Stem cell growth factor-like polypeptides
a stem cell growth factor and polypeptide technology, applied in the field of new stem cell growth factorlike polypeptides, can solve the problems that stem cells in general are extremely difficult to culture and maintain in vitro, and achieve the effect of reducing the side effects of such an agen
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Isolation of SEQ ID NO:1-21 from a cDNA Libraries of Human Cells
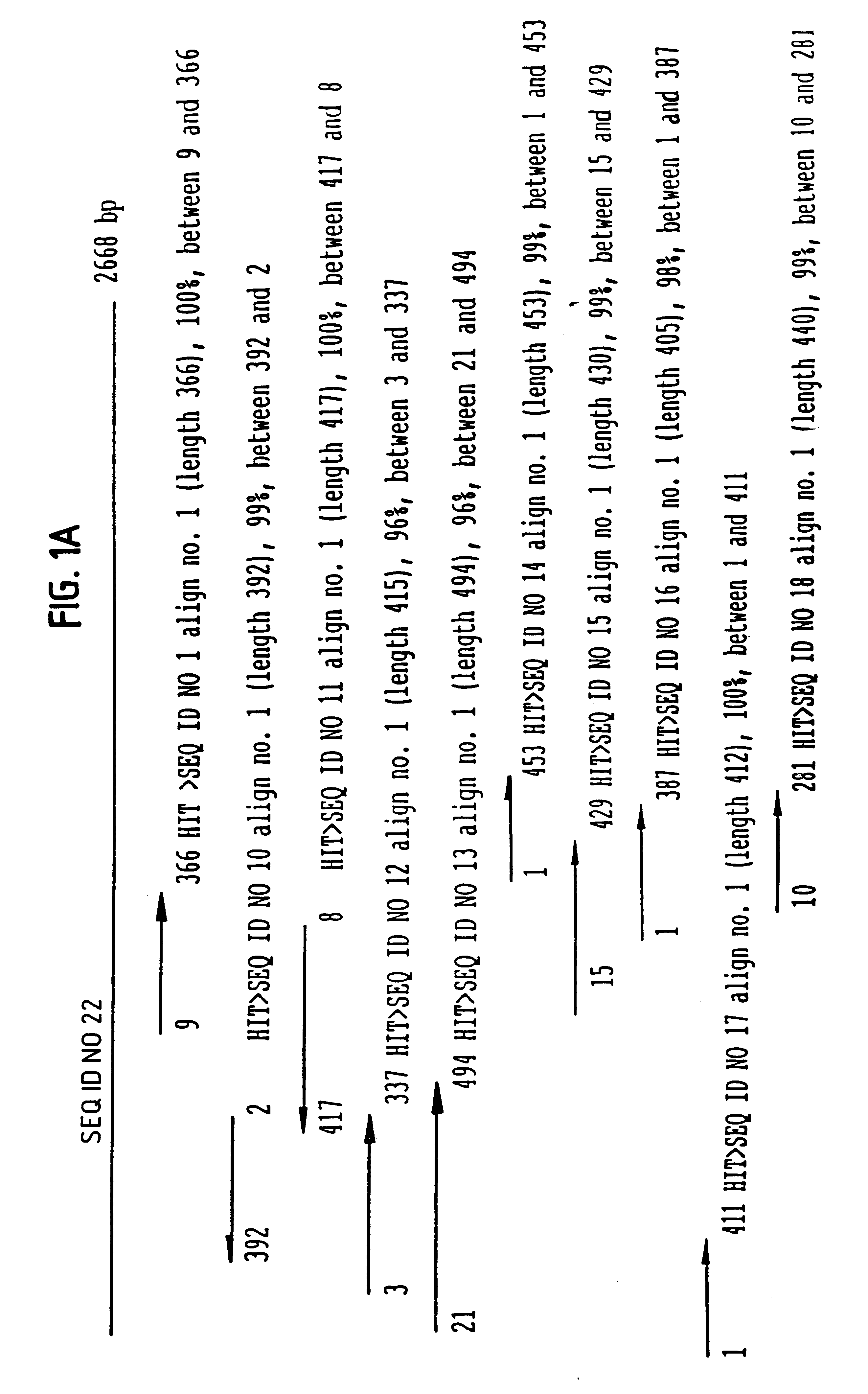
A plurality of novel nucleic acids were obtained from a cDNA library prepared from human fetal liver spleen, ovary, adult brain, lung tumor, spinal cord, cervix, ovary, endothelial cells, umbilical cord, lymphocyte, lung fibroblast, fetal brain, and testis, using standard PCR, sequencing by hybridization sequence signature analysis, and Sanger sequencing techniques. The inserts of the library were amplified with PCR using primers specific for vector sequences flanking the inserts. These samples were spotted onto nylon membranes and interrogated with oligonucleotide probes to give sequence signatures. The clones were clustered into groups of similar or identical sequences, and single representative clones were selected from each group for gel sequencing. The 5' sequence of the amplified inserts was then deduced using the reverse M13 sequencing primer in a typical Sanger sequencing protocol. PCR products were purified and...
example 2
Assemblage of SEQ ID NO: 22 and 24
The novel nucleic acids (SEQ ID NO: 22 and 24) of the invention were assembled from sequences that were obtained from a cDNA library by methods described in Example 1 above. The final sequences were assembled using the EST sequences as seed. Then a recursive algorithm was used to extend the seed into an extended assemblage, by pulling additional sequences from Hyseq's database containing EST sequences that belong to this assemblage. The algorithm terminated when a complete contig was assembled. Inclusion of component sequences into the assemblage was based on a BLASTN hit to the extending assemblage with BLAST score greater than 300 and percent identity greater than 95%.
The nearest neighbor result for the assembled sequence (SEQ ID NO. 22 or 24) was obtained by a FASTA version 3 search against Genpept release 114, using Fastxy algorithm. Fastxy is an improved version of FASTA alignment which allows in-codon frame shifts. The nearest neighbor result ...
example 3
Assemblage of SEQ ID NO: 27
The novel nucleic acid (SEQ ID NO: 27) of the invention was initially assembled from sequences that were obtained from a cDNA library by methods described in Example 1 above. The final sequence was assembled using the EST sequences as seed. Then a recursive algorithm was used to extend the seed into an extended assemblage, by pulling additional sequences from Hyseq's database containing EST sequences that belong to this assemblage. The algorithm terminated when a complete contig was assembled. Inclusion of component sequences into the assemblage was based on a BLASTN hit to the extending assemblage with BLAST score greater than 300 and percent identity greater than 95%.
Using this initial sequence, suitable primers were designed for amplification of ESTs that comprise the initial sequence. The products were cloned. The DNA was isolated, cut with appropriate restriction enzymes, ligated, and recloned to generate the full-length contig. The full-length produc...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular mass | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
| weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com