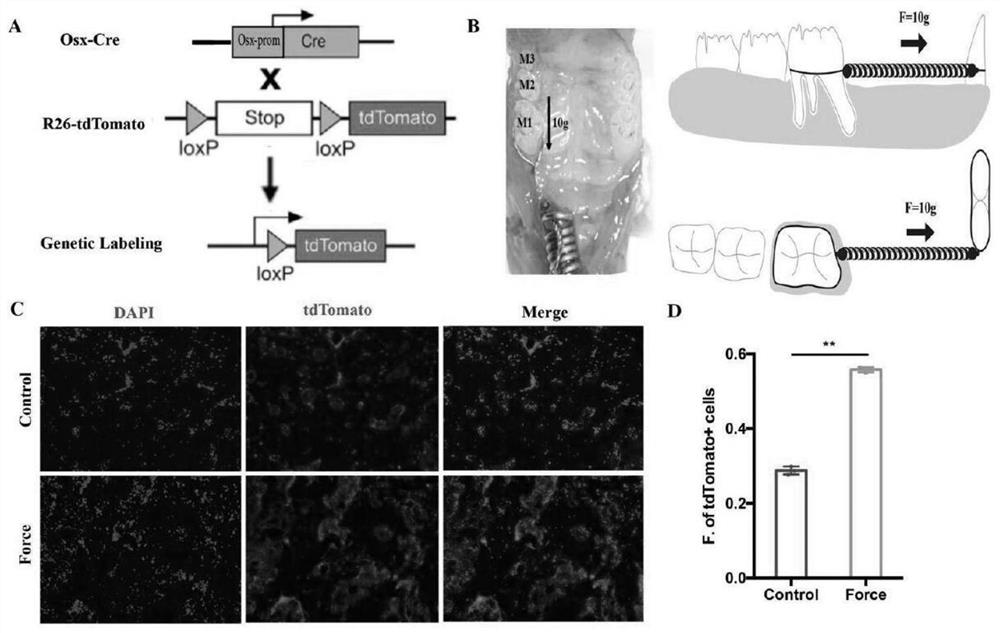
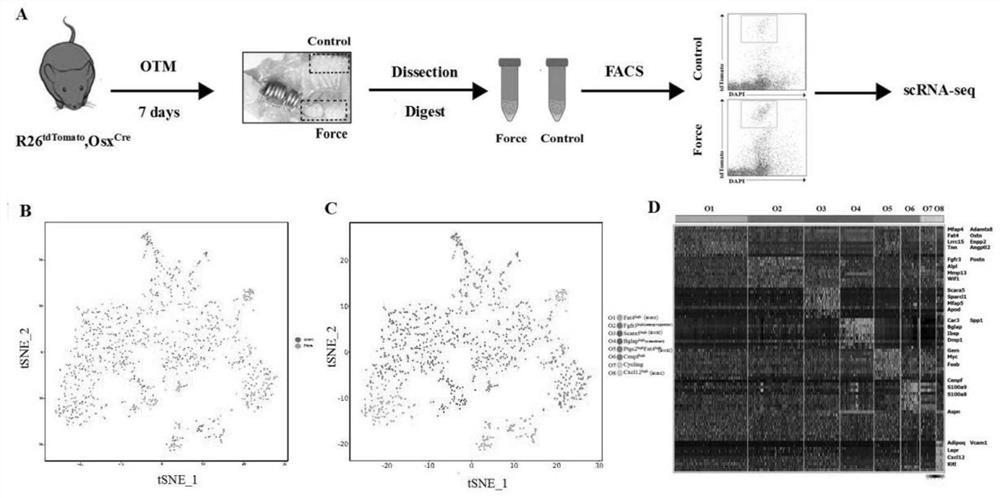
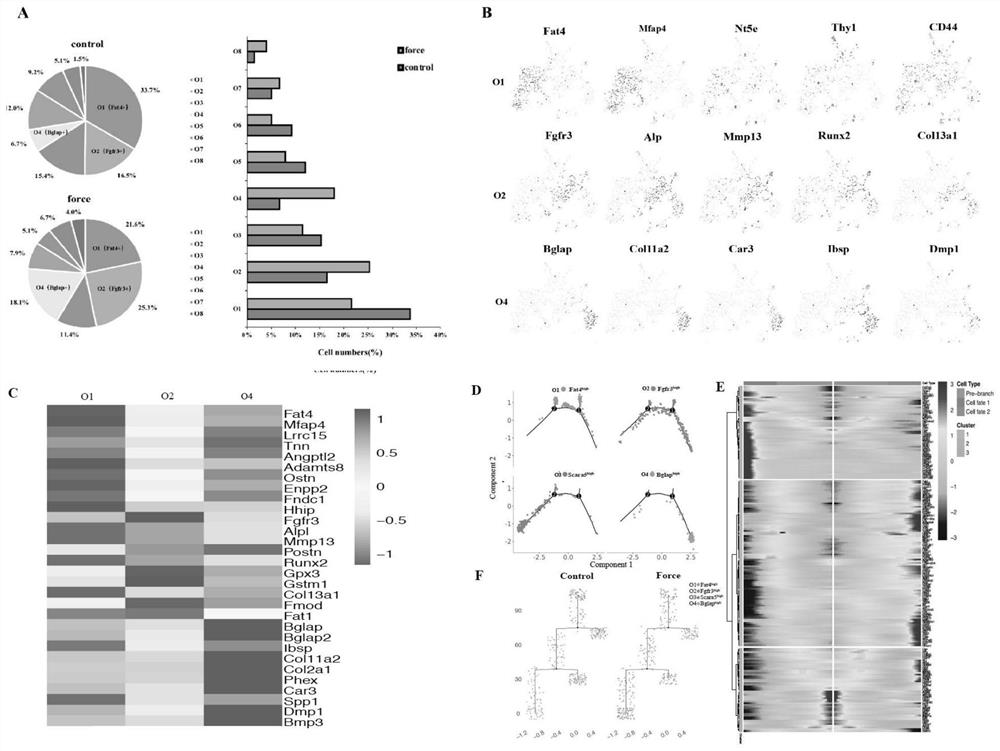
The invention discloses a screening method and application of a direct mechanical response cell subpopulation. The screened cell subpopulation comprises a Fat <4+> subpopulation, a Fgfr <3 > subpopulation and a Scara <5+> subpopulation, and the invention also discloses application of the mechanical response cell subpopulation in preparation of drugs for local bone defect repair and regeneration and stress-related bone metabolic diseases. Direct mechanical response cells are screened through a single-cell sequencing technology, fine analysis of a cell gene expression profile under stress stimulation is carried out through a single-cell transcriptome level, a key cell subpopulation with fate change under stress regulation is searched for,and in-vitro flow type sorting and in-vitro stress loading are carried out through marker genes. In combination with construction of a CreER-loxP inducible reporter gene system, in-vivo lineage tracing is performed to verify the mechanical responsivenessof the cells, so that the purpose of identifying the direct mechanical response cells is achieved.
Owner:SHANGHAI NINTH PEOPLES HOSPITAL SHANGHAI JIAO TONG UNIV SCHOOL OF MEDICINE
Differential gene analysis method based on BD single cell transcriptome and proteome sequencing data
PendingCN113470743ACharacter and pattern recognitionProteomicsProtein regulationSingle cell transcriptome
The invention discloses a differential gene analysis method based on BD single cell transcriptome and proteome sequencing data, and belongs to the technical field of transcriptome data analysis. In order to solve the problem that no complete data analysis method can combine single cell transcriptome and proteome sequencing data for analysis in the prior art, differential gene analysis of cell subset classification in transcription and protein levels is carried out through a single cell transcriptome and proteome sequencing technology, cell characteristics are represented, and a biological mechanism transcribed to a protein regulation expression process is obtained, and a more accurate and richer BD single cell transcriptome and proteome sequencing data combined analysis method is provided. According to the analysis method, deep understanding of differential expression can be obtained, key mRNA or protein regulated after transcription is excavated, some important regulatory pathways are found and verified, and the analysis method can be applied to the field of tumors.
Owner:哈尔滨星云医学检验所有限公司
Single cell transcriptome sequencing method based on third-generation sequencing
ActiveCN111549099ASolve the technical problem of high demand for original samplesAchieving Precise SequencingMicrobiological testing/measurementSingle cell transcriptomeComplementary deoxyribonucleic acid
The invention provides a single-cell transcriptome sequencing method based on third-generation sequencing, which comprises the following steps: (1) carrying out reverse transcription on single-cell RNA by using a reverse transcription primer to obtain cDNA with a bar code; (2) carrying out PCR (Polymerase Chain Reaction) amplification on the cDNA with the bar code, and mixing obtained PCR amplification products with different single cell sources; or mixing cDNA (complementary deoxyribonucleic acid) with barcodes from different single cell sources and then carrying out PCR (polymerase chain reaction) amplification; the reverse transcription primer sequentially comprises an anchoring sequence, a bar code sequence and poly dT from the 5'end to the 3 'end. According to the invention, a bar code sequence is added into a reverse transcription primer to carry out reverse transcription on single-cell full-length RNA; sequences of different single cell sources are marked, DNA amplification products of different sources are mixed to meet the requirement of a third-generation sequencing platform for the template amount, and accurate sequencing of a full-length transcript is achieved through the third-generation sequencing platform.
Owner:BIOISLAND LAB
Integrated device for integrated analysis of high-throughput single-cell transcriptome and gene mutation
ActiveCN110951580AIntegrated Analysis RealizationAmplifyBioreactor/fermenter combinationsBiological substance pretreatmentsGenes mutationSingle cell transcriptome
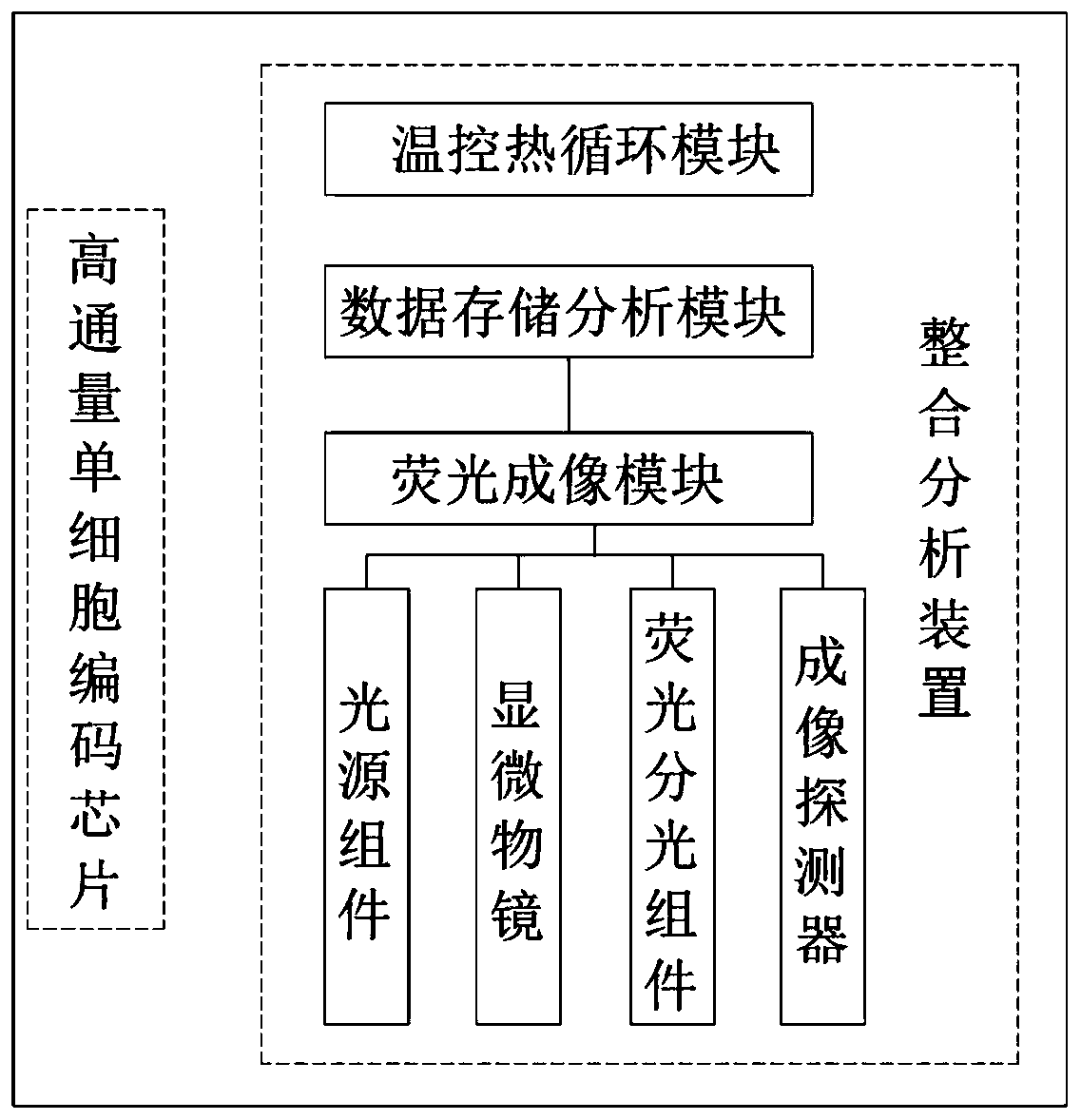
The invention discloses an integrated device for integrated analysis of high-throughput single-cell transcriptome and gene mutation. The integrated device includes a high-throughput single-cell codingchip and an integrated analysis device body, wherein the integrated analysis device body includes a housing and a temperature-control thermal cycle module, a fluorescence imaging module and a data storage analysis module which are disposed in the housing, and the fluorescence imaging module includes a light source component, a microscope objective, a fluorescence splitting component and an imaging detector. One-to-one correspondence of gene mutation, transcriptome and protein expression information of single cells can be achieved through the designed high-throughput single-cell coding chip which is provided with triple-encoding functions of micropore spatial coordinates, cellular nucleic acid tags and molecular nucleic acid tags, and PCR amplification can be realized through the temperature-control thermal cycle module; and the fluorescence imaging module is used for collecting fluorescence images of a sample, and the fluorescence images are stored and analyzed through the data storage analysis module, so that integrated analysis of single-cell transcriptome and gene mutation is achieved.
Owner:SUZHOU INST OF BIOMEDICAL ENG & TECH CHINESE ACADEMY OF SCI
Multi-omics method for single cell transcriptome and translation group combined sequencing
ActiveCN112831552AQuality improvementMicrobiological testing/measurementCDNA librarySingle cell transcriptome
The invention provides a method for simultaneously carrying out transcriptome sequencing and transscriptome sequencing, which is characterized by comprising the following steps: carrying out first-step lysis treatment on cells and a lysis buffer solution to obtain a first lysis solution; after uniformly mixing and centrifuging, taking a part of the first lysis solution in the supernate as a transcriptome template, and taking the rest of the supernate and the lysis buffer solution to carry out second lysis treatment so as to obtain a second lysis solution; centrifuging the second lysate, and collecting supernate as a translation group template; obtaining a transcriptome cDNA library by using the transcriptome template, and sequencing the transcriptome cDNA library; and extracting a ribosome protected RNA fragment from the translation group template, and carrying out library establishment and sequencing on the ribosome protected RNA fragment. According to the method, high-quality transcriptome and translation group data in a whole genome range can be obtained at the same time, and the method is particularly suitable for single cells, so that possibility is provided for knowing collaborative regulation of single cell transcription and translation levels.
Owner:TSINGHUA UNIV
Sequencing method for single cell transcriptome
ActiveCN109971843AGet rid of application limitationsImprove stabilityMicrobiological testing/measurementEucaryotic cellMagnetic bead
The present invention belongs to the field of biotechnology and discloses a sequencing method for a single cell transcriptome. PAP / PGP is used to add poly A / G to a tail part of a RNA strand to increase RNA stability and at the same time produces an extended strand used for reverse transcription and cDNA synthesis. When effective RNA enrichment is carried out downstream, a small-fragment PCR non-specific product is purified by magnetic beads labeled with ribosomal DNA probes, a half-annealed splint hybridization is used to remove rRNA in a cDNA form having a size of 5s, 5.8s, 18s and 28s, and besides, a large amount of invalid Reads during sequencing are reduced. The sequencing method can conduct one-stop non-coding RNA, microRNA and mRNA total transcriptome sequencing of a single eukaryocyte, produces an expression matrix of the transcriptome and frees an application limitation that one cell can only perform one sequencing.
Owner:复旦大学泰州健康科学研究院
Bladder cancer exhausted T cell subset as well as characteristic genes and application thereof
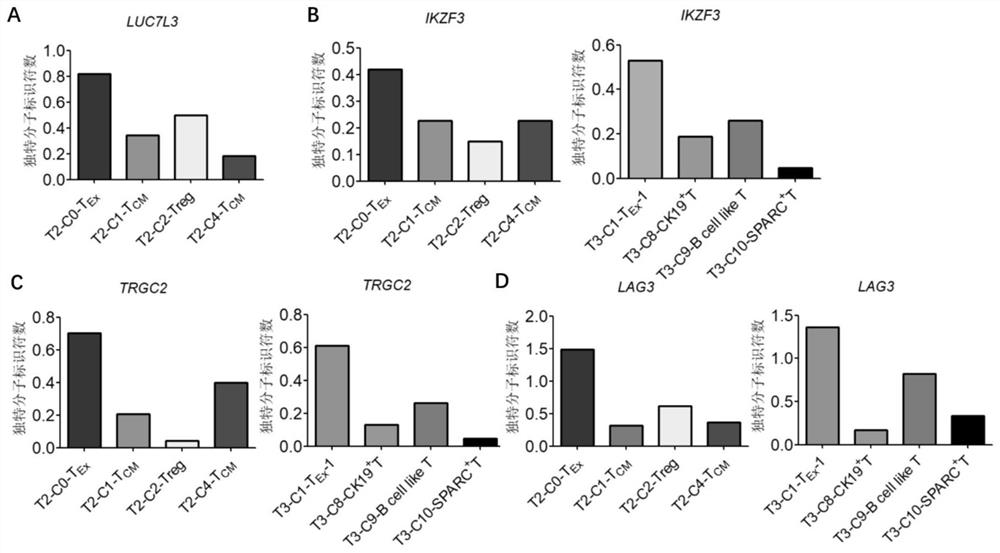
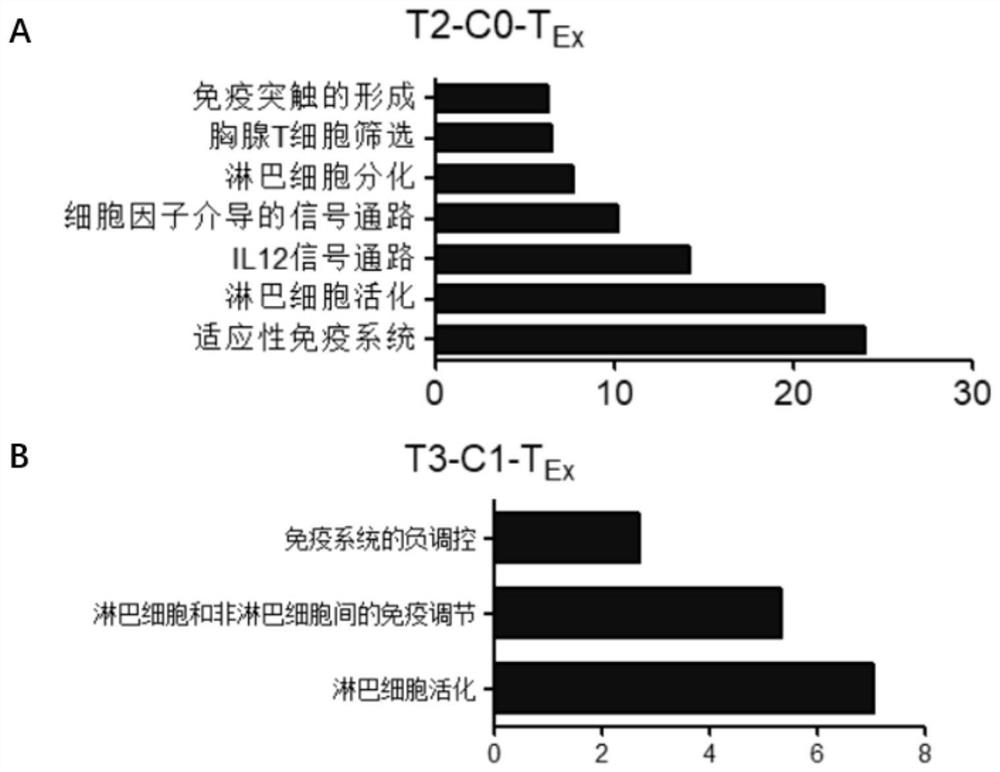
InactiveCN111748627AMicrobiological testing/measurementBlood/immune system cellsSingle cell transcriptomeCD8
According to the invention, single-cell transcriptome sequencing is carried out on peripheral blood, normal bladder tissues and bladder cancer tissue-derived T cells of an infiltrated bladder cancer patient by utilizing a single-cell transcriptome sequencing analysis technology, a bladder cancer specific T cell subset, namely CD8<+> exhausted T cells, is identified, and characteristic genes, namely genes LUC7L3, IKZF3 and TRGC2, expressed by the CD8<+> exhausted T cells are determined. The characteristic genes can be used for identifying or authenticating CD8<+> exhausted T cells from bladdercancer, can be further used for auxiliary diagnosis, prognosis and immunotherapy of bladder cancer, and provide related detection reagents or kits.
Owner:BEIJING UNIV OF CHEM TECH
Popular searches
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com