Tumor neoantigen screening method fused with single cell TCR sequencing data
A technology of sequencing data and screening methods, applied in biochemical equipment and methods, biological systems, sequence analysis, etc., can solve the problems of low interaction affinity, inability to cause immune response, time-consuming, etc., and achieve rapid and accurate identification and narrow down of candidates range effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
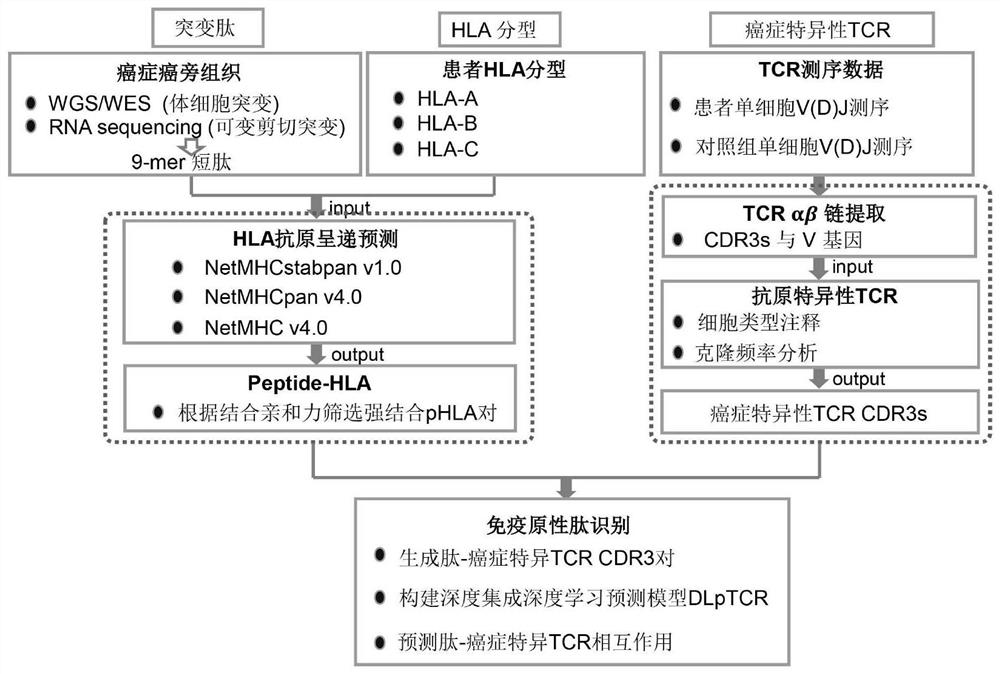
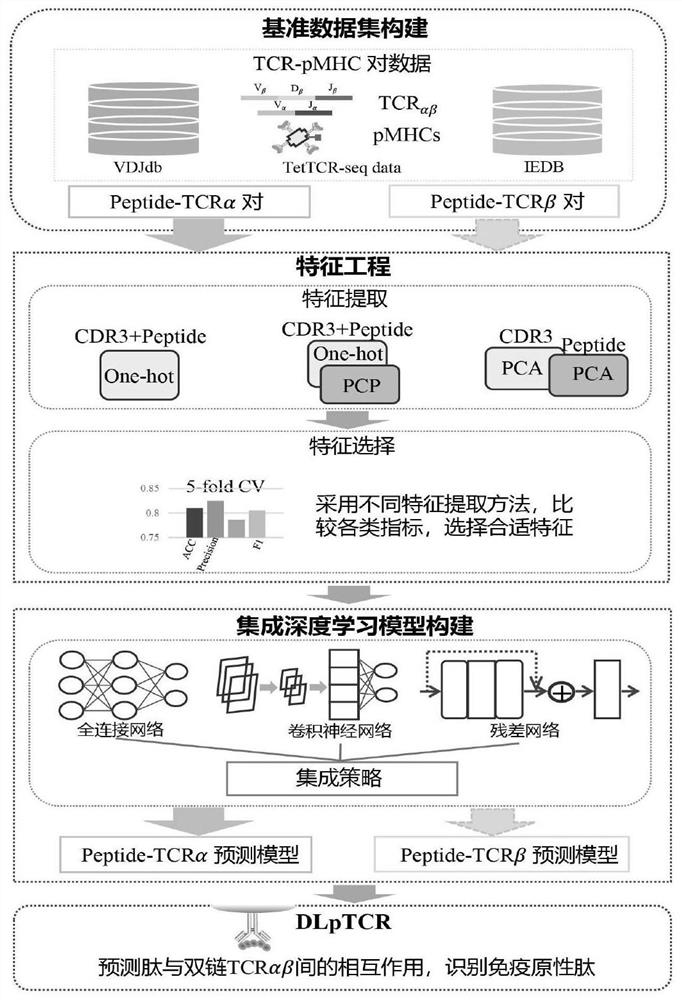
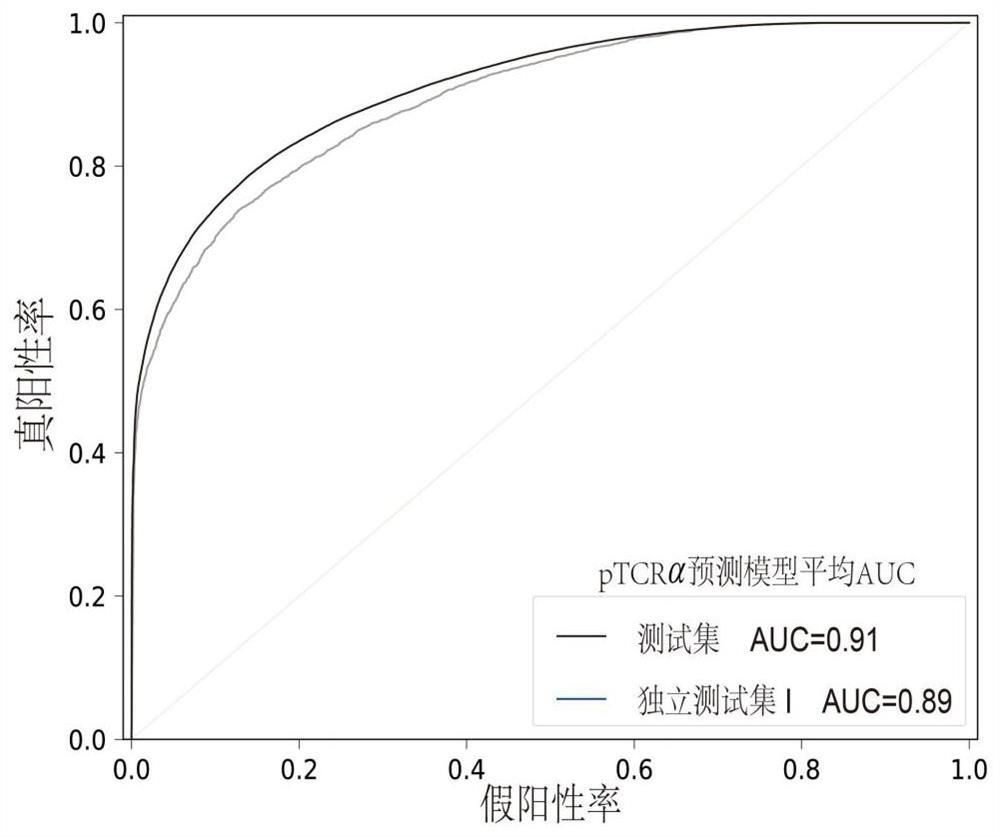
[0044] Such as figure 1 As shown, a tumor neoantigen screening method that combines single-cell TCR sequencing data includes the following steps:
[0045] S1: Construction of personalized nascent mutant peptide library for tumor patients. Obtain whole exome sequencing (WES) data and transcriptome sequencing (RNA-seq) data of paired tumor tissues and paracancerous tissues, perform quality control analysis, single nucleotide variation analysis, and construct individualized nascent mutant peptide libraries, using In downstream antigen screening;
[0046] S1.1. Obtain whole exome sequencing (WES) data and transcriptome sequencing (RNA-seq) data of paired tumor tissues and paracancerous tissues;
[0047] S1.2. Use Trimmomatic-0.36 software to perform quality control analysis on tumor tissue and patient-matched paracancerous tissue WES data, remove reads with an average Phred score lower than 20, and cut off standard adapters;
[0048]S1.3. Use the bwa 0.5.9 software to compare t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com