Method for determining gene expression regulation mechanism based on unicellular transcriptome data
A technology for gene expression and determination method, applied in the field of bioinformatics, which can solve the problems of the complexity of cell signaling network, ignoring cell specificity, and unable to comprehensively and systematically elucidate gene expression.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0039] The technical solutions in the embodiments of the present application will be described below with reference to the accompanying drawings in the embodiments of the present application.
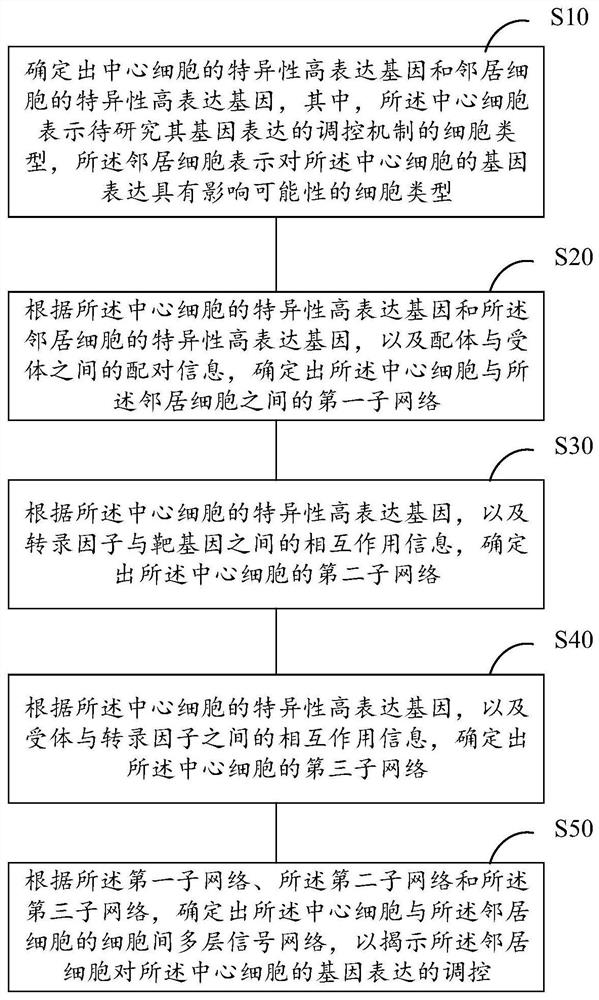
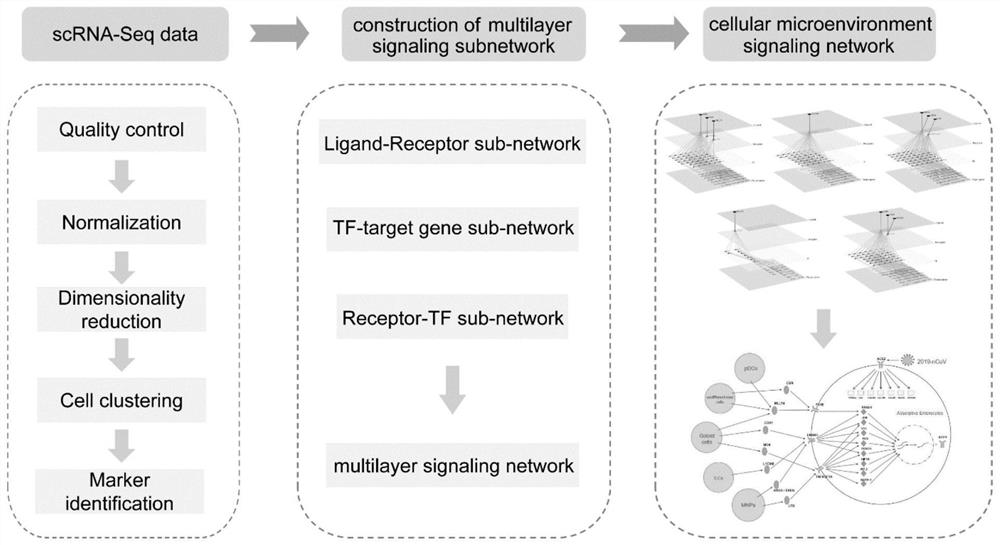
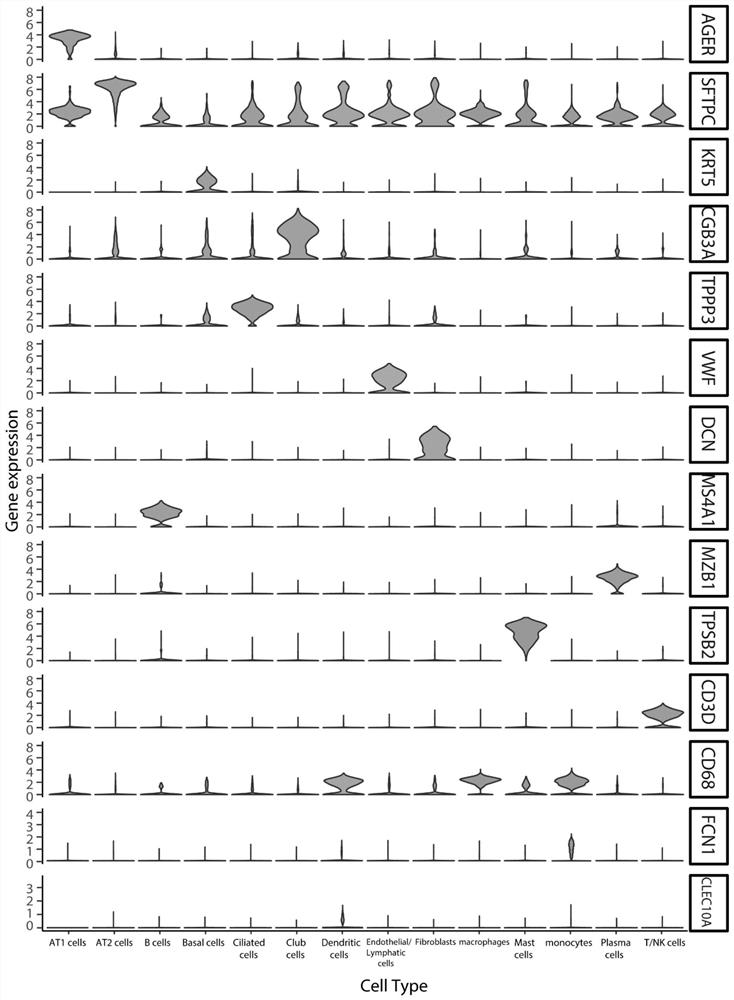
[0040] The regulation of gene expression is regulated by a very complex signaling network. Signaling networks are usually intertwined by signaling pathways composed of functional molecules such as ligands-receptors-transcription factors-target genes. Because there is still mutual influence between signaling pathways, there is no strict one-to-one correspondence between functional molecules: for example, the same receptor can bind to multiple ligands with similar structures, and after the receptor is activated, it can change multiple ligands. Downstream molecular conformation, transcription factors can also activate the expression of multiple target genes; or, the same functional molecule may appear in different pathways and play different biological functions, and different ligands acti...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com