Primer and kit for using RPA for detecting procambarus clarki picornaviridae and detection method
A technology for RNA virus and Crayfish is applied in the field of special primers for detection, which can solve the problems of low timeliness, long time, complicated and tedious operations, etc., and achieve the effects of high sensitivity and strong specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
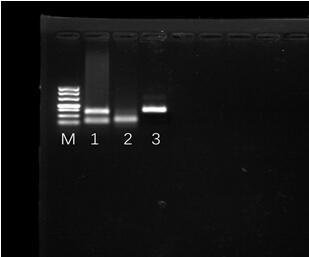
[0037] According to the sequence of Procambarus clarkii picornavirus, an RPA primer pair consisting of upstream and downstream primers with the nucleotide sequences shown in SEQ ID NO.1 and SEQ ID NO.2 was designed.
[0038] Upstream primer RPA PcPV-1F: 5'-CTGATGAGGAATTAGACGATCTTATCGTTG-3' (SEQ ID NO.1);
[0039] Downstream primer RPA PcPV-1R: 5'-CAGAATTAAAGACCGACGTTAGTAGATGAC-3' (SEQ ID NO.2).
Embodiment 2
[0041] Using the TRIzol method, viral RNA was extracted from Procambarus clarkii known to be infected with picornaviruses and reverse transcribed into cDNA. The specific operation process is as follows.
[0042]Take an appropriate amount (0.02g-0.05g) of the hepatopancreas of Procambarus clarkii into an EP tube, and put small magnetic beads in the tube (2 in each EP tube), add 1ml of RNA Plus, and put it into the homogenate In the machine for 2-3 minutes, after the end, let it stand for 5 minutes.
[0043] Put the sample into a refrigerated centrifuge at 4°C, centrifuge at 12000rpm for 5min, and take 600μL of supernatant.
[0044] Add 200 μL of chloroform, shake vigorously for 15 seconds, and let stand at room temperature for 2-3 minutes.
[0045] Centrifuge at 12000rpm for 15min at 4°C, take 200μL of the supernatant,
[0046] Add 500 μL of 100% isopropanol, invert up and down 5 times to mix well, and place at room temperature for 10 minutes.
[0047] Centrifuge at 12000rp...
Embodiment 3
[0053] Using the cDNA obtained in Example 2 as a template and the RPA primers designed in Example 1, the RPA reaction was carried out in the following reaction system.
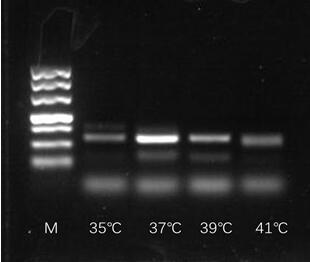
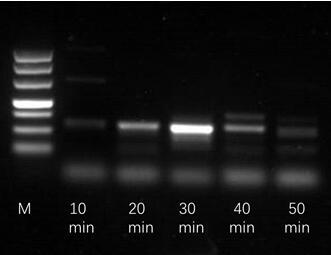
[0054] Sample detection: Prepare 47.5 μl of mixed reaction solution, including 29.5 μl Rehydration Buffer, 2.4 μl of 10 μM upstream primer, 2.4 μl of 10 μM downstream primer, 1 μl of template cDNA and ddH 2 O 12.5 μl. The mixed solution is mixed and transferred to the reaction unit tube containing lyophilized enzyme powder, wherein the lyophilized enzyme powder includes recombinase capable of binding single-stranded nucleic acid (oligonucleotide primer), single-stranded DNA binding protein (SSB) and Strand displacement DNA polymerase three enzymes, mix with a pipette gun to suspend all the particles in the tube, add 2.5 μl of 280 mM magnesium acetate solution, cover the tube cap, mix quickly, and react in a metal bath at 37°C for 4min, 4min After the end, take it out, turn it vigorously 8-10 times, mix well, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com