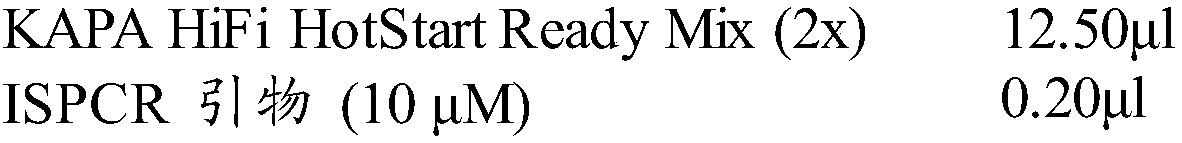Sequencing method for single cell transcriptome
A single-cell, transcriptome technology, applied in the biological field, can solve problems such as undetectable, no total RNA or long non-coding RNA single-cell RNA sequencing methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] For the single-cell total RNA sequencing of five common cell lines, the specific implementation steps are as follows.
[0037] 1. Use 1% Trypsin to digest LNCaP, T24, PC3, HEK293t, and C2C12 cells for 10-15 minutes to a single cell state, resuspend in PBS, and absorb 6 cells of each type with a capillary tube, and then lyse them in CLB.
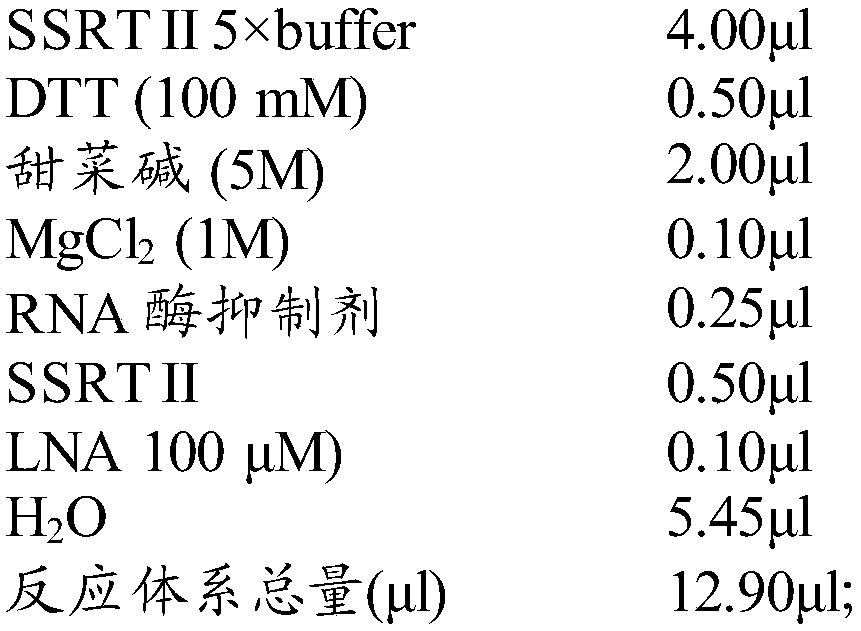
[0038] 2. PolyA polymerase reaction: configure the cell lysis system according to Table 1, first CLB and H 2 O, DTT, MnCl 2 , ATP mixed and reacted at 37°C for 3min, then added Poly(A) Polymerase and 0.5% BSA and reacted at 37°C for 5min or 15min. Prepare the PolyA Polymerase reaction solution according to Table 2, and place it on ice after reacting at 72° C. for 3 minutes.
[0039] Table 1 Cell Lysis System
[0040]
[0041] Table 2 PolyA Polymerase reaction liquid formula
[0042] SINGLE-CELL CUSTOM SMARTER 1T PAP reaction buffer 5μl dNTPs mix (10mM) 1.00μl oligo dT (10μM) 1.00μl ERCC spike-i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com