Methods and compositions for cDNA synthesis and single-cell transcriptome profiling using template switching reaction
一种模板转换、DNA链的技术,应用在用于使用模板转换反应进行cDNA合成和单细胞转录组概况分析和组合物领域,能够解决尚未报道cDNA文库收率和平均长度的系统性努力等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0077] method
[0078] Experiments using total RNA
[0079] Using DNA extracted from mouse brain and used for Illumina sequencing The control total RNA provided with the UltraLowRNA kit (Clontech Company) was used for RNA experiments. One microliter of 1 ng / μl solution was used in each experiment and combined with 1 μl of anchored oligo-dT primer (10 mM, 5′-AAGCAGTGGTATCAACGCAGAGTACT 30 VN-3', wherein "N" is any base and "V" is "A", "C", or "G") is mixed with 1 μl of dNTP mixture (10 mM, Fermentas), Denature at 72°C for 3 minutes and place on ice immediately thereafter. Add 7 μl of the first-strand reaction mixture to each sample, the first-strand reaction mixture contains 0.50 μl SuperScriptIIRT (200Uml-1, Invitrogen), 0.25 μl RNase inhibitor (20Uml-1, Clontech Company), 2 μl SuperscriptII first-strand buffer solution (5x, Invitrogen), 0.25 μl DTT (100 mM, Invitrogen), 2 μl betaine (5M, Sigma (Sigma)), 0.9 μl MgCl 2 (100 mM, Sigma), 1 μl TSO (10 μM, a full list of oligo...
example 2
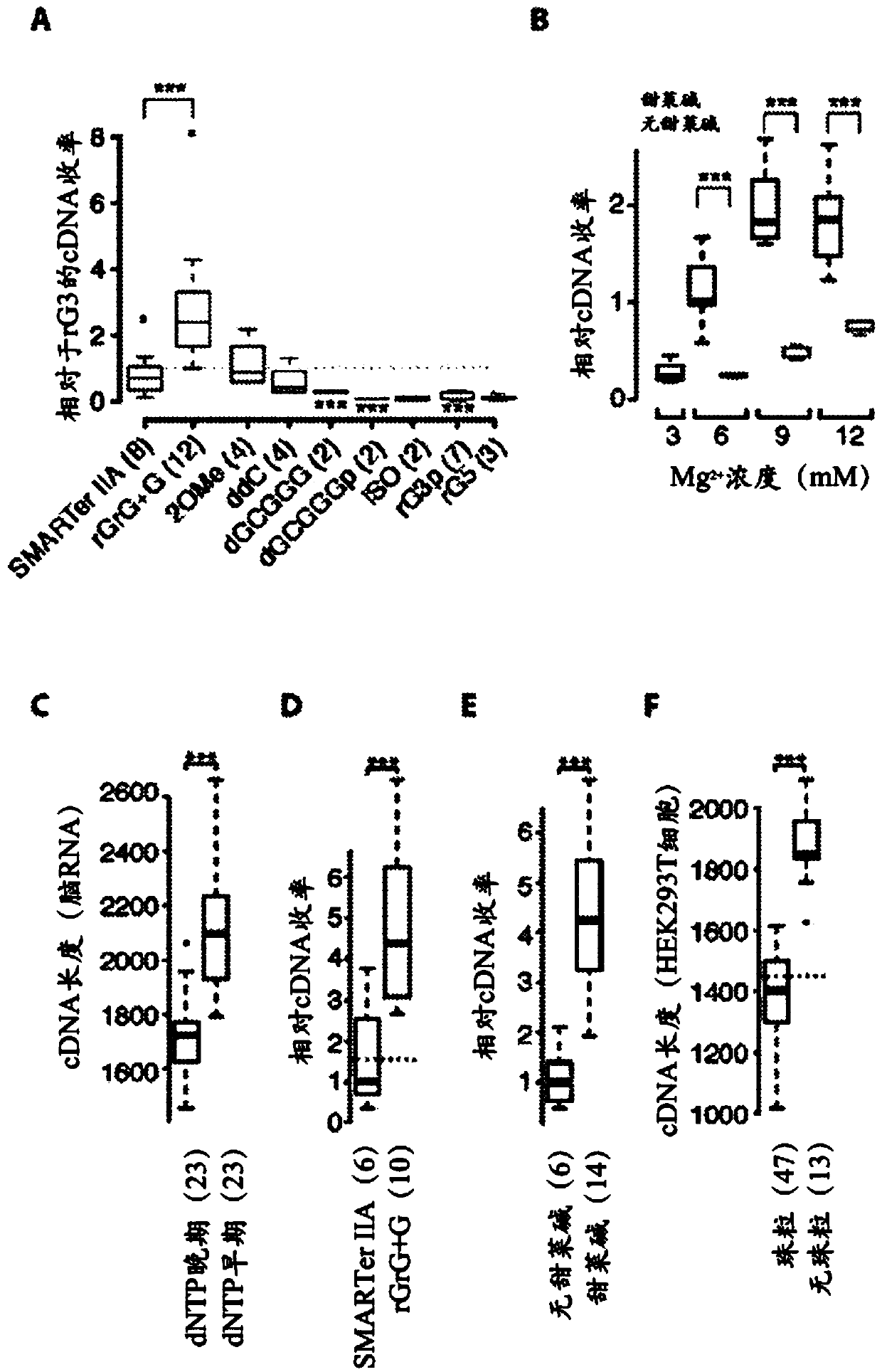
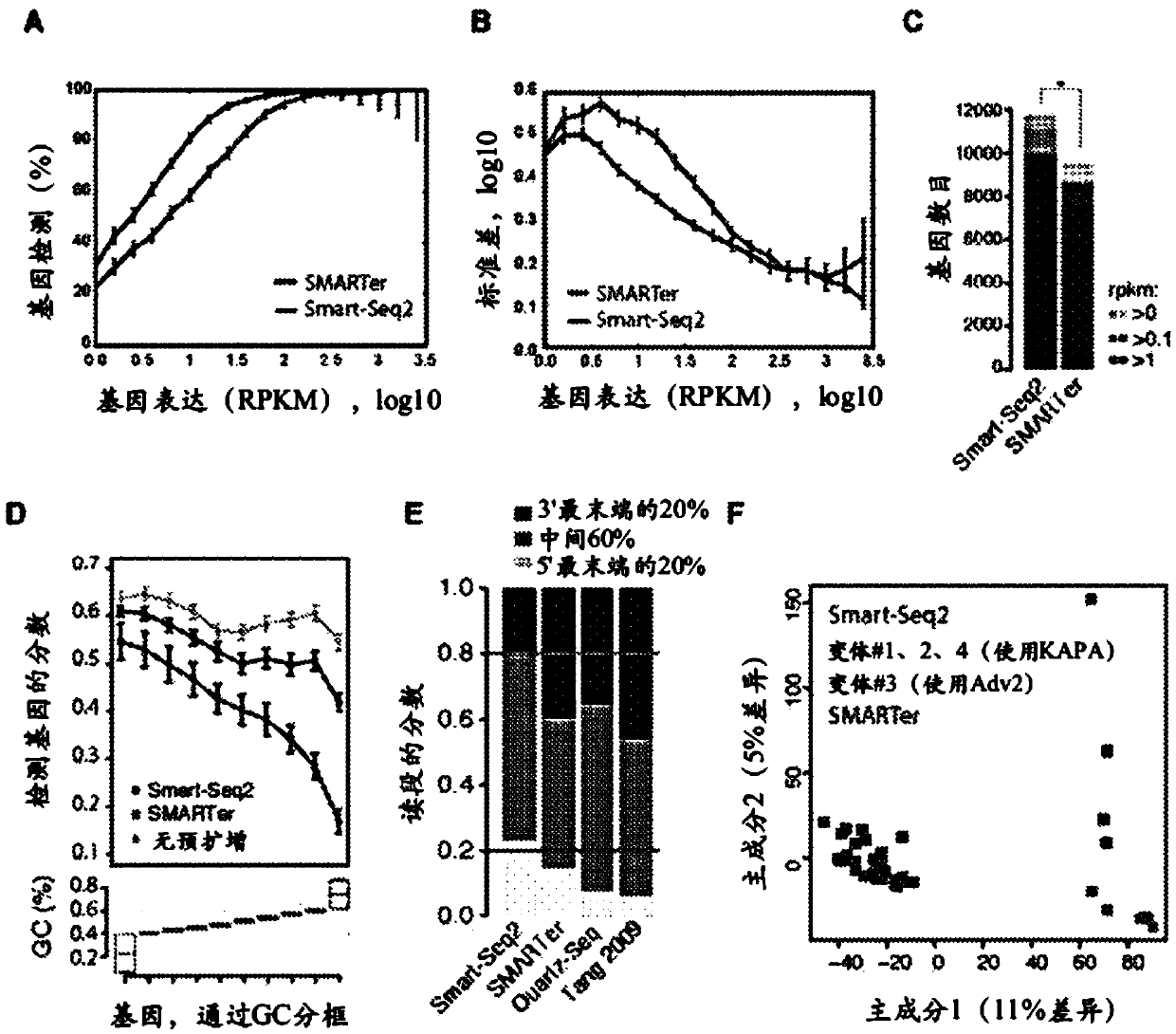
[0103] To improve the profiling of full-length transcriptomes obtained from single cells, a large number of variants (457 experiments in total) of reverse transcription, template-switching oligonucleotides (TSOs), and PCR preamplification were performed in terms of cDNA library yield and length evaluation, and compared these results with commercial Smart-Seq (hereinafter referred to as ) compared with (Table 1). Importantly, improvements were identified that significantly increased cDNA yield and length obtained from 1 ng of total promoter RNA (Table 1).
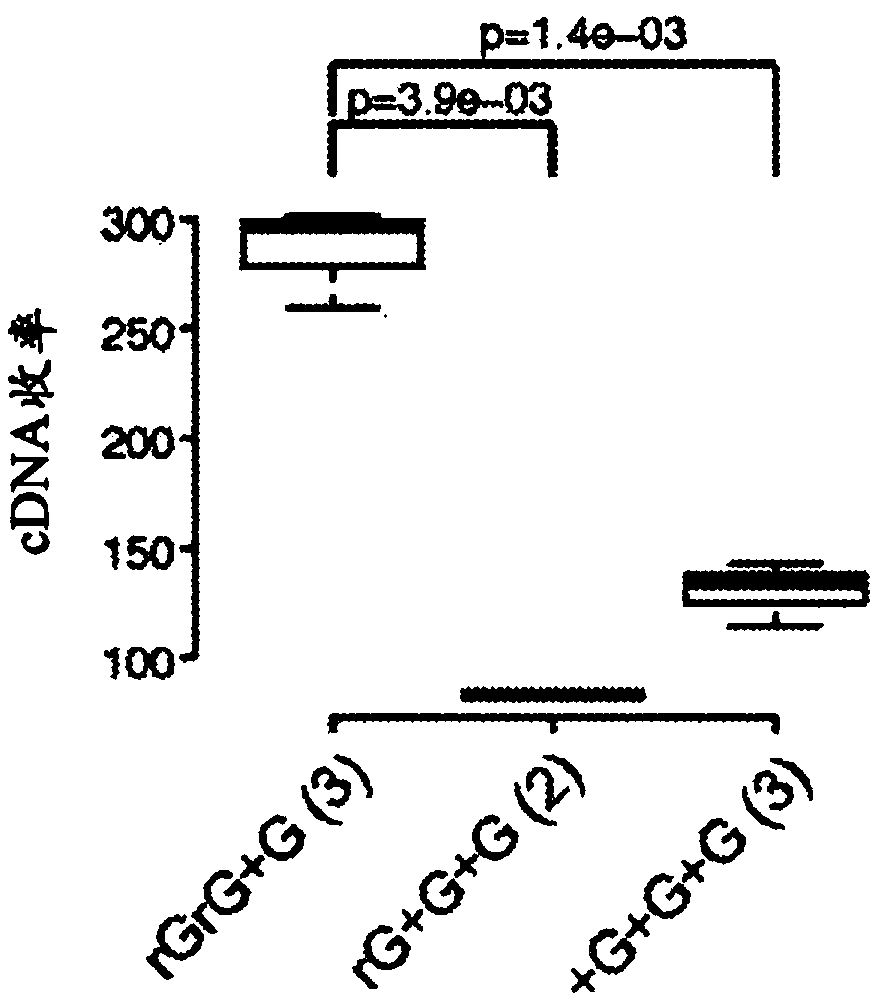
[0104] Specifically, compared to the commercial Smart-Seq used IIAoligo, only a single guanylate at the 3′ end (rGrG+G) of TSO was exchanged for a locked nucleic acid (LNA) guanylate resulting in a 2-fold increase in cDNA yield (p=0.003, Student's t-test; figure 1 a. Table 2 and image 3 ).
[0105] Additionally, the methyl group donor betaine was found to be associated with higher MgCl 2 Concentration binding had a s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com