Integrated device for integrated analysis of high-throughput single-cell transcriptome and gene mutation
An integrated analysis and single-cell technology, applied in biochemical cleaning devices, enzymology/microbiology devices, bioreactor/fermenter combinations, etc. Troubleshooting problems such as cumbersome operation to achieve the optimization of treatment plan and optimization of new target discovery
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
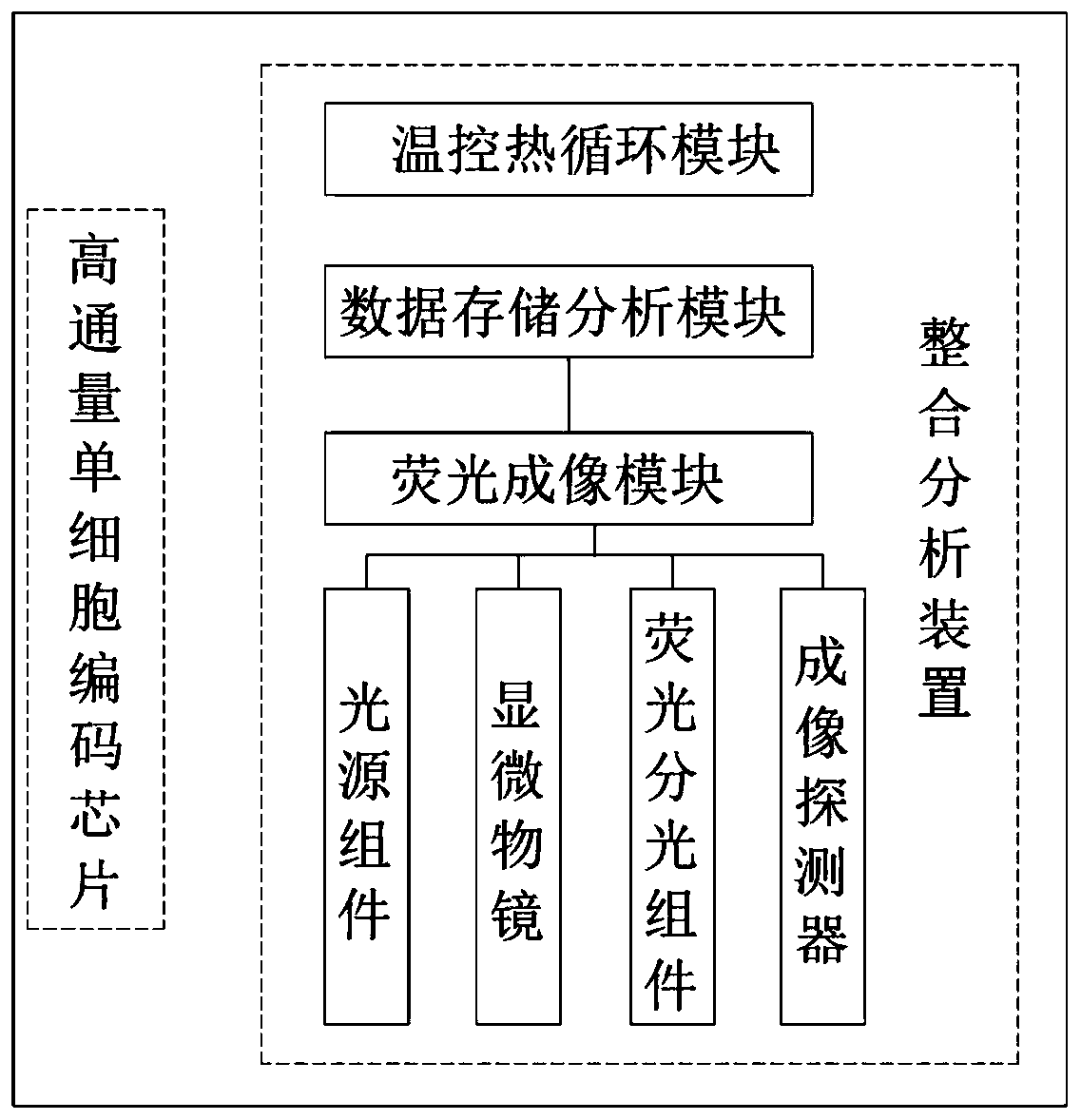
[0055] Based on the above, a specific integrated analysis device 2 is provided in this embodiment.
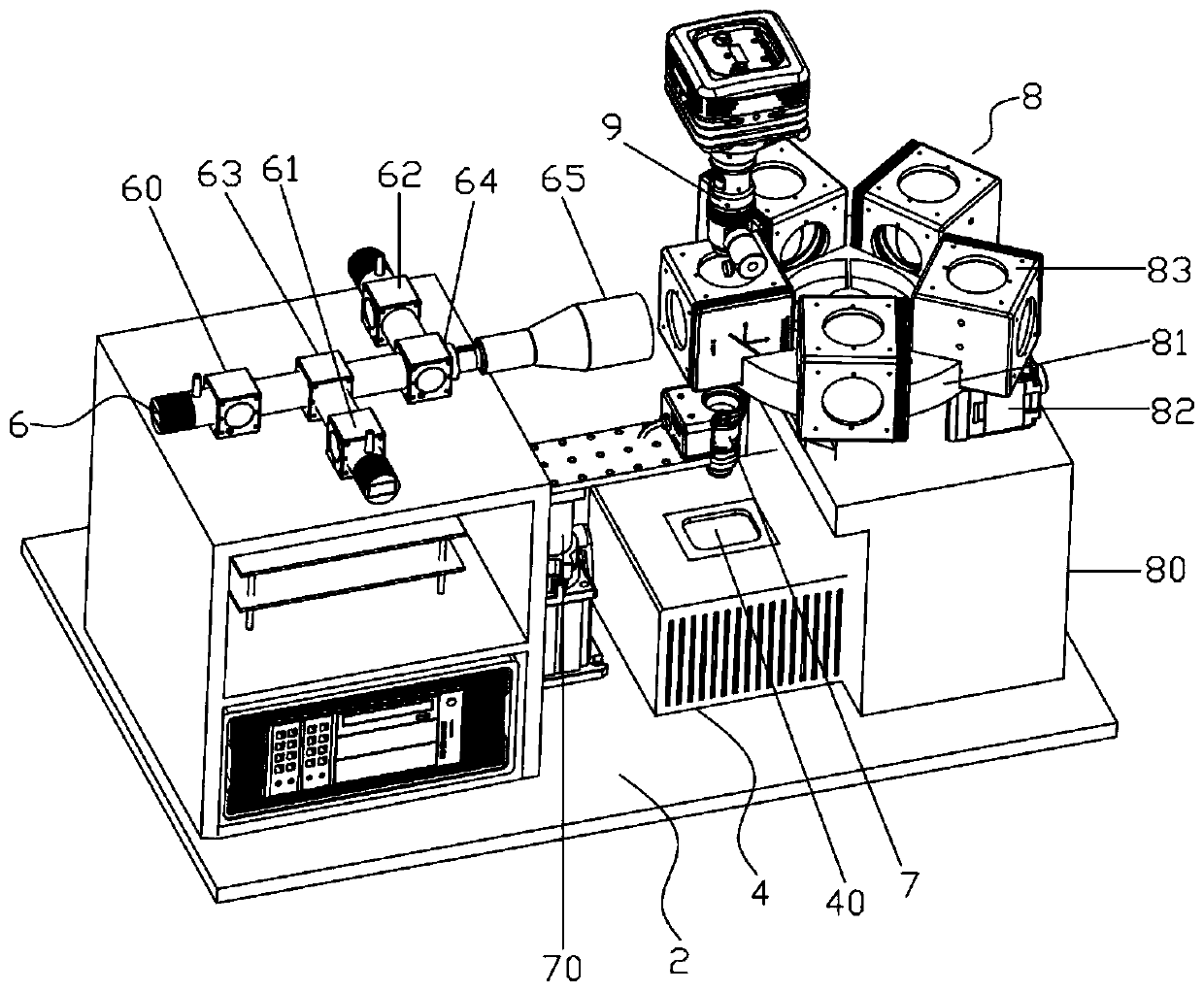
[0056] refer to Figure 2-8 , wherein the light source assembly 6 includes a first LED light source 60, a second LED light source 61, a third LED light source 62, a first dichroic mirror 63, a second dichroic mirror 64 and a beam expander lens group 65, the first LED The light emitted by the light source 60 is transmitted through the first dichroic mirror 63 and the second dichroic mirror 64 in turn and reaches the beam expander lens group 65; the light emitted by the second LED light source 61 is reflected by the first dichroic mirror 63, and the second dichroic mirror The dichroic mirror 64 transmits and reaches the beam expander lens group 65; the light emitted by the third LED light source 62 reaches the beam expander lens group 65 after being reflected by the second dichroic mirror 64; the first LED light source 60, the second LED light source 61 , The third LED light sou...
Embodiment 2
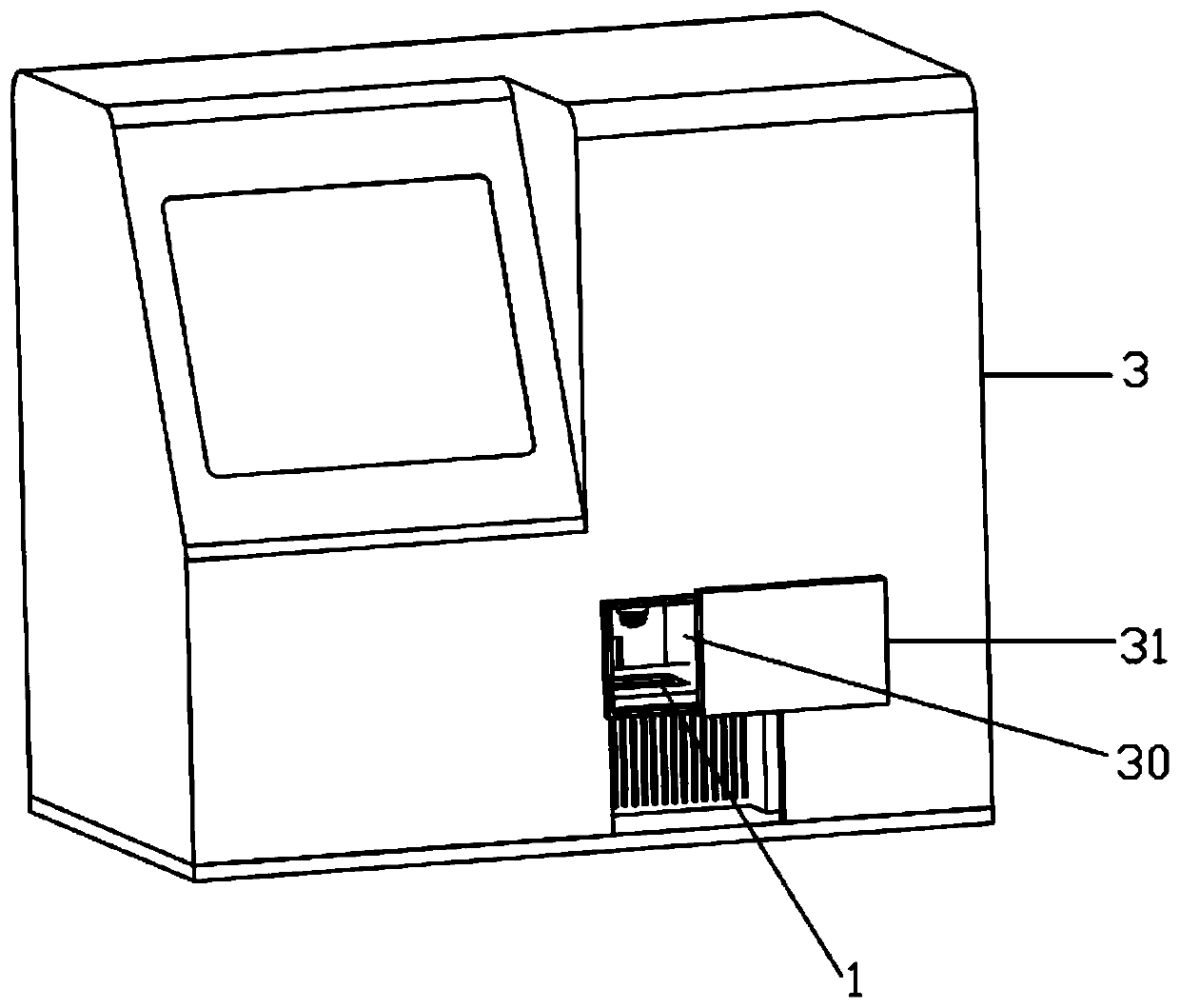
[0063] Based on the above, a high-throughput single-cell encoding chip 1 is provided.
[0064] Among them, the chip is provided with a plurality of microwells on its substrate, each microwell has a unique spatial coordinate code, and several known nucleic acid sequences for capturing target RNA are modified in the microwells, and the nucleic acid sequences include A cell label indicating the cell from which the RNA originated and a molecular label used to identify the bound RNA, the cell label of each microwell corresponds to the spatial coordinate code one-to-one.
[0065] Wherein, the nucleic acid sequence also includes a Spacer sequence, a universal primer sequence as a primer binding region during PCR amplification, and Ploy T. In a further preferred embodiment, the modified nucleic acid sequence in each microwell is not less than 10 6 strip. A molecular tag is a known random nucleic acid sequence.
[0066] Here, the microwells have a size and shape that can accommodate...
Embodiment 3
[0077] An integrated high-throughput single-cell transcriptome and gene mutation integrated analysis device is provided, which is obtained by combining the integrated analysis device 2 of Example 1 and the high-throughput single-cell coding chip 1 of Example 2.
[0078] The analysis steps of the high-throughput single-cell transcriptome and gene mutation integrated analysis integrated device in this embodiment include:
[0079] 1) Fluorescently label the target gene of the cells in advance, then add the sample to the high-throughput single-cell coding chip 1, capture single cells through the micropores on it, and place the high-throughput single-cell coding chip 1 on the loading heat On stage 40, light source assembly 6, microscope objective lens 7, fluorescence spectroscopic assembly 8, and imaging detector 9 are activated, and fluorescence imaging is performed on high-throughput single-cell encoding chip 1 through fluorescence imaging module 5, and then the data storage and a...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com