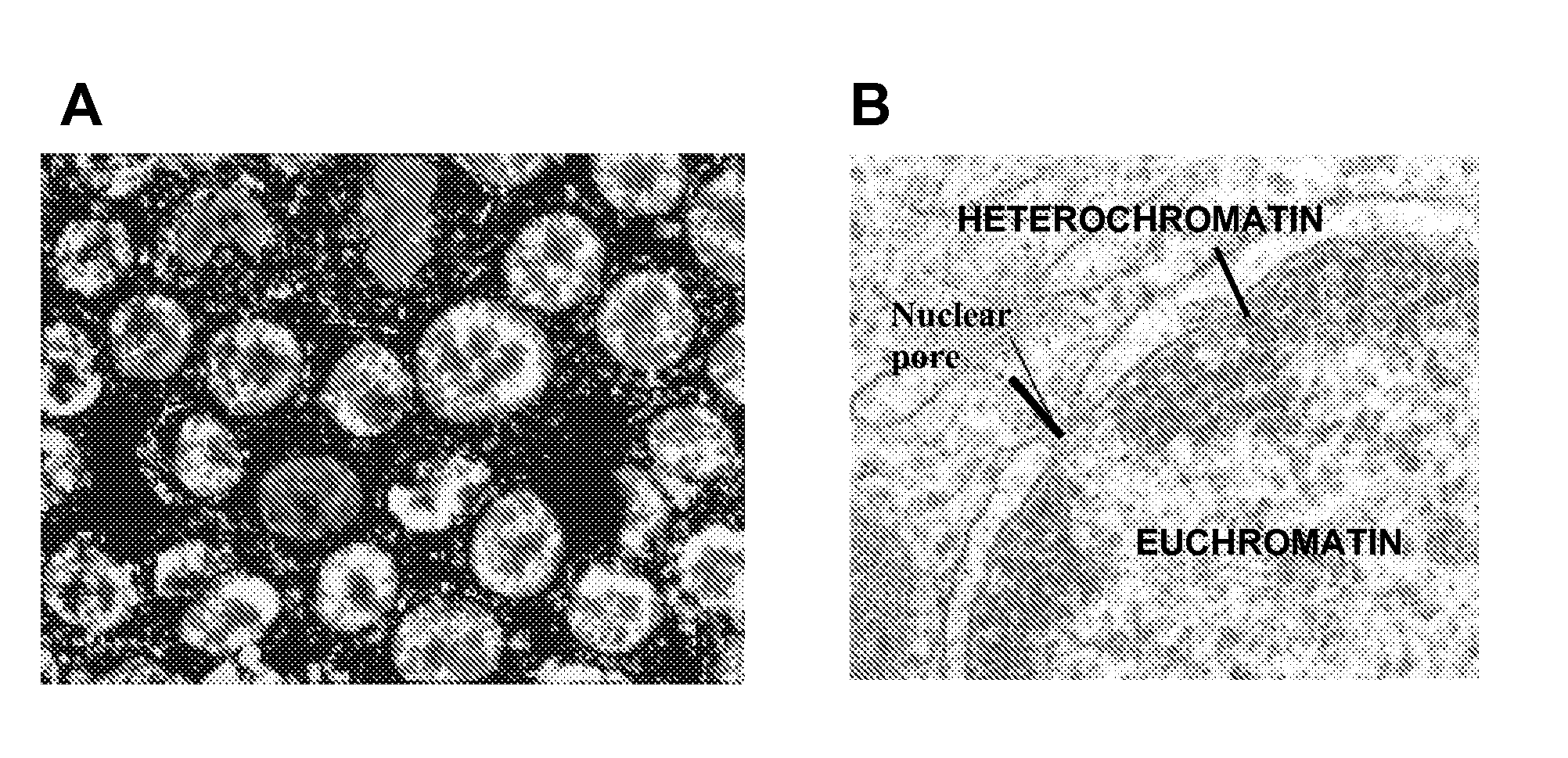
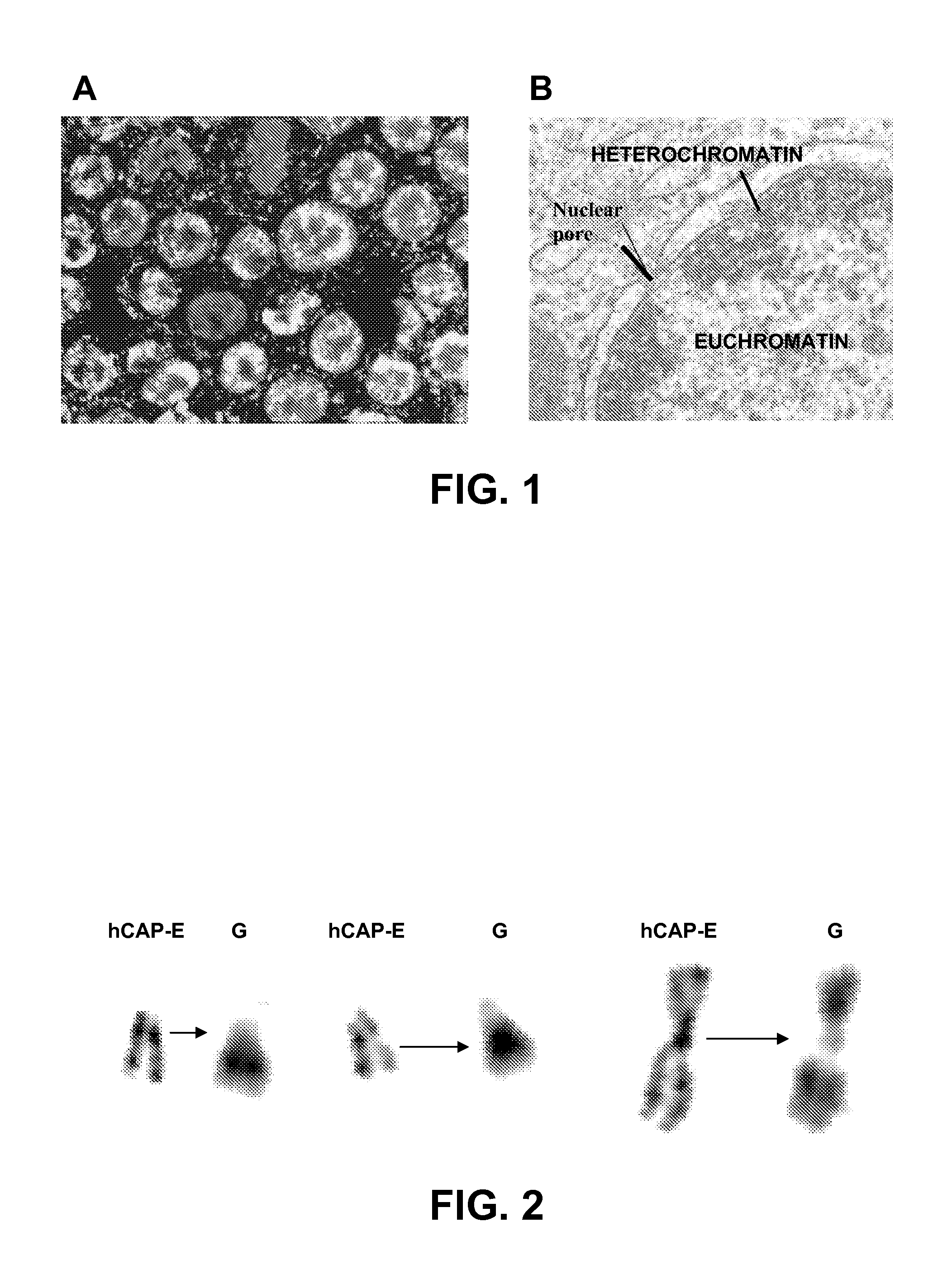
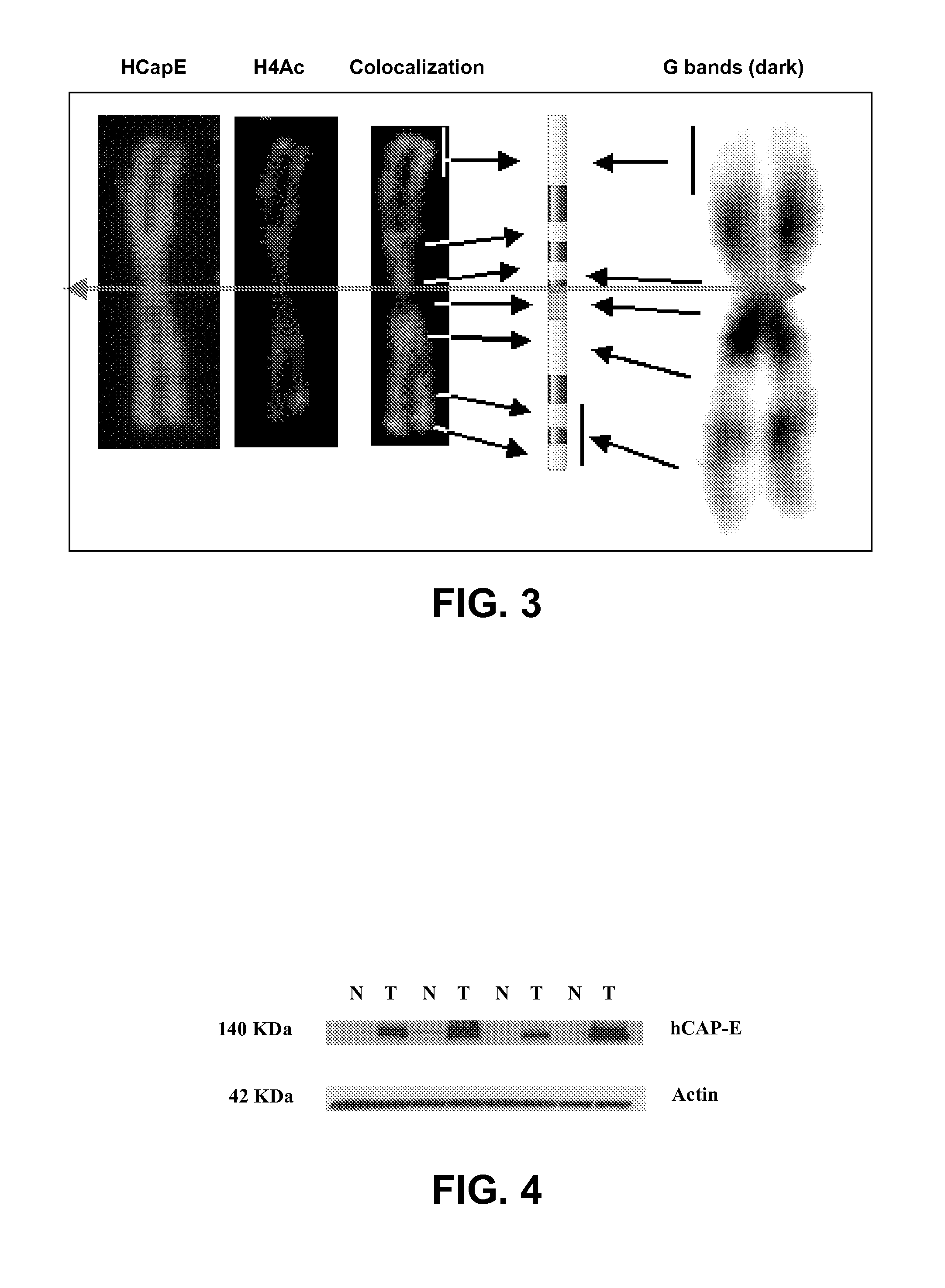
Patents
Literature
758 results about "Gene expression level" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Gene expression levels result from the dynamic interplay of activators and repressors. These factors may influence the basal transcriptional machinery directly or indirectly, through interactions that govern each other's activity or access to DNA.
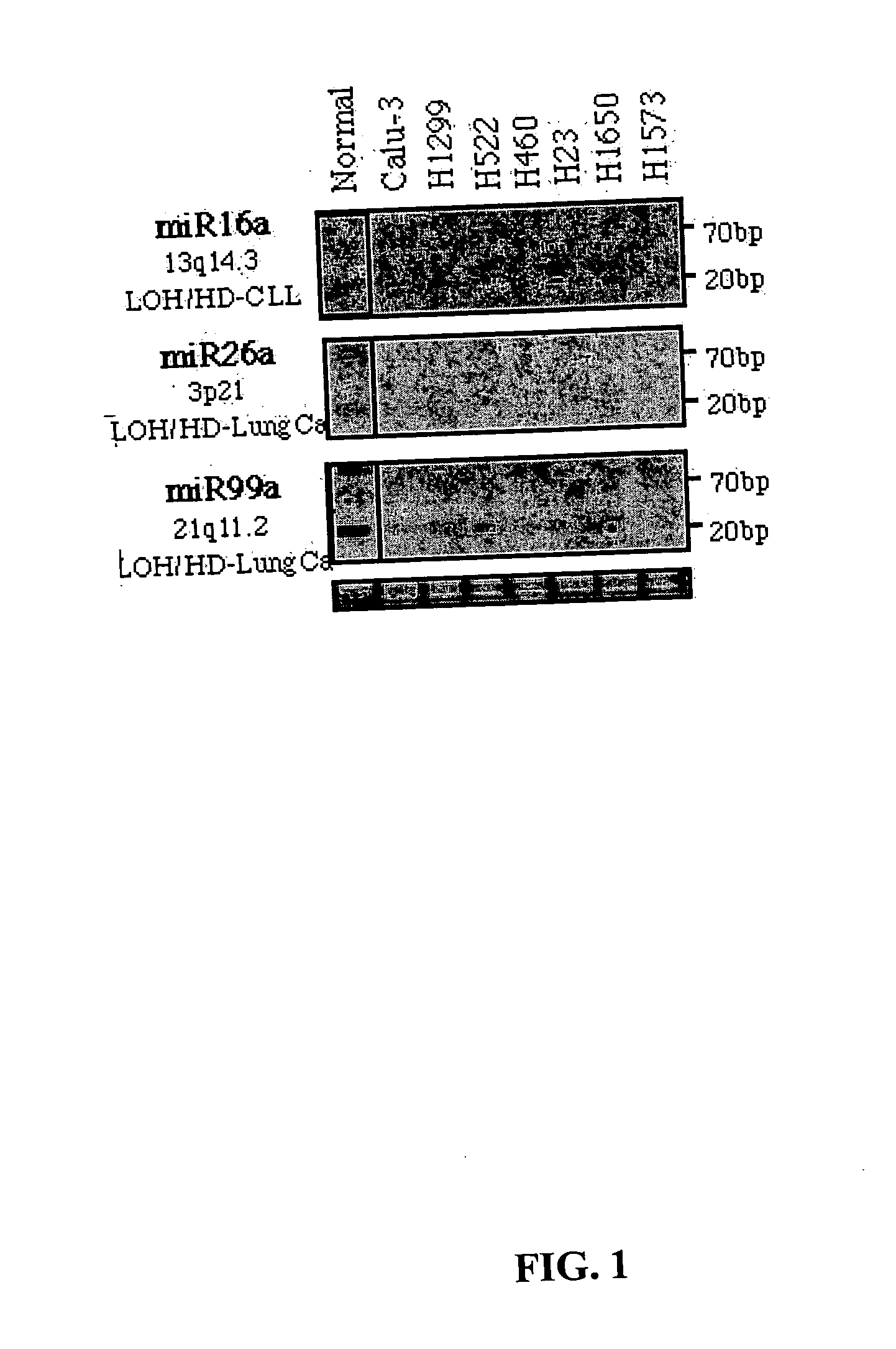
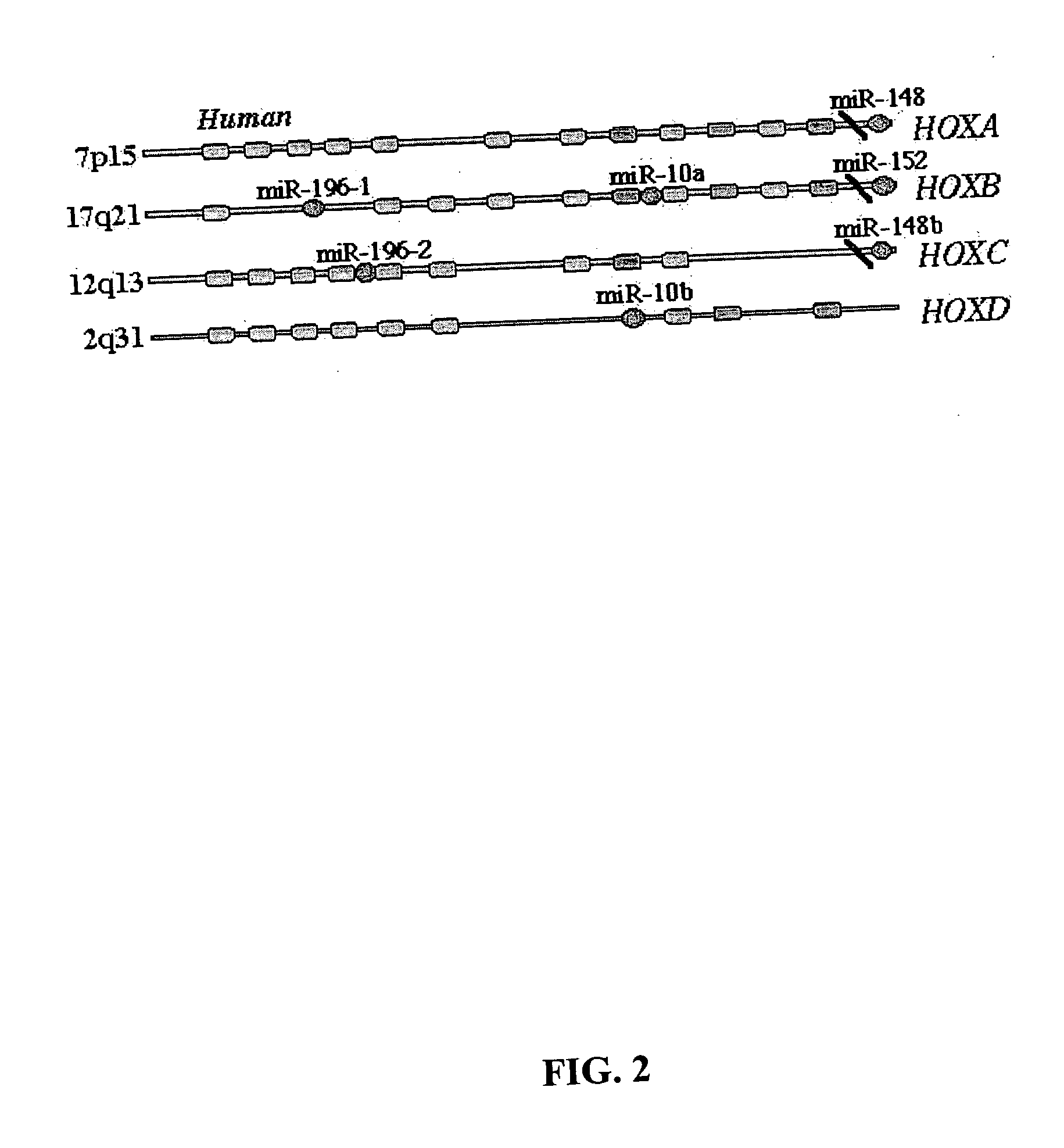
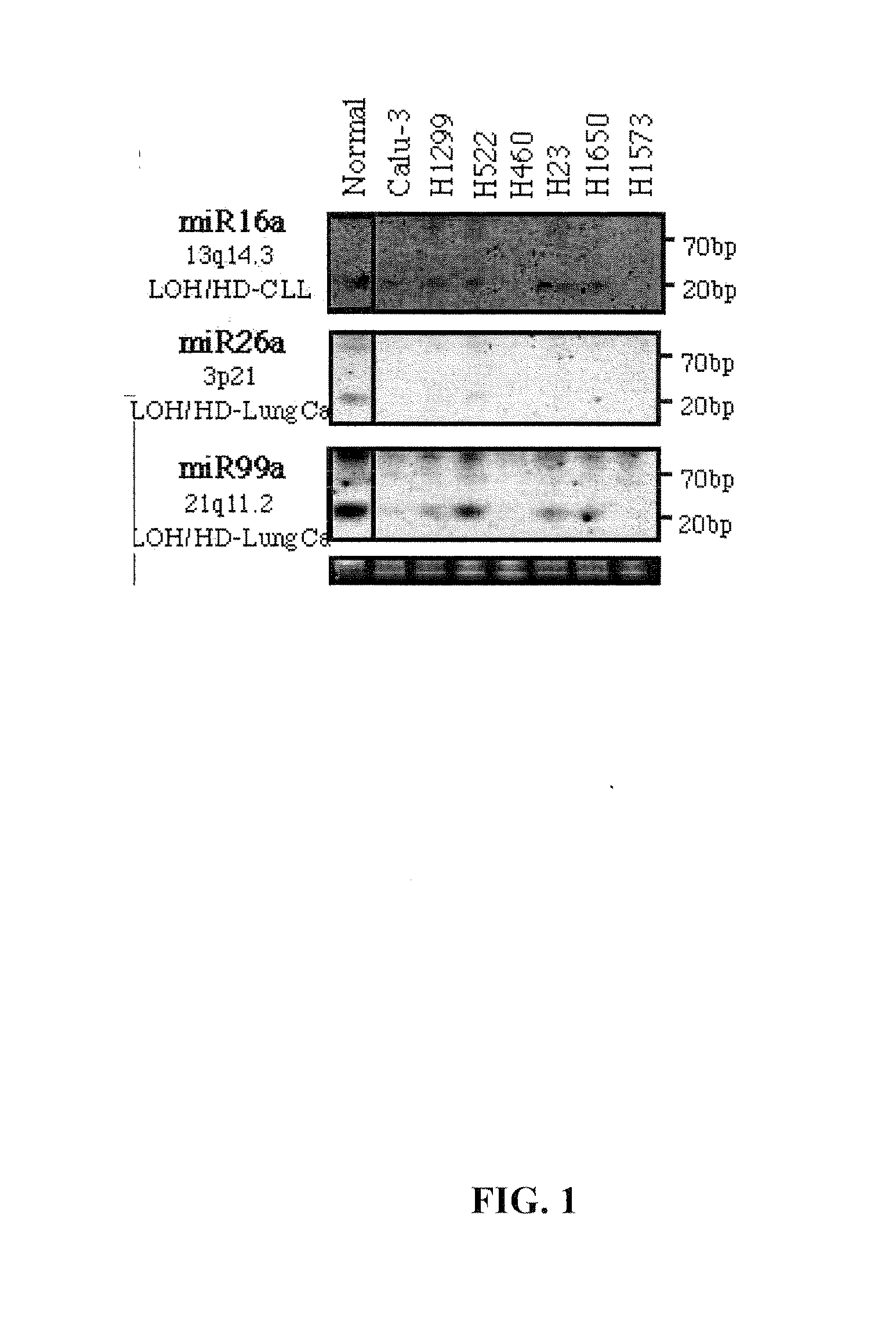
Diagnosis and treatment of cancers with microRNA located in or near cancer associated chromosomal features
InactiveUS20060105360A1Restore levelOrganic active ingredientsMicrobiological testing/measurementEtiologyMicroRNA Gene
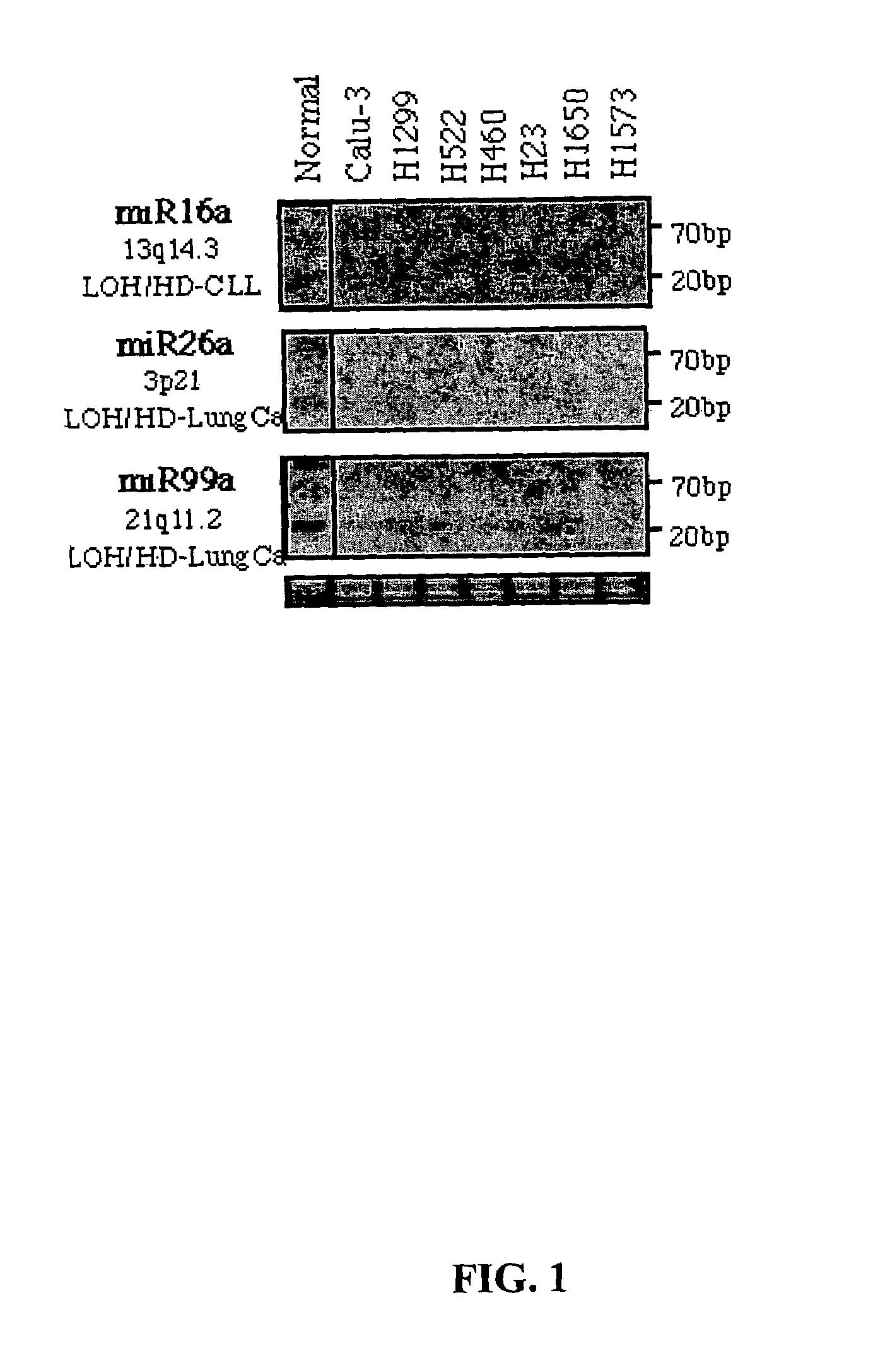
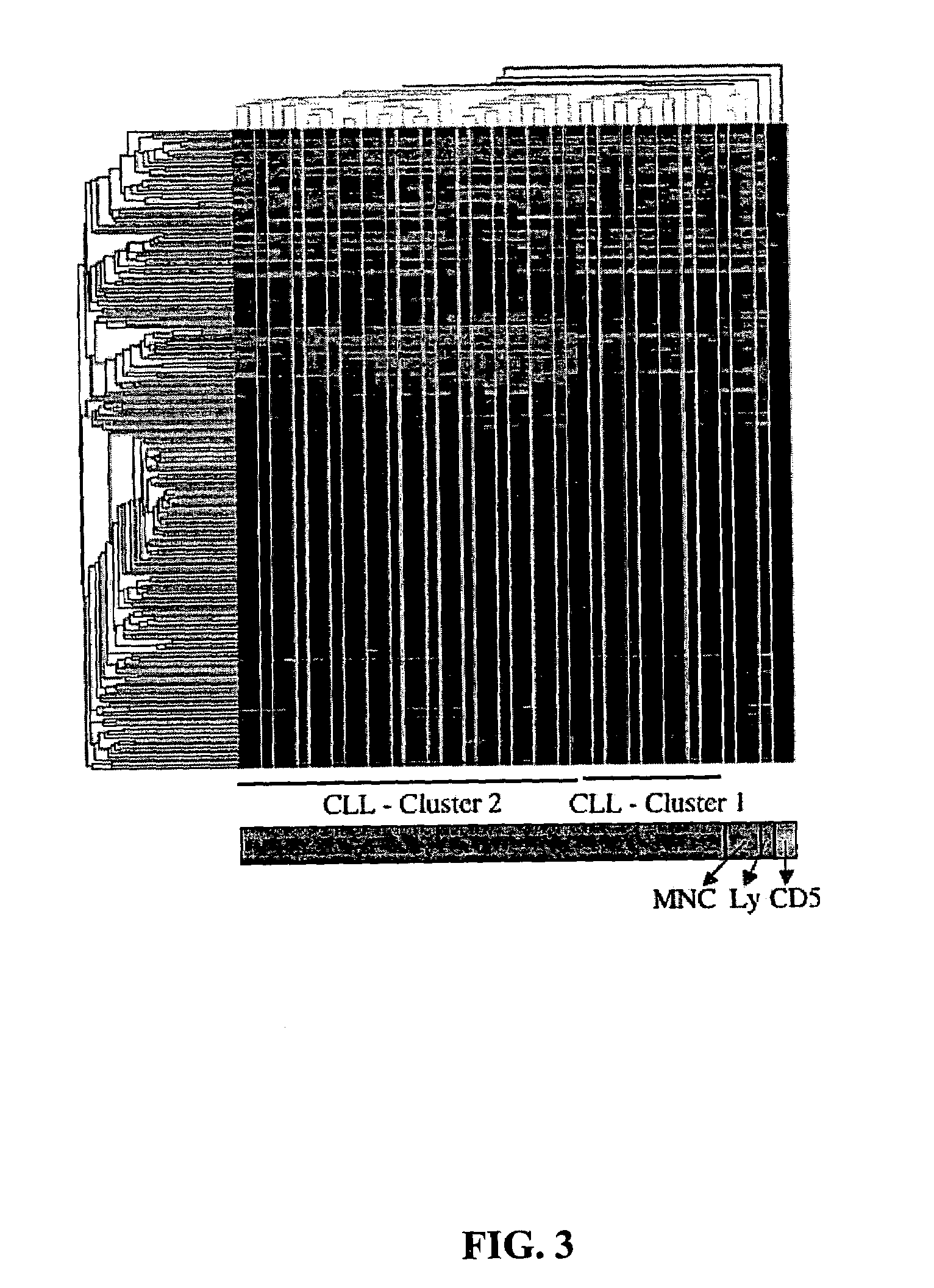
MicroRNA genes are highly associated with chromosomal features involved in the etiology of different cancers. The perturbations in the genomic structure or chromosomal architecture of a cell caused by these cancer-associated chromosomal features can affect the expression of the miR gene(s) located in close proximity to that chromosomal feature. Evaluation of miR gene expression can therefore be used to indicate the presence of a cancer-causing chromosomal lesion in a subject. As the change in miR gene expression level caused by a cancer-associated chromosomal feature may also contribute to cancerigenesis, a given cancer can be treated by restoring the level of miR gene expression to normal. microRNA expression profiling can be used to diagnose cancer and predict whether a particular cancer is associated with an adverse prognosis. The identification of specific mutations associated with genomic regions that harbor miR genes in CLL patients provides a means for diagnosing CLL and possibly other cancers.
Owner:THOMAS JEFFERSON UNIV
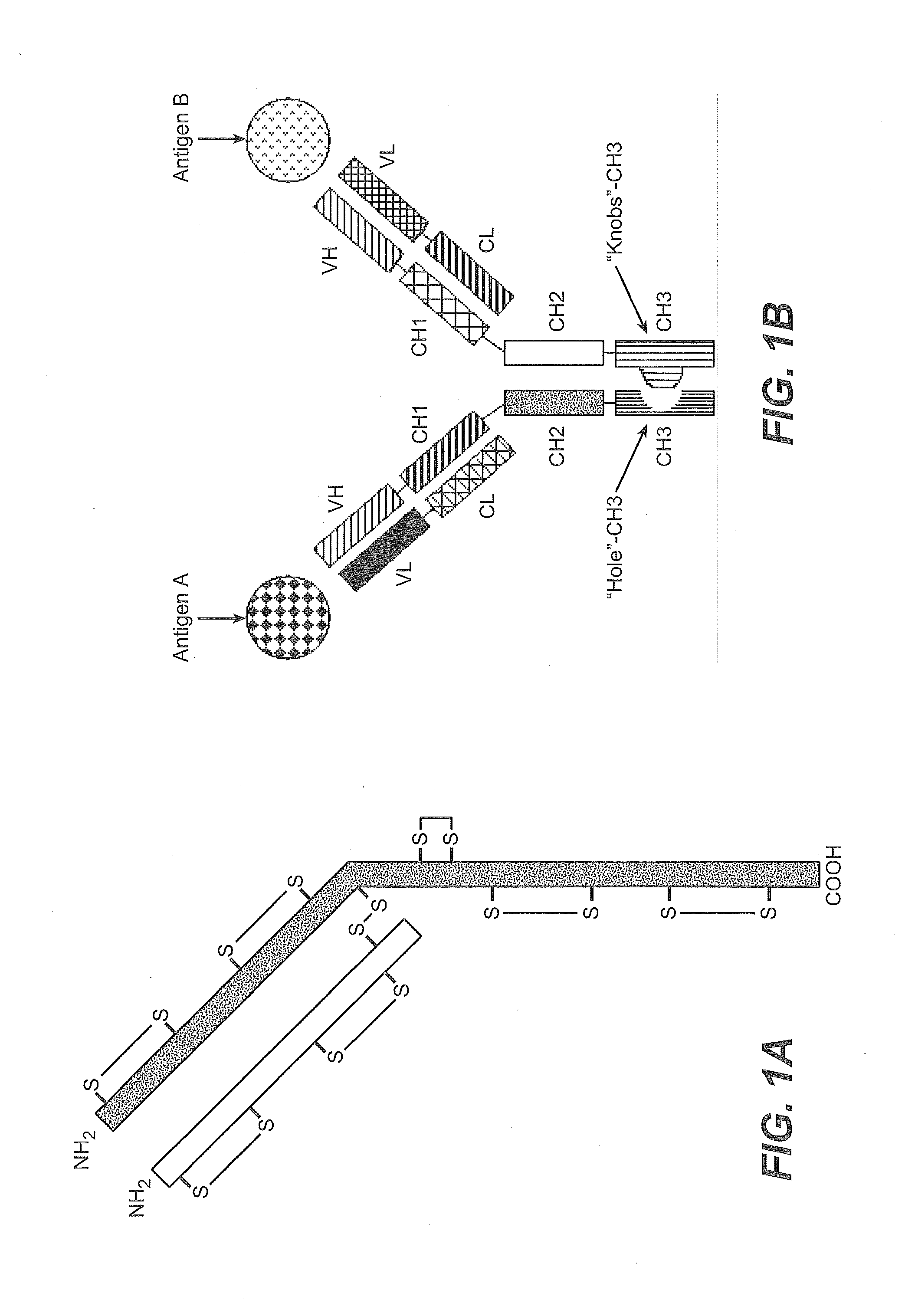
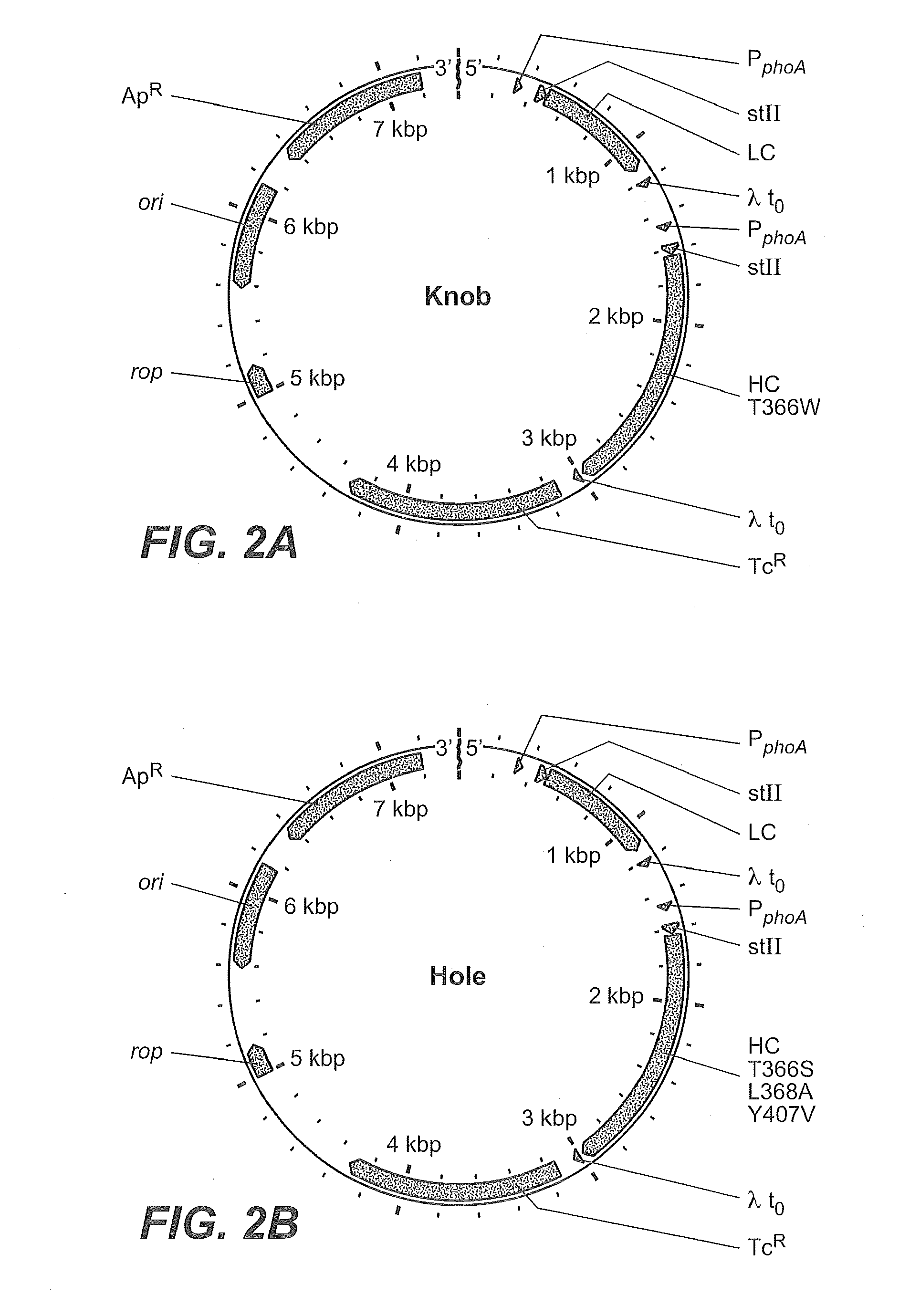
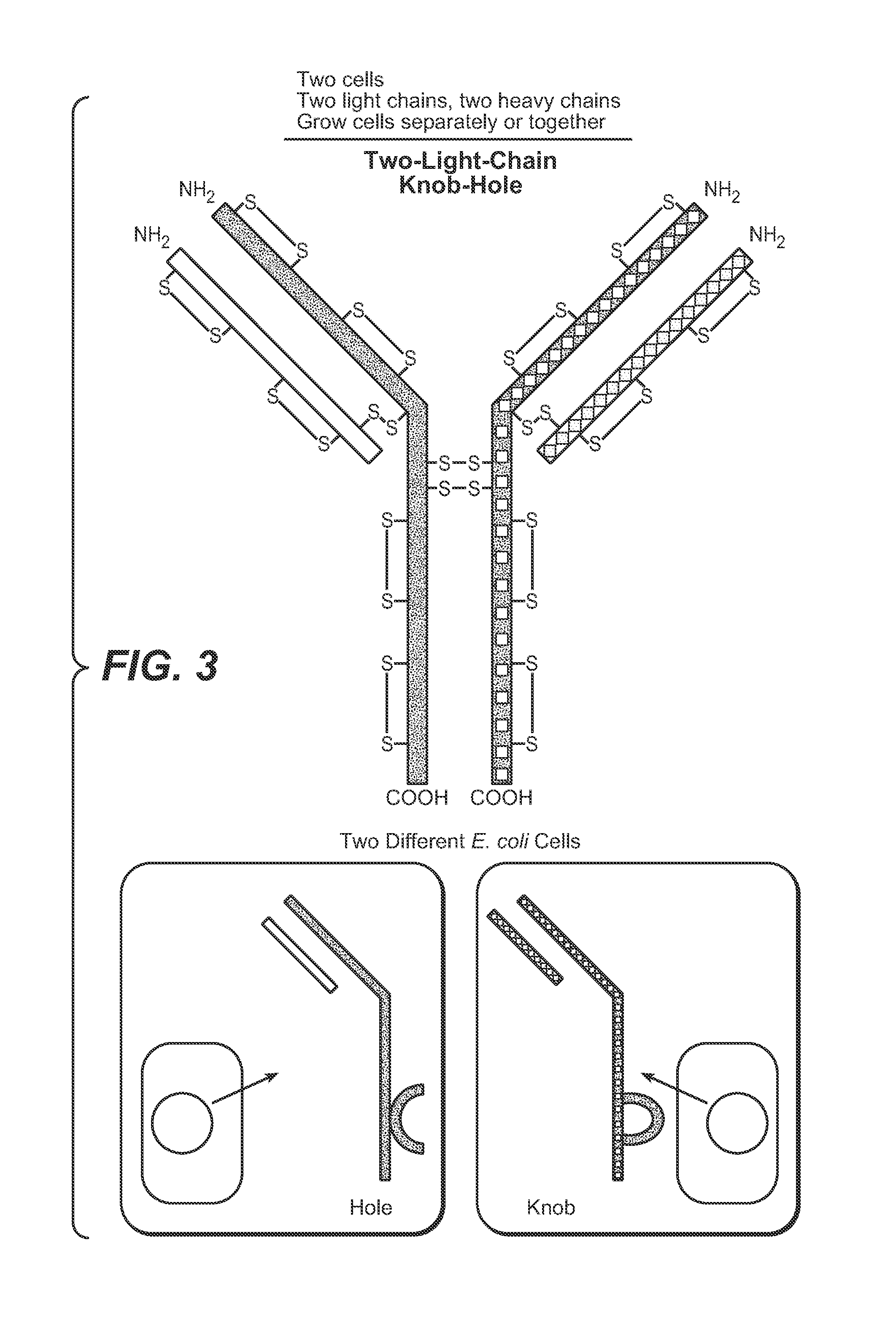
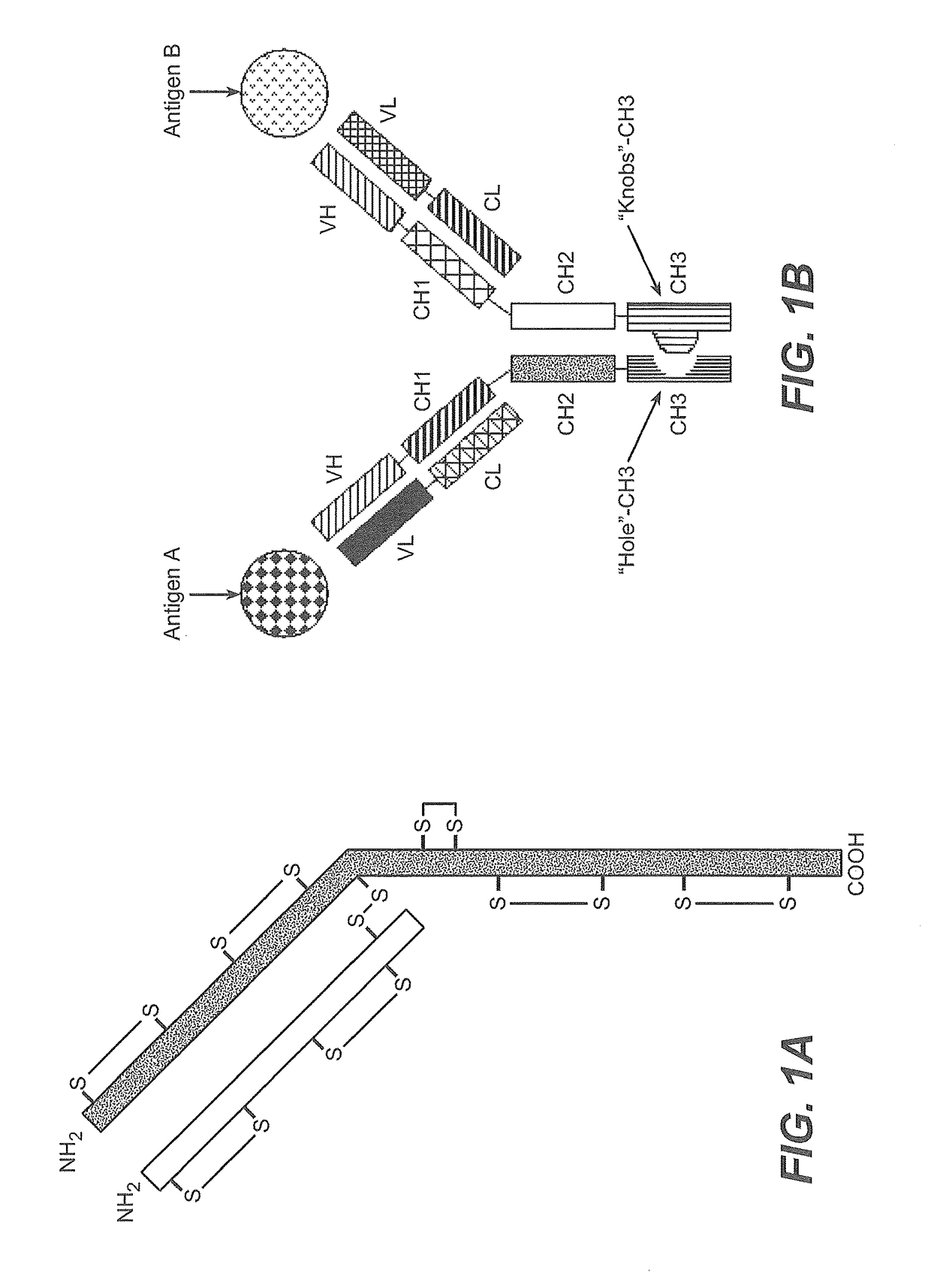
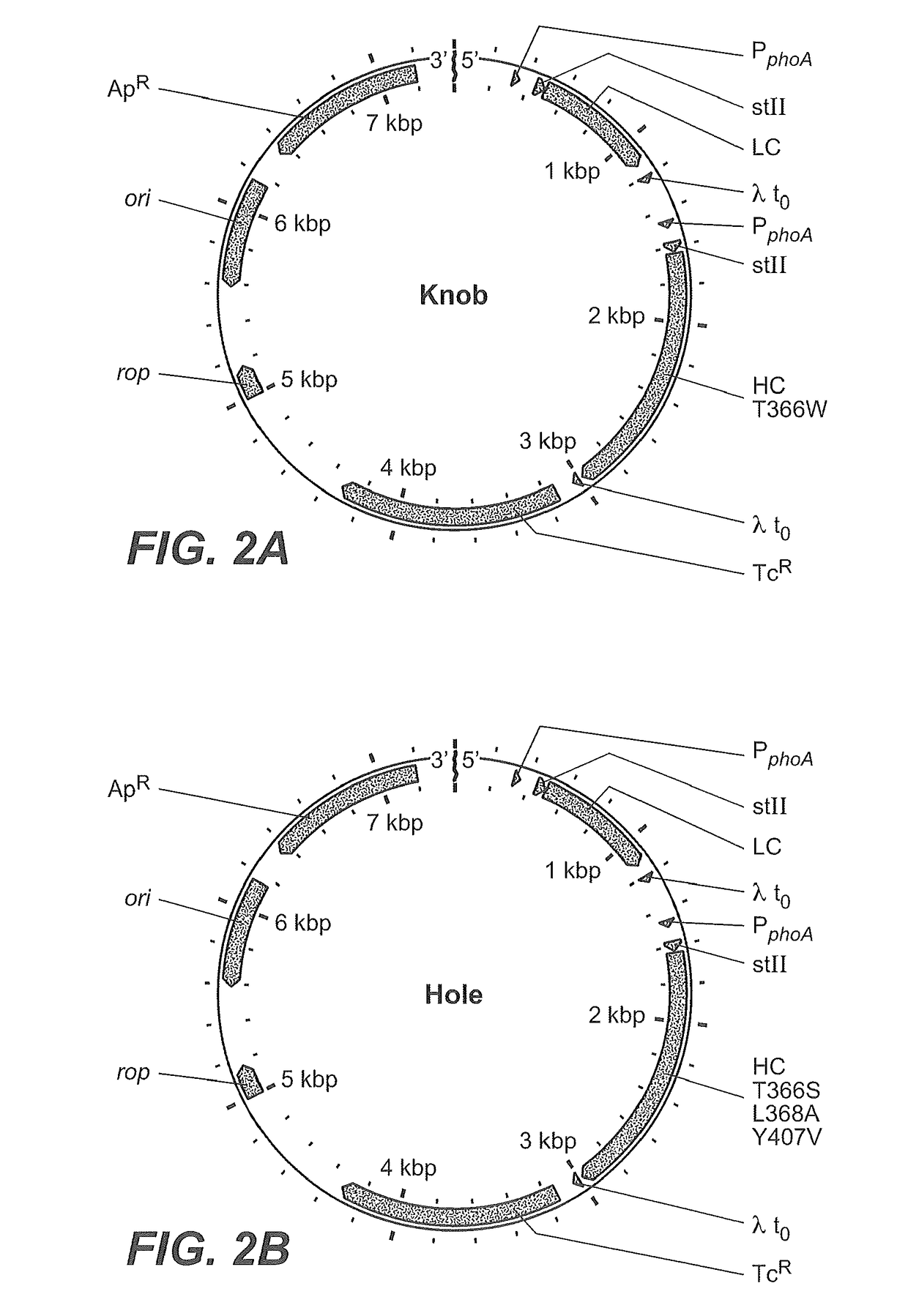
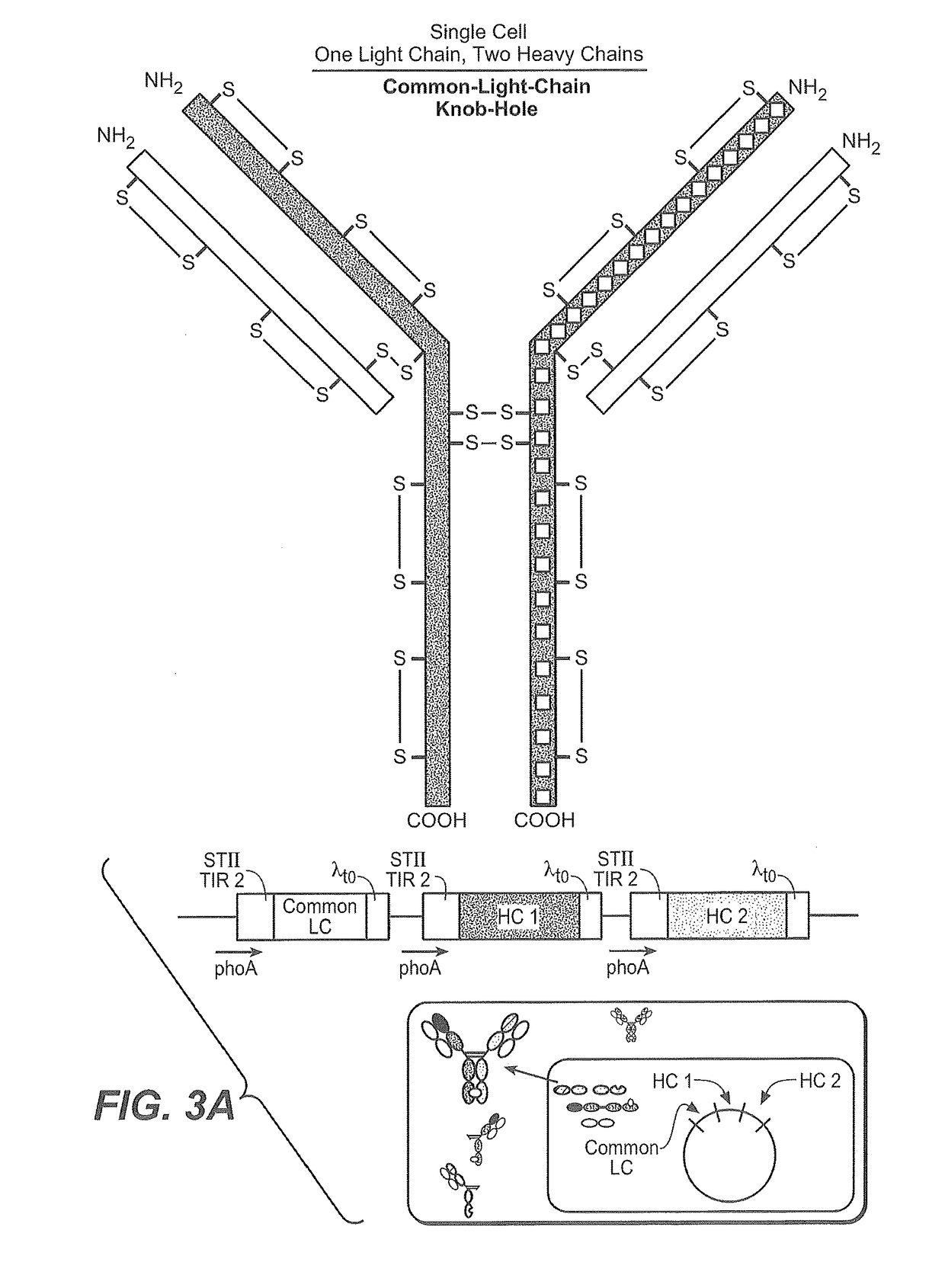
Production of Heteromultimeric Proteins
ActiveUS20110287009A1Reduction in yieldDecreased/elimination of effector functionAntipyreticAnalgesicsEpitopeBiochemistry
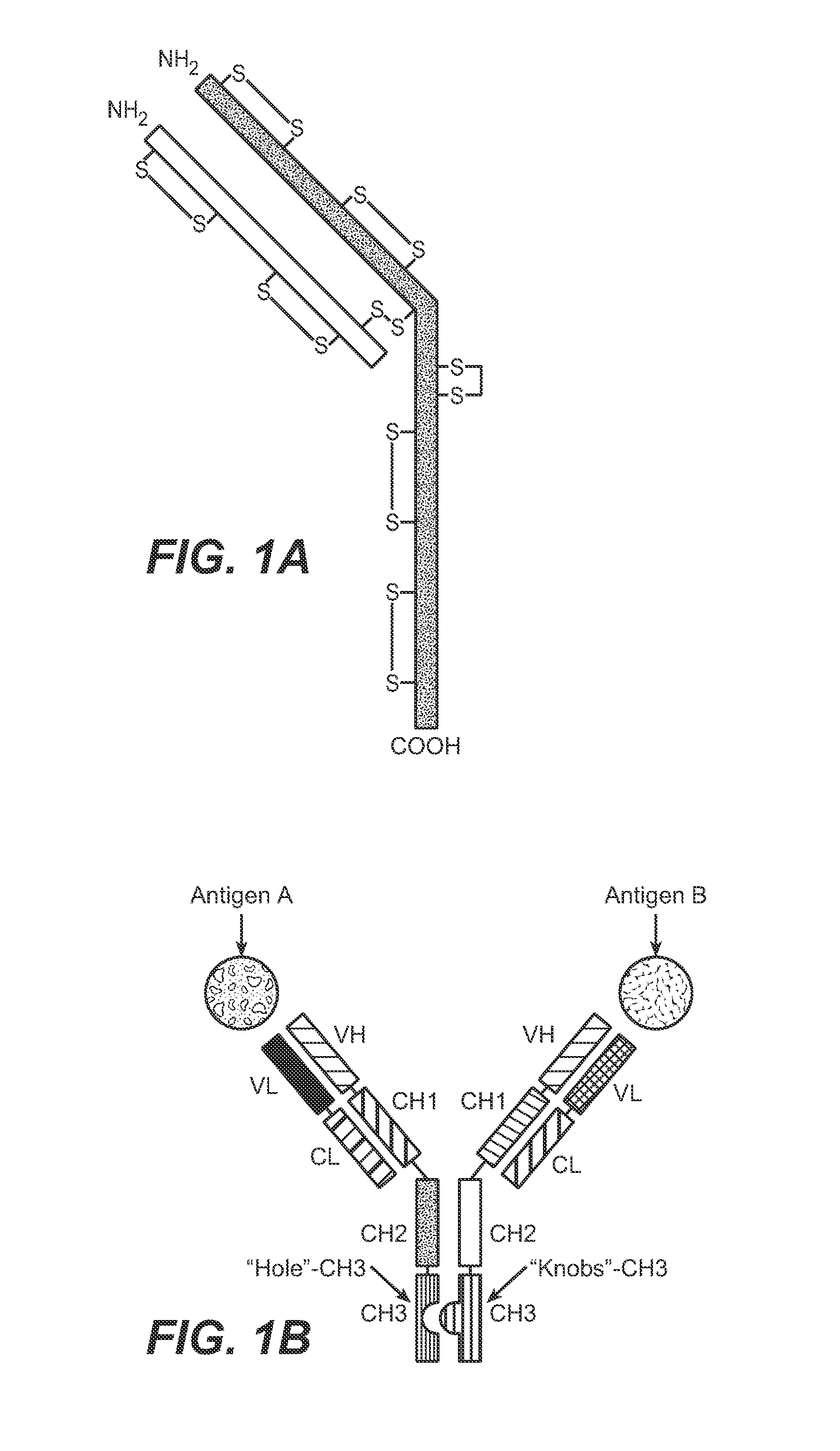
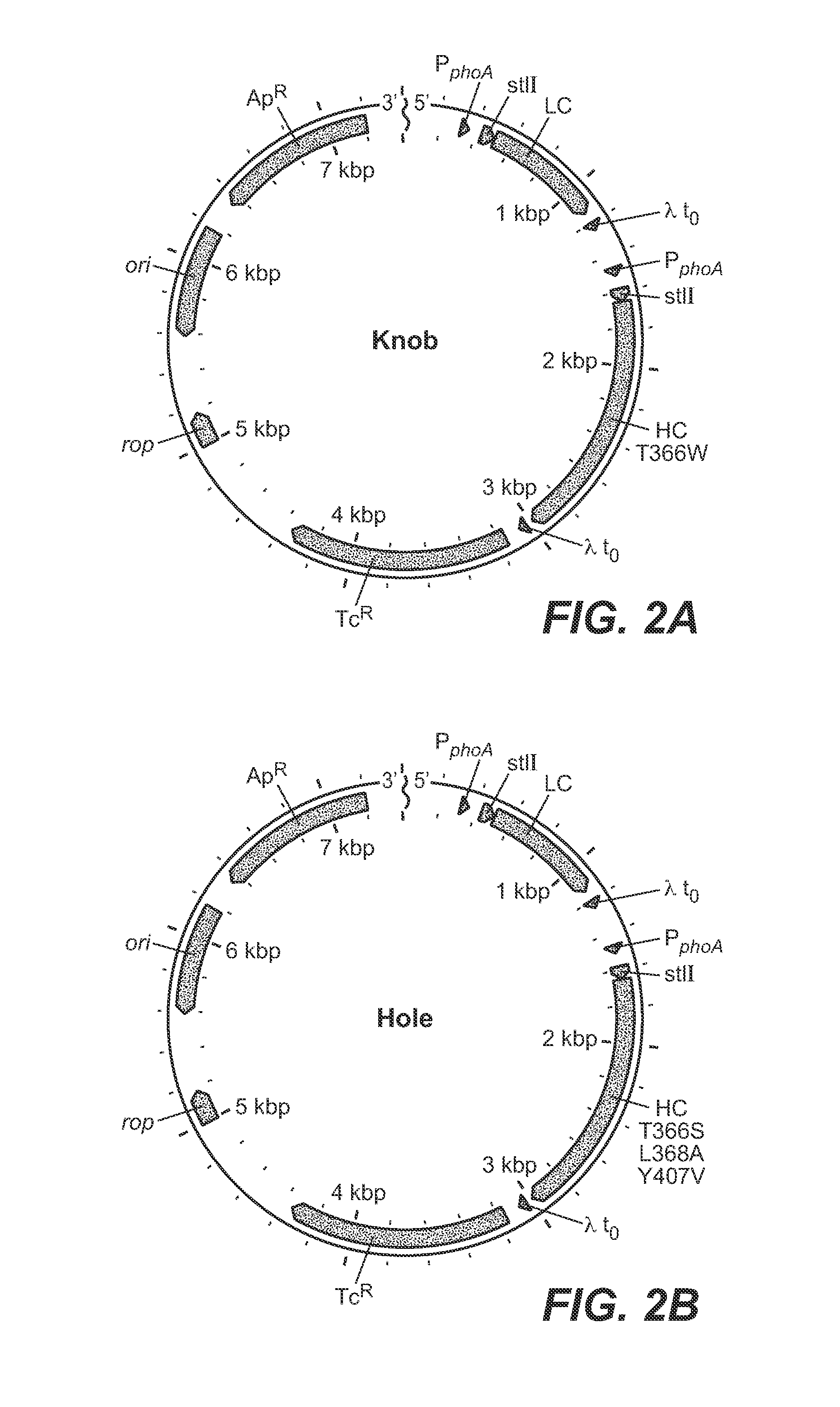
Described herein are methods for the efficient production of antibodies and other multimeric protein complexes (collectively referred to herein as heteromultimeric proteins) capable of specifically binding to more than one target. The targets may be, for example, different epitopes on a single molecule or located on different molecules. The methods combine efficient, high gene expression level, appropriate assembly, and ease of purification for the heteromultimeric proteins. The invention also provides methods of using these heteromultimeric proteins, and compositions, kits and articles of manufacture comprising these antibodies.
Owner:F HOFFMANN LA ROCHE & CO AG
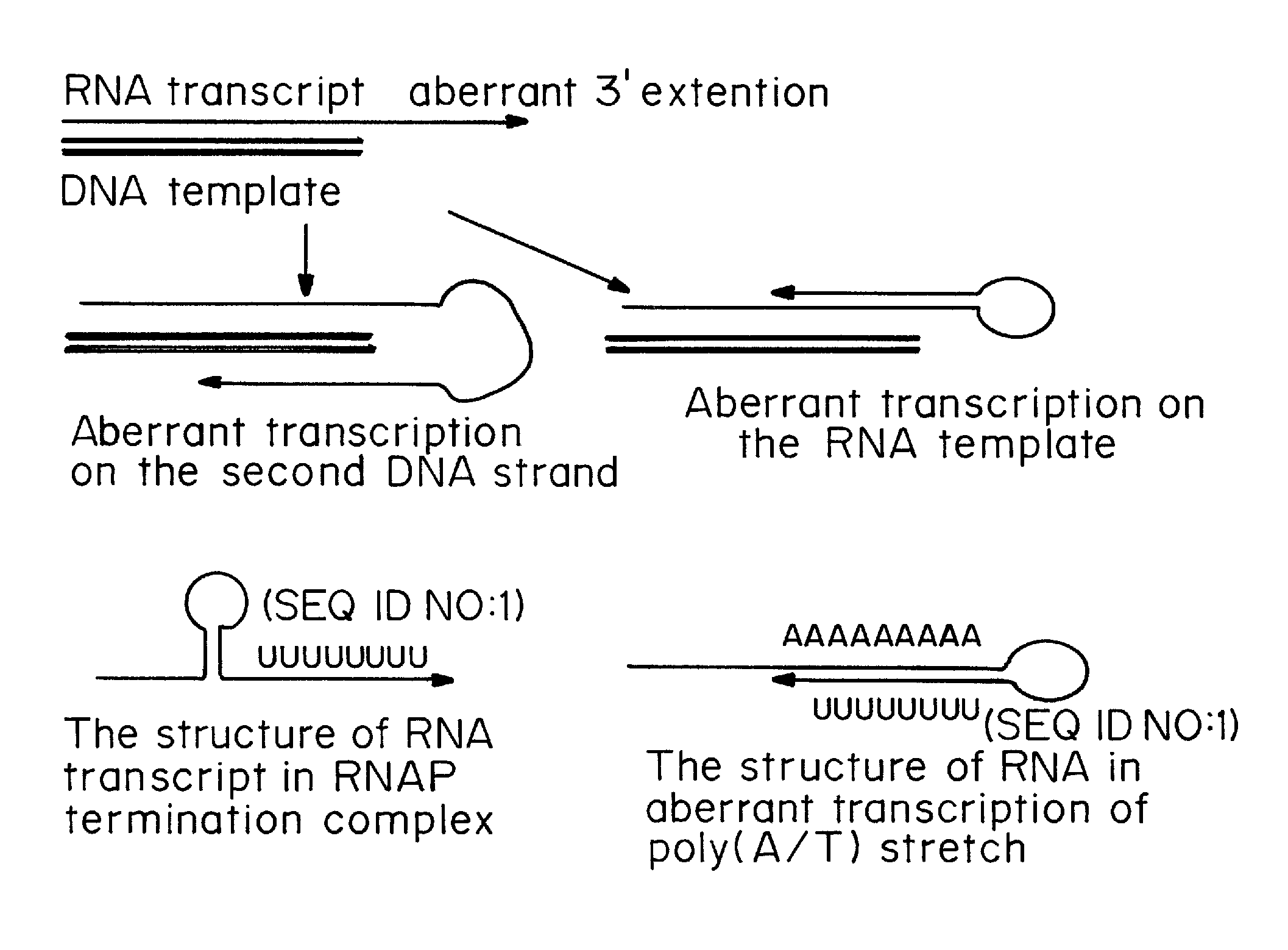
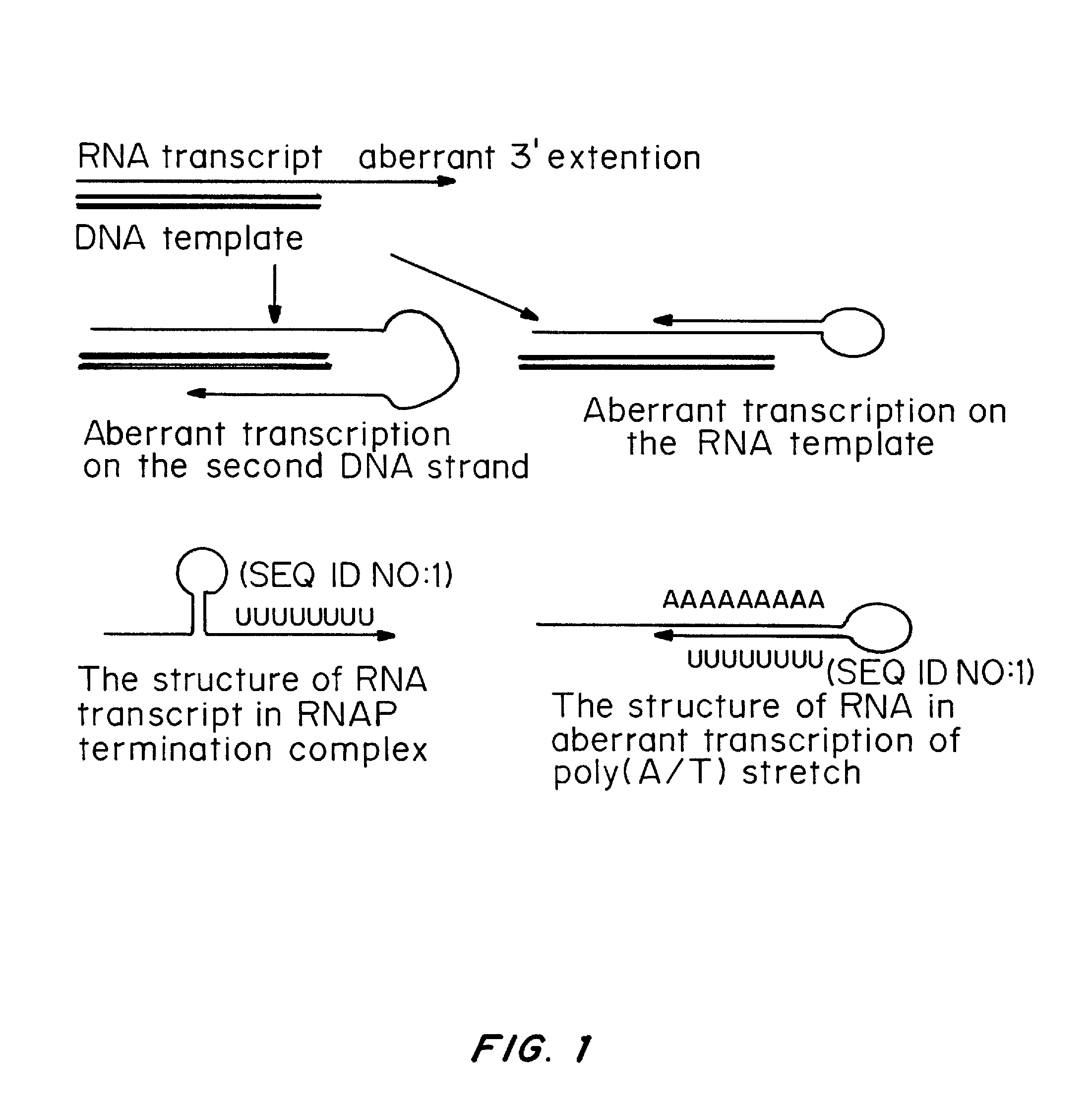
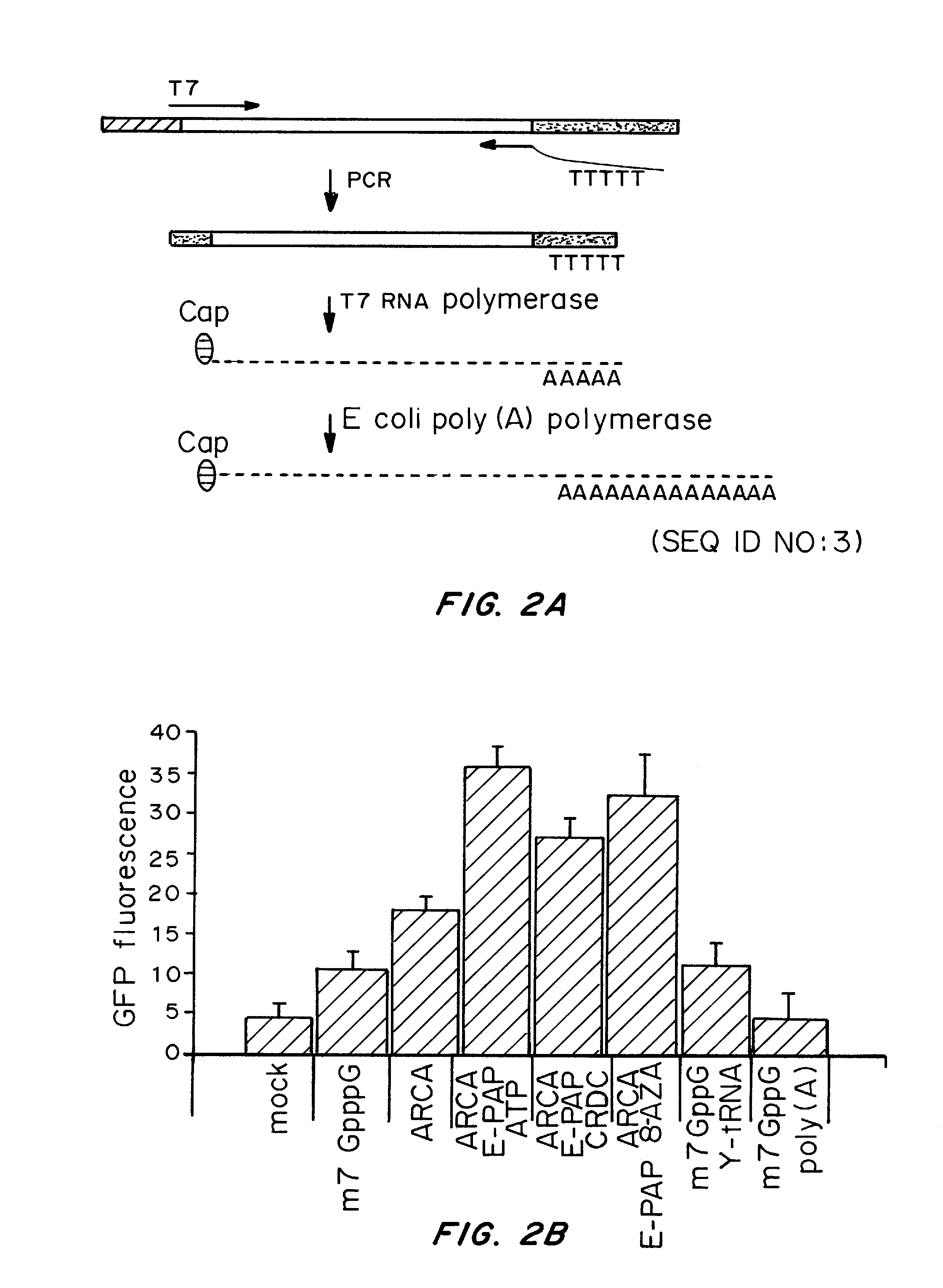
Transient Transfection with RNA
ActiveUS20080260706A1Lymphocyte transfectabilitySimilar efficiencyBiocideGenetic material ingredientsGene deliveryDNA construct
A method of mRNA production for use in transfection is provided, that involves in vitro transcription of PCR generated templates with specially designed primers, followed by polyA addition, to produce a construct containing 3′ and 5′ untranslated sequence (“UTR”), a 5′ cap and / or Internal Ribosome Entry Site (IRES), the gene to be expressed, and a polyA tail, typically 50-2000 bases in length. This RNA can efficiently transfect different kinds of cells. This approach results in increased efficiency (fidelity and productivity) of mRNA synthesis and is less time consuming because it does not require cloning, and also consequently eliminates the unwanted errors and effects related to RNA made on DNA templates obtained with cloning techniques. The results of transfection of RNAs demonstrate that RNA transfection can be very effective in cells that are exceedingly difficult to transfect efficiently with DNA constructs. Further, the levels of gene expression following mRNA transfection are consistent from cell to cell in an experiment and these levels can be controlled over a wide range simply by changing the amount of mRNA that is transfected, and without obvious cytotoxic effects due to the levels of RNA per se. Due to high efficiency the cells can be simultaneously transfected with multiple genetic constructs. The method can be used to deliver genes into cells not- or only poorly transfectable for DNA, in vitro and in vivo.
Owner:YALE UNIV
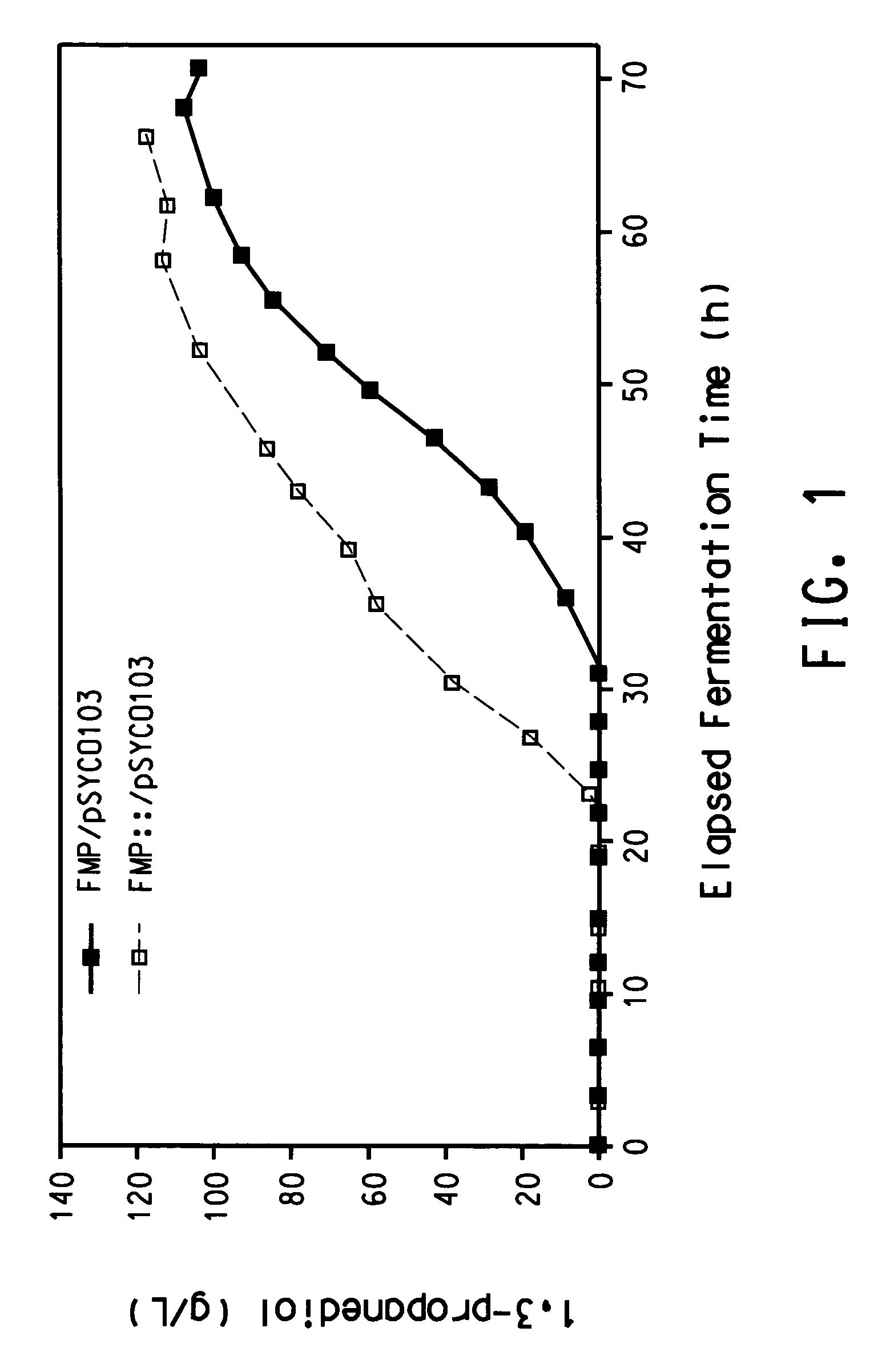
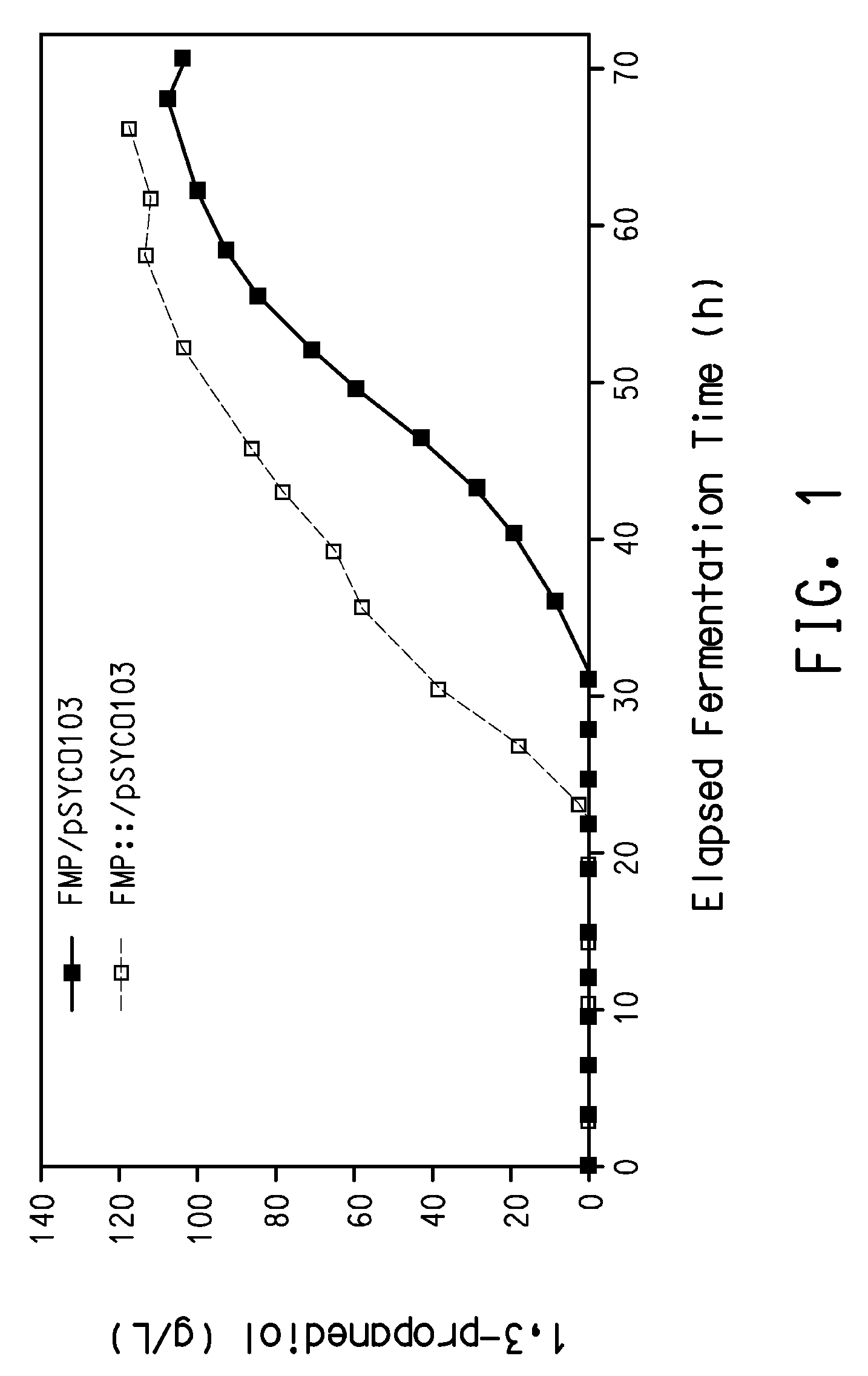
Process for the biological production of 1,3-propanediol with high yield
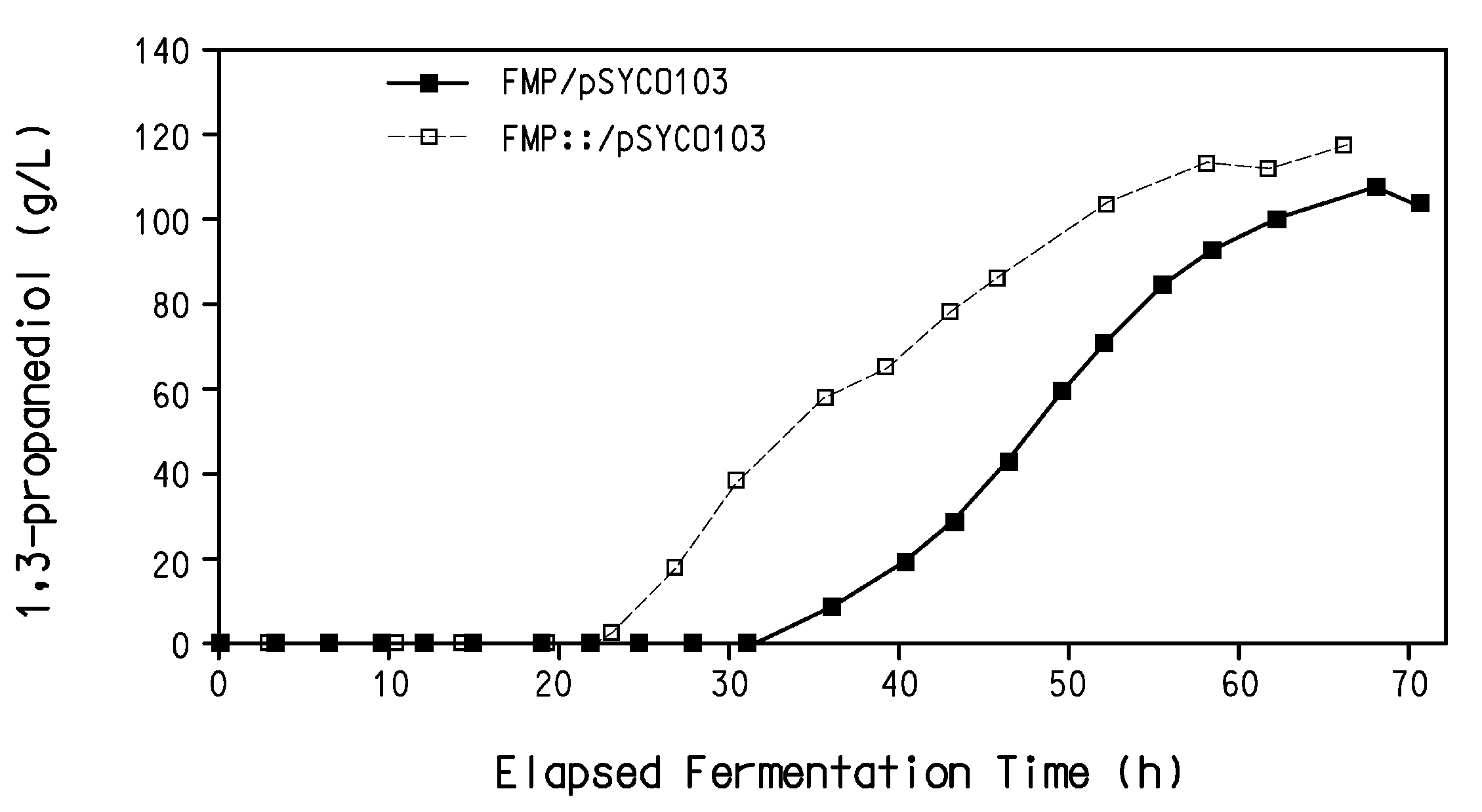
The present invention provides a microorganism useful for biologically producing 1,3-propanediol from a fermentable carbon source at higher yield than was previously known. The complexity of the cofactor requirements necessitates the use of a whole cell catalyst for an industrial process that utilizes this reaction sequence to produce 1,3-propanediol. The invention provides a microorganism with disruptions in specified genes and alterations in the expression levels of specified genes that is useful in a higher yielding process to produce 1,3-propanediol.
Owner:NUTRITION & BIOSCIENCES USA 4 INC +1
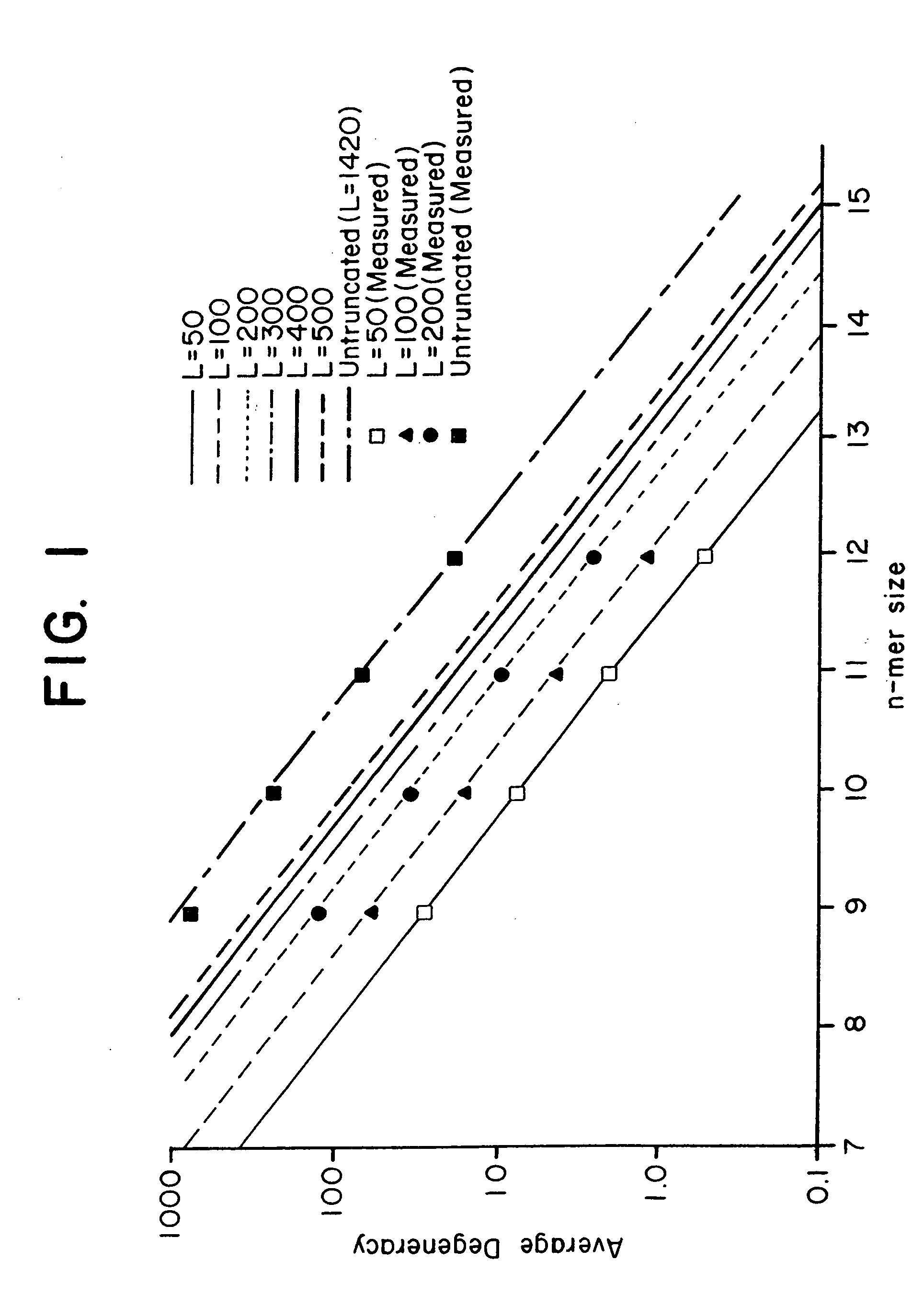
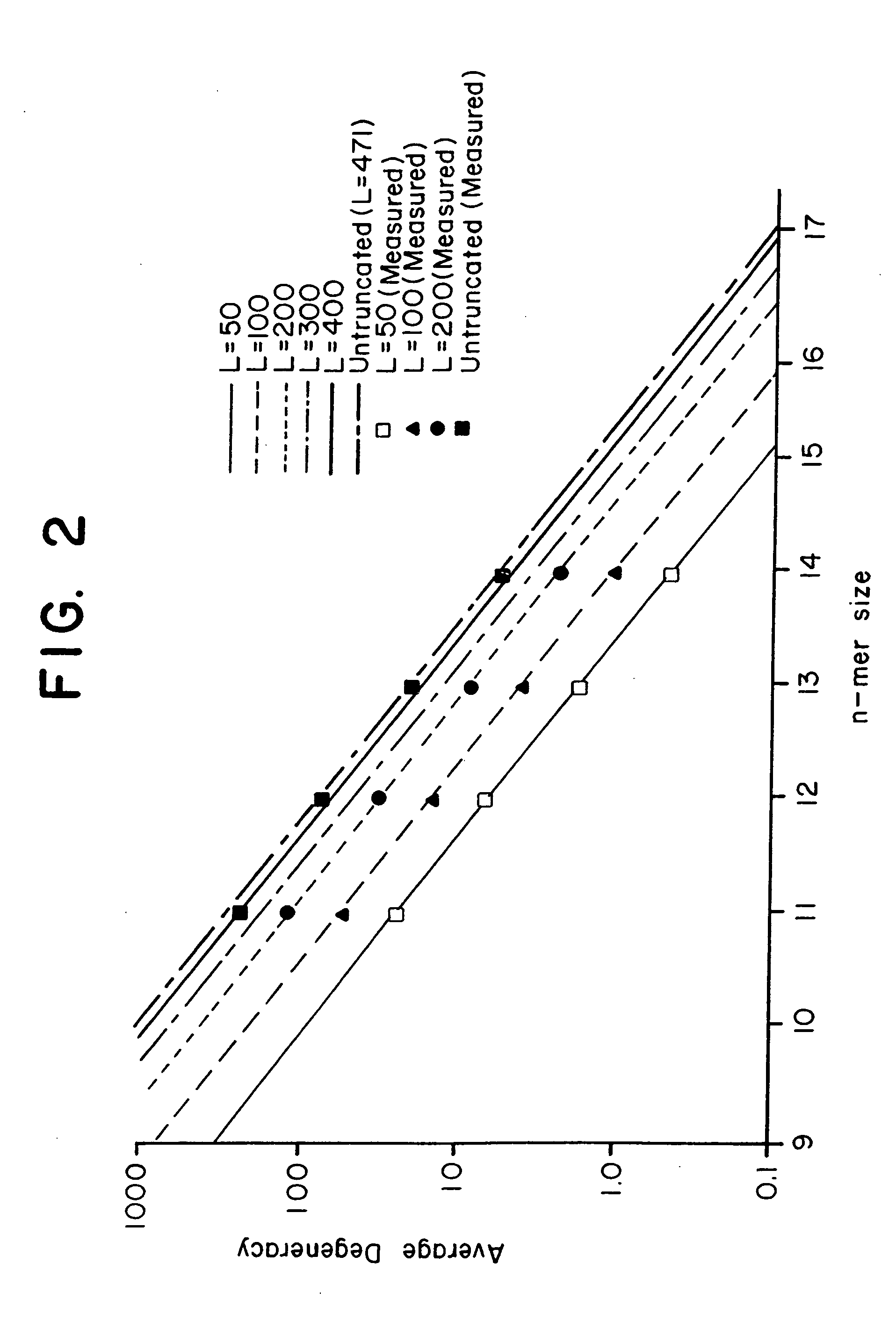
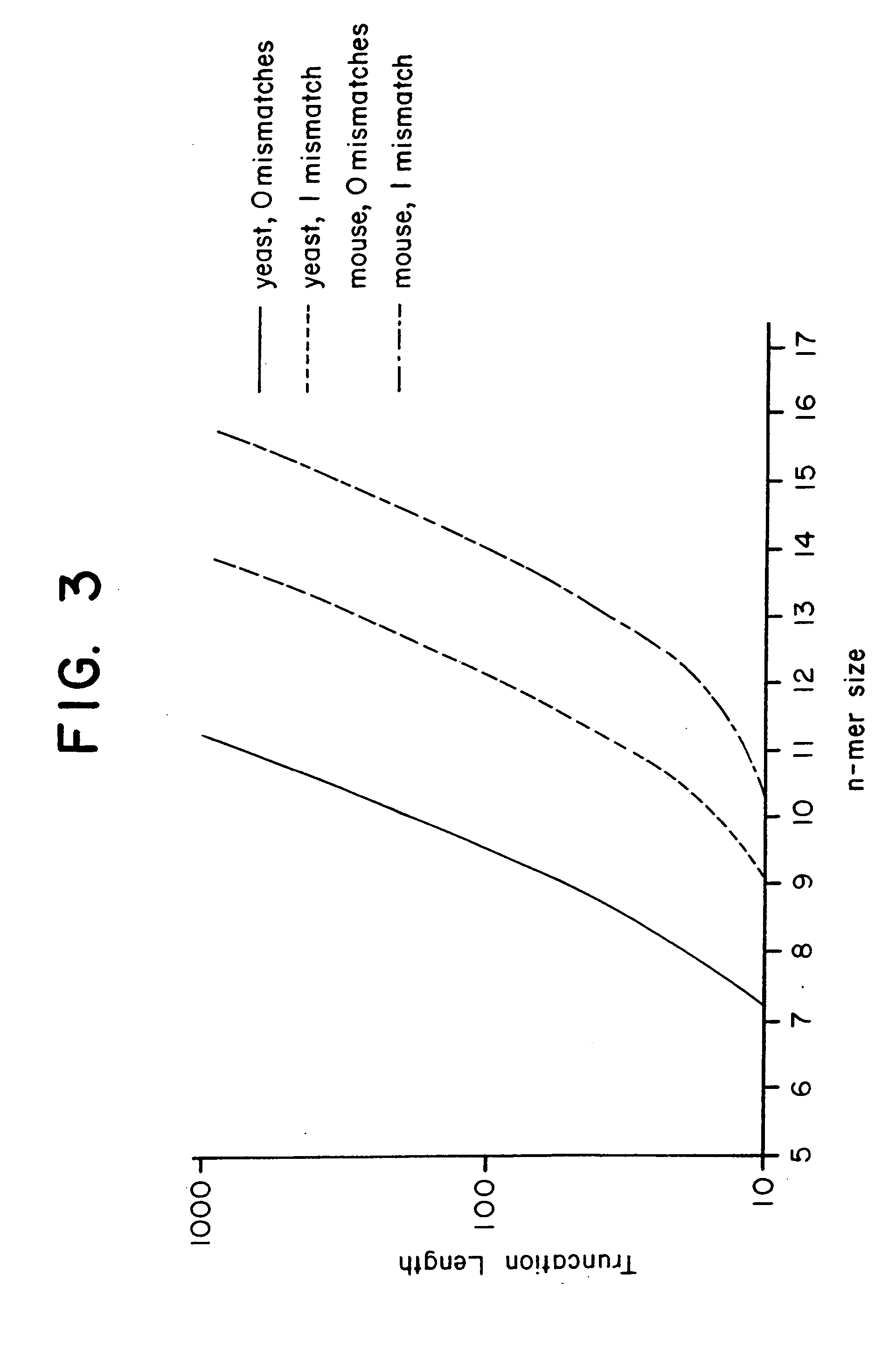
Combinational array for nucleic acid analysis
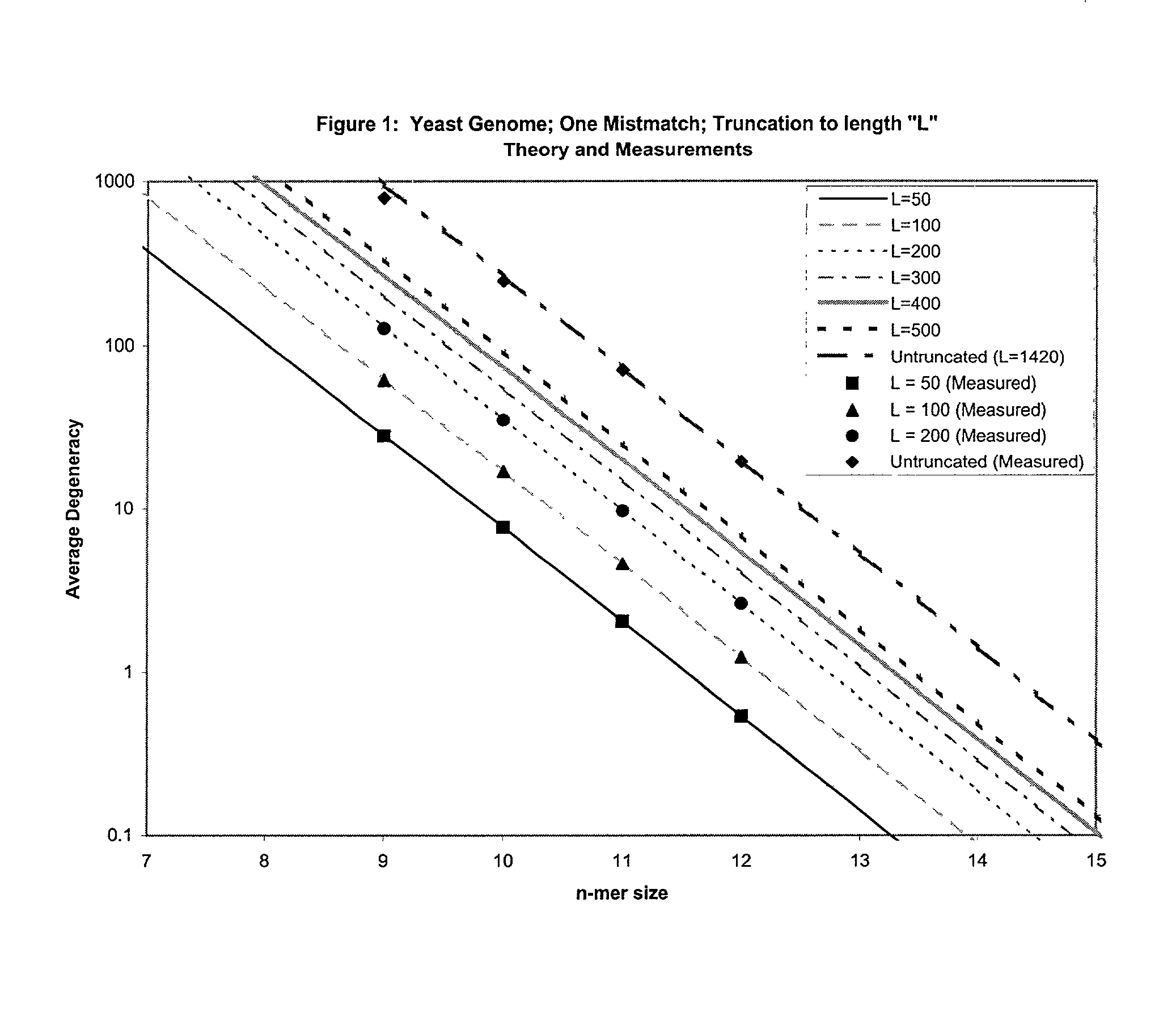
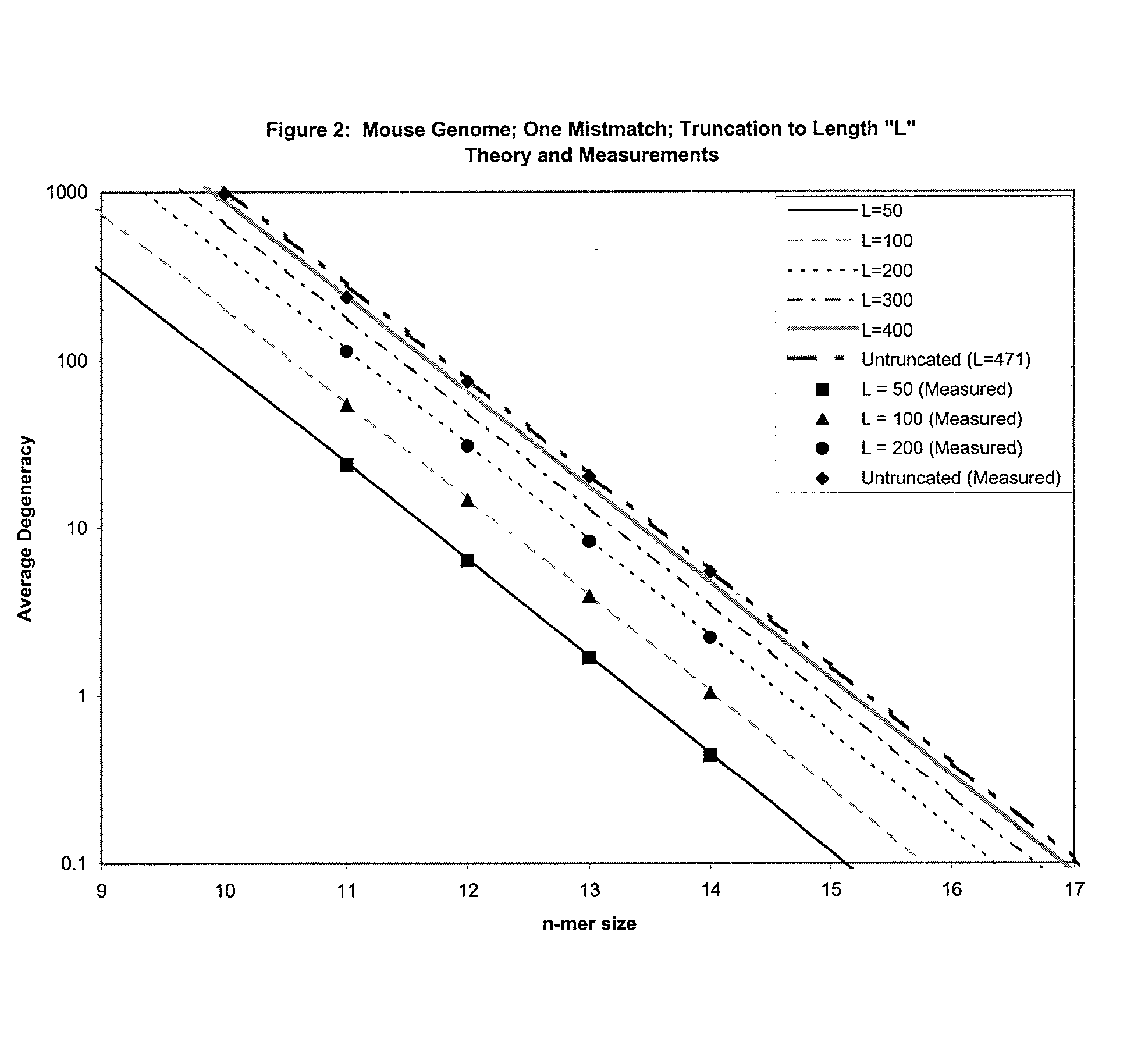
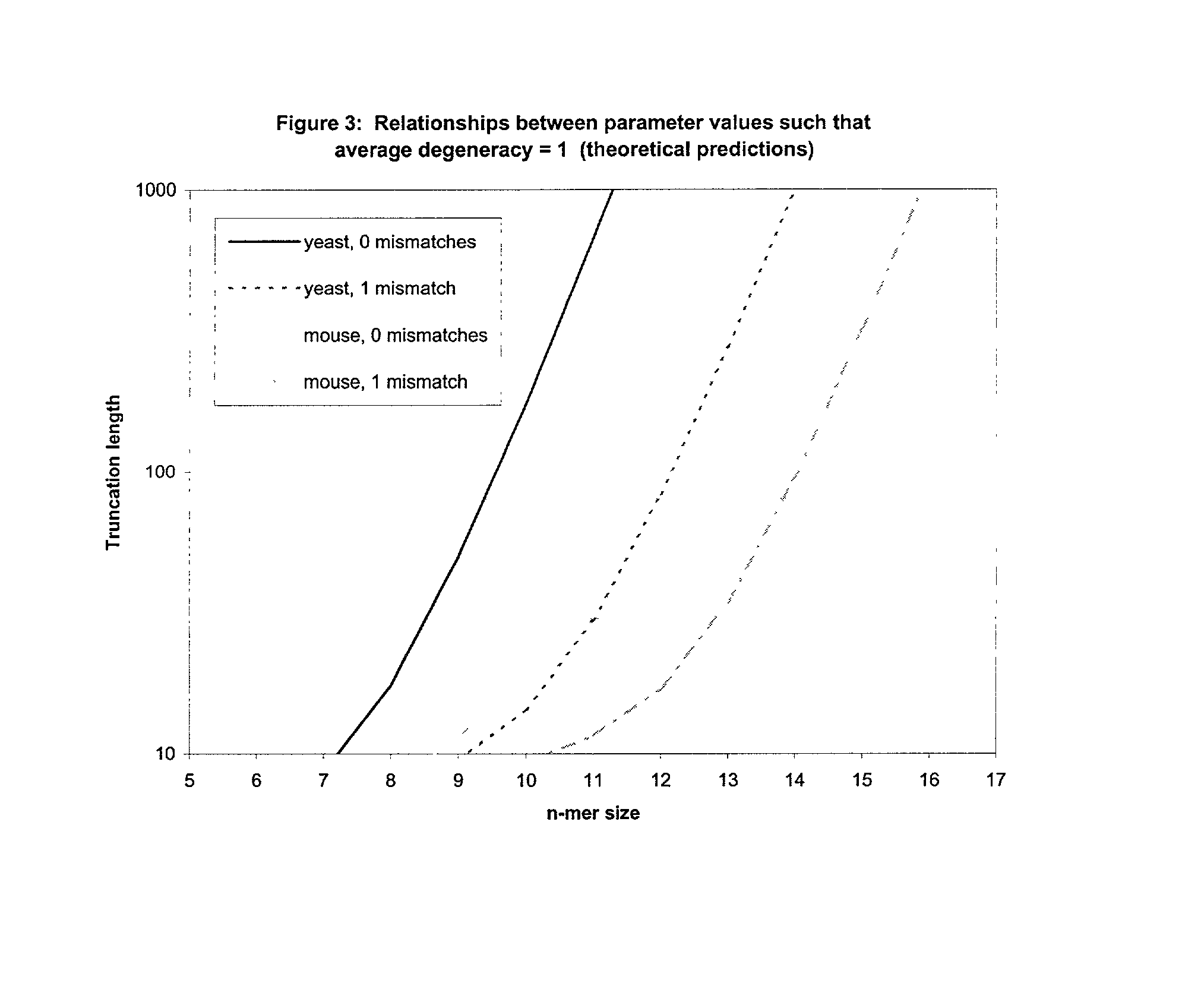
This invention relates to an array, including a universal micro-array, for the analysis of nucleic acids, such as DNA. The devices and methods of the invention can be used for identifying gene expression patterns in any organism. More specifically, all possible oligonucleotides (n-mers) necessary for the identification of gene expression patterns are synthesized. According to the invention, n is large enough to give the specificity to uniquely identify the expression pattern of each gene in an organism of interest, and is small enough that the method and device can be easily and efficiently practiced and made. The invention provides a method of analyzing molecules, such as polynucleotides (e.g., DNA), by measuring the signal of an optically-detectable (e.g., fluorescent, ultraviolet, radioactive or color change) reporter associated with the molecules. In a polynucleotide analysis device according to the invention, levels of gene expression are correlated to a signal from an optically-detectable (e.g. fluorescent) reporter associated with a hybridized polynucleotide. The invention includes an algorithm and method to interpret data derived from a micro-array or other device, including techniques to decode or deconvolve potentially ambiguous signals into unambiguous or reliable gene expression data.
Owner:CALIFORNIA INST OF TECH
Combinatorial array for nucleic acid analysis
InactiveUS20020012926A1Microbiological testing/measurementBiological testingBiological bodyFluorescence
This invention relates to an array, including a universal micro-array, for the analysis of nucleic acids, such as DNA. The devices and methods of the invention can be used for identifying gene expression patterns in any organism. More specifically, all possible oligonucleotides (n-mers) necessary for the identification of gene expression patterns are synthesized. According to the invention, n is large enough to give the specificity to uniquely identify the expression pattern of each gene in an organism of interest, and is small enough that the method and device can be easily and efficiently practiced and made. The invention provides a method of analyzing molecules, such as polynucleotides (e.g., DNA), by measuring the signal of an optically-detectable (e.g., fluorescent, ultraviolet, radioactive or color change) reporter associated with the molecules. In a polynucleotide analysis device according to the invention, levels of gene expression are correlated to a signal from an optically-detectable (e.g. fluorescent) reporter associated with a hybridized polynucleotide. The invention includes an algorithm and method to interpret data derived from a micro-array or other device, including techniques to decode or deconvolve potentially ambiguous signals into unambiguous or reliable gene expression data.
Owner:CALIFORNIA INST OF TECH
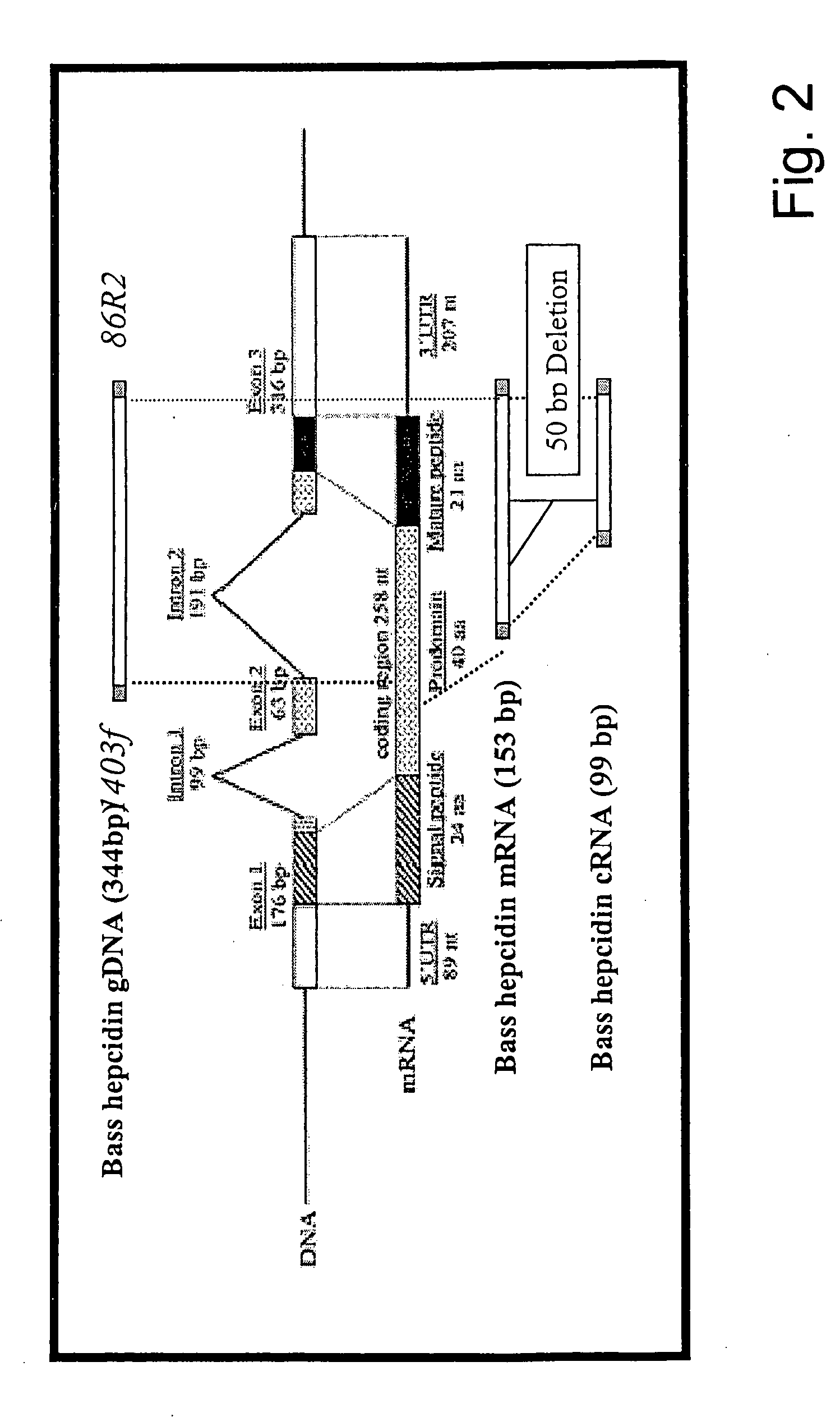
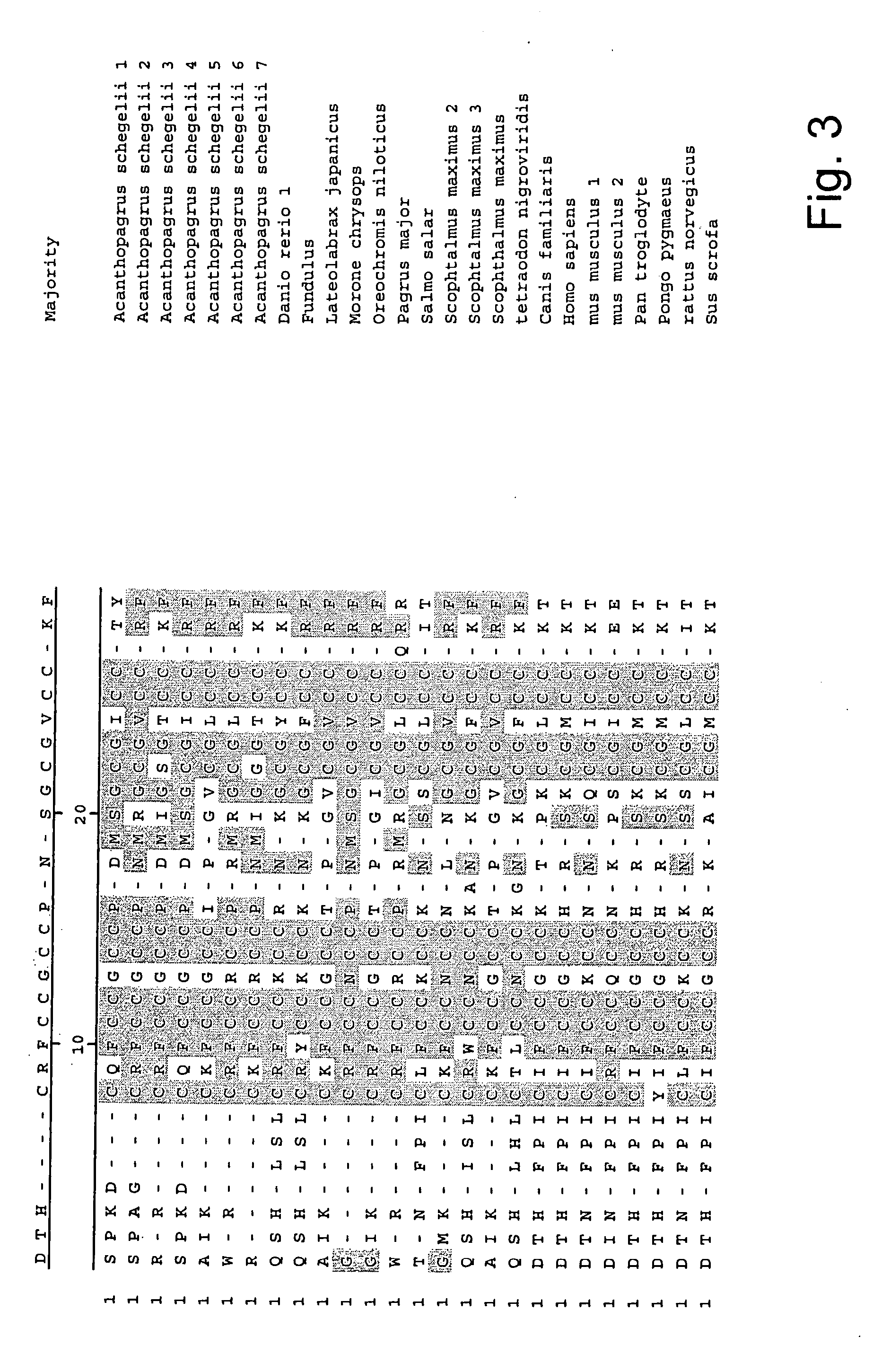
Measurement of bioactive hepcidins
ActiveUS20060019339A1Enhancing innate immunityAntibacterial agentsBiocideMicroorganismVertebrate Animals
The present invention concerns a method for the oxidative refolding of a hepcidin polypeptide to a form that is mature, bioactive and folded as in the native configuration and molecular mass; a method for measuring the level of native, bioactive hepcidin in a vertebrate animal; a method for measuring the level of hepcidin gene expression in a vertebrate animal; and a method for regulating the production of native, bioactive hepcidin in a vertebrate animal in vivo. The present invention also concerns an antibody or fragment thereof that specifically binds to a continuous, discontinuous, and / or conformational epitope of a mature and bioactive hepcidin folded as in the native configuration; and a pharmaceutical composition that includes the antibody or a hepcidin polypeptide and that provides antimicrobial, agonistic, or antagonistic activities in vivo in a vertebrate animal.
Owner:INTRINSIC LIFESCIENCES LLC
Derivatives of fluorene, anthracene, xanthene, dibenzosuberone and acridine and uses thereof
Owner:ALLERGAN SALES LLC
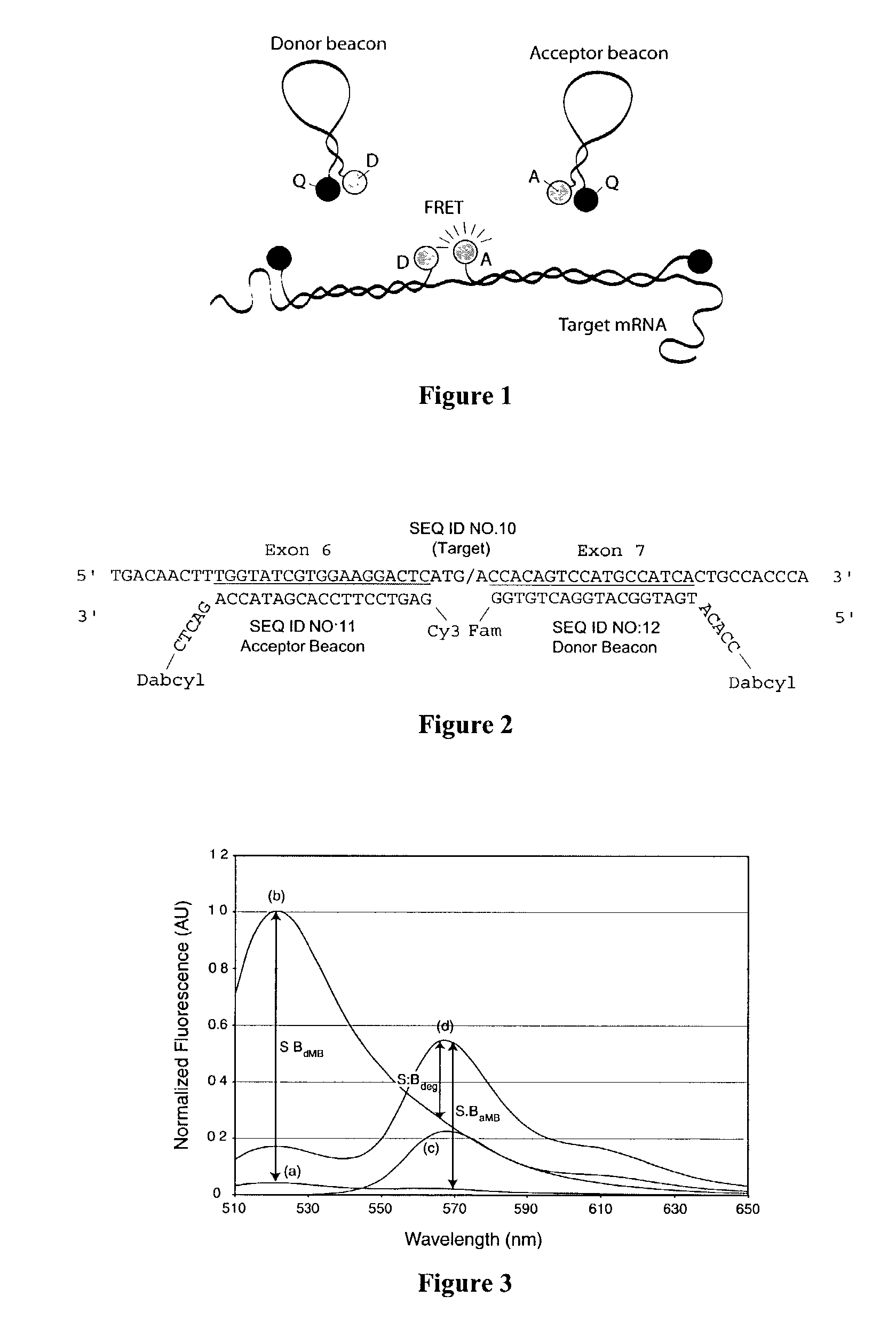
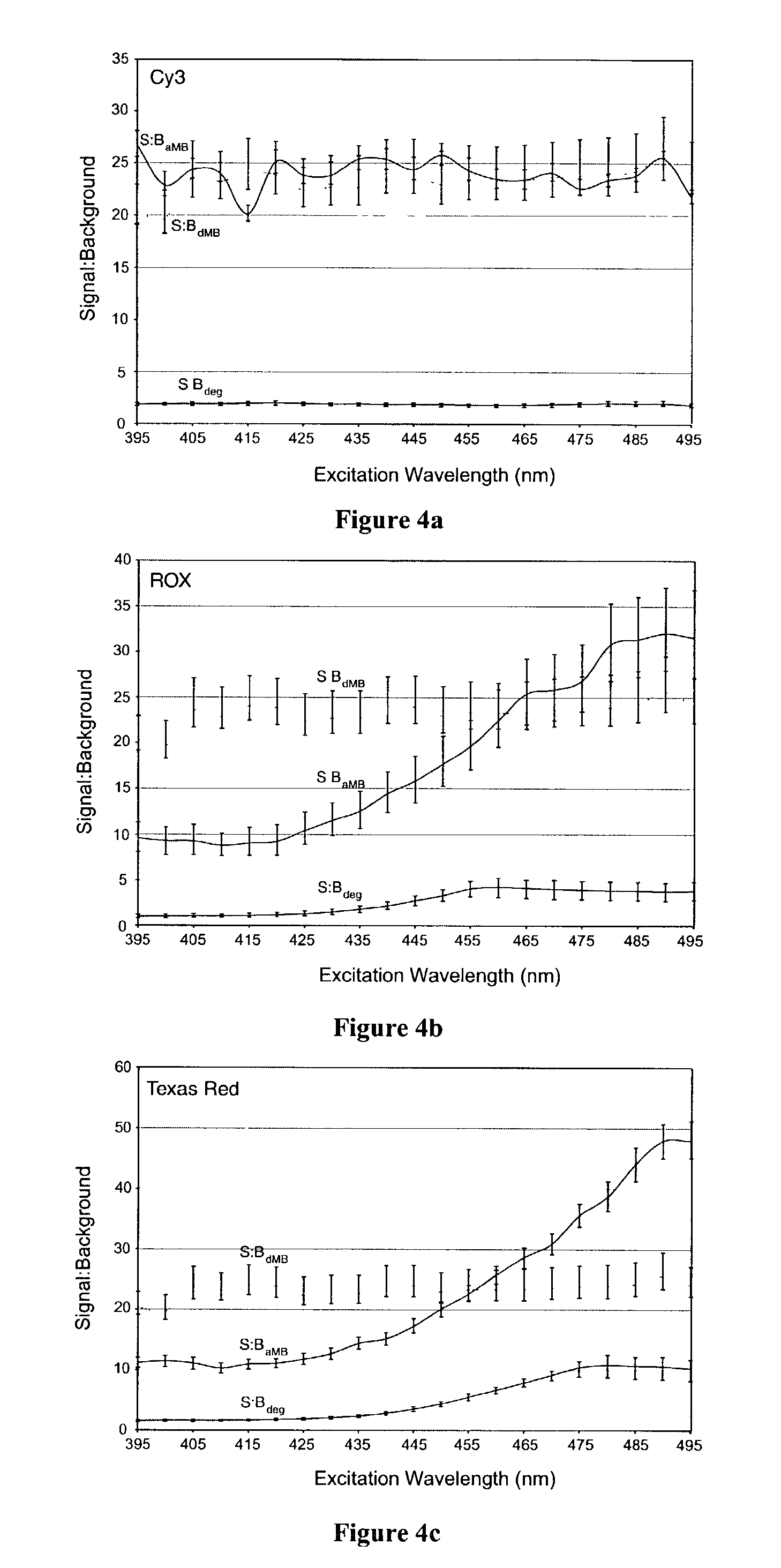
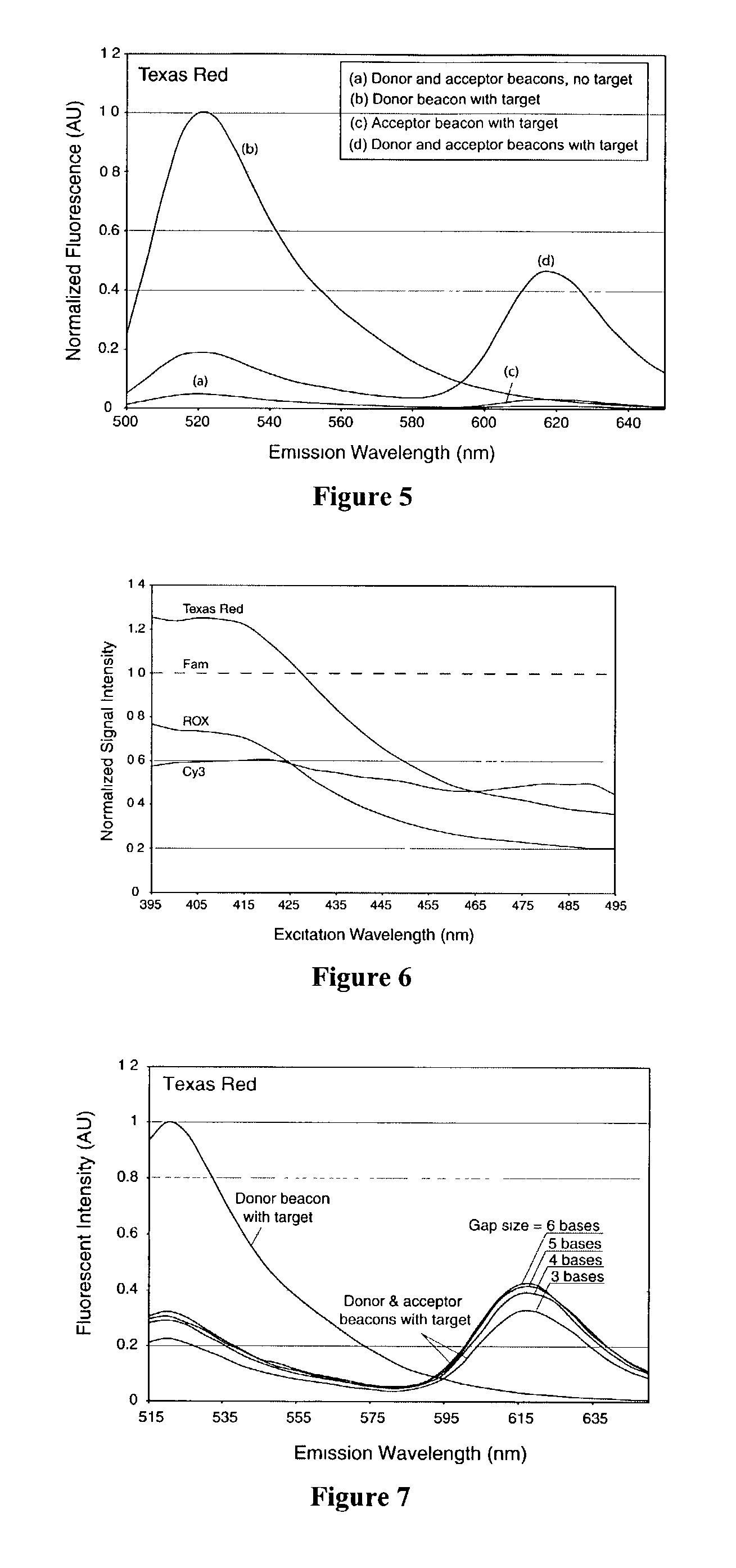
Dual resonance energy transfer nucleic acid probes
InactiveUS7081336B2Quick checkEasy to useSugar derivativesMaterial analysis by observing effect on chemical indicatorDiseaseCell specific
Owner:GEORGIA TECH RES CORP
GENE EXPRESSION ANALYSIS METHOD USING TWO DIMENSIONAL cDNA LIBRARY
InactiveUS20120245053A1Efficient detectionMicrobiological testing/measurementLibrary screeningCDNA libraryGene expression level
The present invention provides a method and / or means for collecting and analyzing an individual cell in a tissue, and at the same time, quantitatively monitoring the expression levels of various genes while keeping two-dimensional information in the tissue. Specifically, the present invention provides a method comprising preparing a cDNA library from mRNA while keeping two-dimensional cellular distribution information and obtaining the gene expression levels at any site or all sites at a level of single cell. More specifically, the present invention provides a method comprising preparing a cDNA library in a sheet-form from mRNA while keeping two-dimensional cellular distribution information and repeatedly using the cDNA library in the detection of the gene expression, thereby allowing measurement of the expression distribution for a number of genes at a high accuracy.
Owner:HITACHI LTD
Compositions of microbiota and methods related thereto
Methods and compositions are generally provided for treating metabolic disorders, e.g., obesity. One aspect discloses methods and compositions for obtaining a biological sample from the subject, evaluating the sample for the presence or absence of a genetic indicator, wherein the genetic indicator is selected from a single nucleotide polymorphism and a level of gene expression, and performing a first metabolic procedure if the genetic indicator is present, or performing an alternative second metabolic procedure if the genetic indicator is absent. One aspect discloses methods and compositions for obtaining a sample including deoxyribonucleic acids (DNA) from the subject, evaluating the DNA for an absence or presence of one or more genetic indicators and performing a first metabolic procedure or an alternative second metabolic procedure based on the absence or presence of the genetic indicator(s). Other aspects are also disclosed.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
Diagnosis and treatment of cancers with microRNA located in or near cancer associated chromosomal features
InactiveUS7723030B2Restore levelOrganic active ingredientsMicrobiological testing/measurementEtiologyMicroRNA Gene
MicroRNA genes are highly associated with chromosomal features involved in the etiology of different cancers. The perturbations in the genomic structure or chromosomal architecture of a cell caused by these cancer-associated chromosomal features can affect the expression of the miR gene(s) located in close proximity to that chromosomal feature. Evaluation of miR gene expression can therefore be used to indicate the presence of a cancer-causing chromosomal lesion in a subject. As the change in miR gene expression level caused by a cancer-associated chromosomal feature may also contribute to cancerigenesis, a given cancer can be treated by restoring the level of miR gene expression to normal. microRNA expression profiling can be used to diagnose cancer and predict whether a particular cancer is associated with an adverse prognosis. The identification of specific mutations associated with genomic regions that harbor miR genes in CLL patients provides a means for diagnosing CLL and possibly other cancers.
Owner:THOMAS JEFFERSON UNIV
Method for the analysis of differential expression in colorectal cancer
InactiveUS20080286801A1Reduced expression levelReduce expressionMicrobiological testing/measurementAntineoplastic agentsGene expression levelGene
A method for the analysis of differential expression in colorectal cancer based on the variation in the expression levels of genes encoding for proteins forming part of the condensin complex or associated proteins that occurs in patients with the disease and that can be used as markers for the diagnosis of the cancers, as well as for the prevention and treatment thereof.
Owner:ORYZON GENOMICS SA
Methods of whole genome or microarray expression profiling using nucleic acids prepared from formalin fixed paraffin embedded tissue
InactiveUS20070254305A1Overcomes shortcomingMicrobiological testing/measurementDNA preparationHigh densityFormalin fixed paraffin embedded
The present invention provides novel methods for analyzing gene expression levels from fresh or aged (more than one year old) formalin-fixed, paraffin-embedded tissue (“FFPET”) samples that comprise pre-hybridizing a labeled nucleic acid sample prepared from the formalin-fixed, paraffin-embedded tissue sample with a first microarray, hybridizing the unbound labeled nucleic acid sample with a second microarray, and detecting the labeled nucleic acid sample bound to the second microarray. The pre-hybridization step results in an increase in the specific gene signals in subsequent hybridizations with high density gene expression arrays. The first microarray used for the pre-hybridization step can be either a new or used microarray. Importantly, from a cost-savings perspective, the inventors determined that when the first microarray used for the pre-hybridization step is a previously used microarray, the results of the subsequent hybridization on a second microarray are nearly identical to the results obtained when the pre-hybridization was carried out using a new or previously unused microarray.
Owner:NSABP FOUND INC
DIAGNOSIS AND TREATMENT OF CANCERS WITH MicroRNA LOCATED IN OR NEAR CANCER ASSOCIATED CHROMOSOMAL FEATURES
InactiveUS20100203544A1Restore levelOrganic active ingredientsMicrobiological testing/measurementEtiologyMicroRNA Gene
MicroRNA genes are highly associated with chromosomal features involved in the etiology of different cancers. The perturbations in the genomic structure or chromosomal architecture of a cell caused by these cancer-associated chromosomal features can affect the expression of the miR gene(s) located in close proximity to that chromosomal feature. Evaluation of miR gene expression can therefore be used to indicate the presence of a cancer-causing chromosomal lesion in a subject. As the change in miR gene expression level caused by a cancer-associated chromosomal feature may also contribute to cancerigenesis, a given cancer can be treated by restoring the level of miR gene expression to normal. microRNA expression profiling can be used to diagnose cancer and predict whether a particular cancer is associated with an adverse prognosis. The identification of specific mutations associated with genomic regions that harbor miR genes in CLL patients provides a means for diagnosing CLL and possibly other cancers.
Owner:THOMAS JEFFERSON UNIV
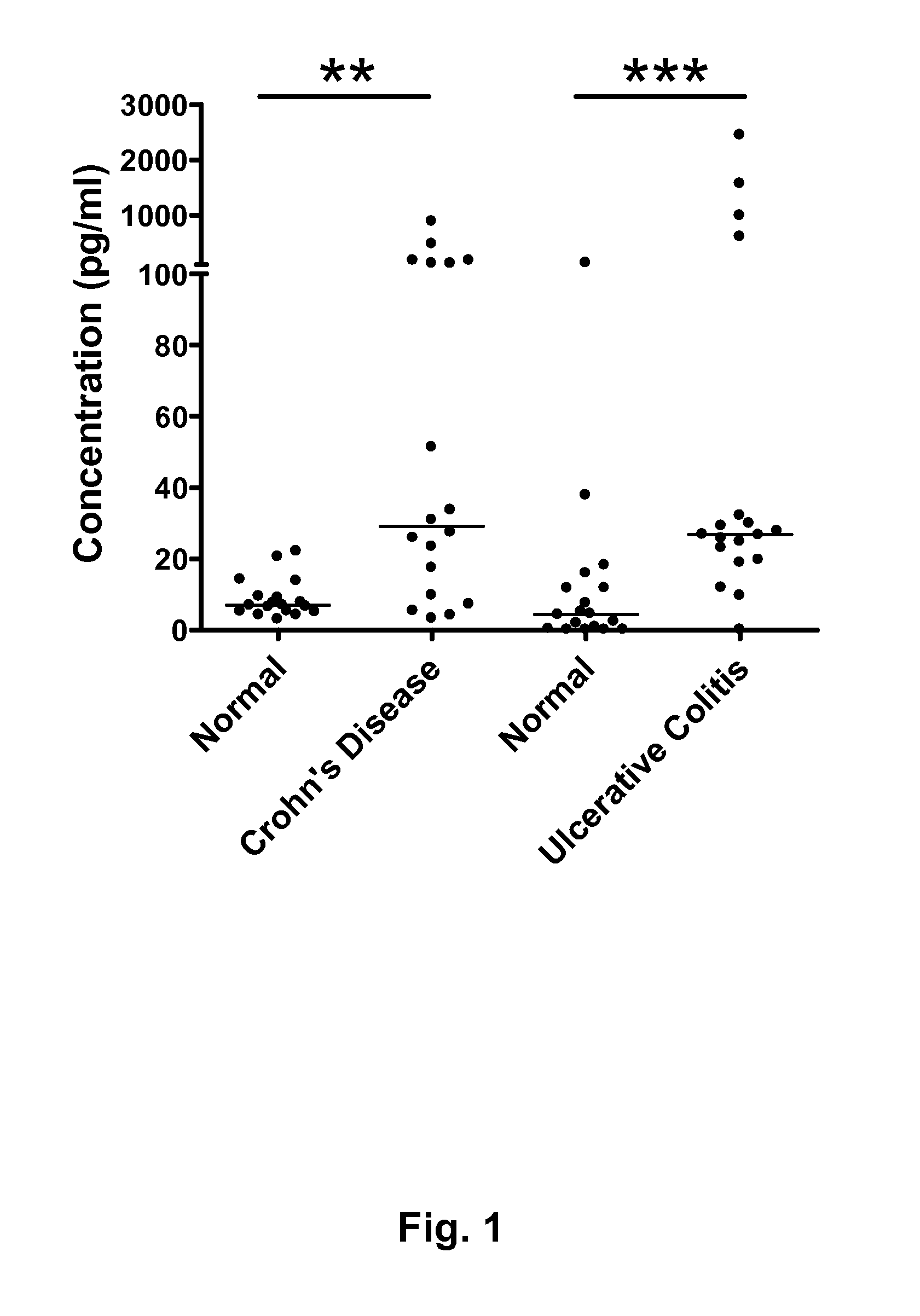
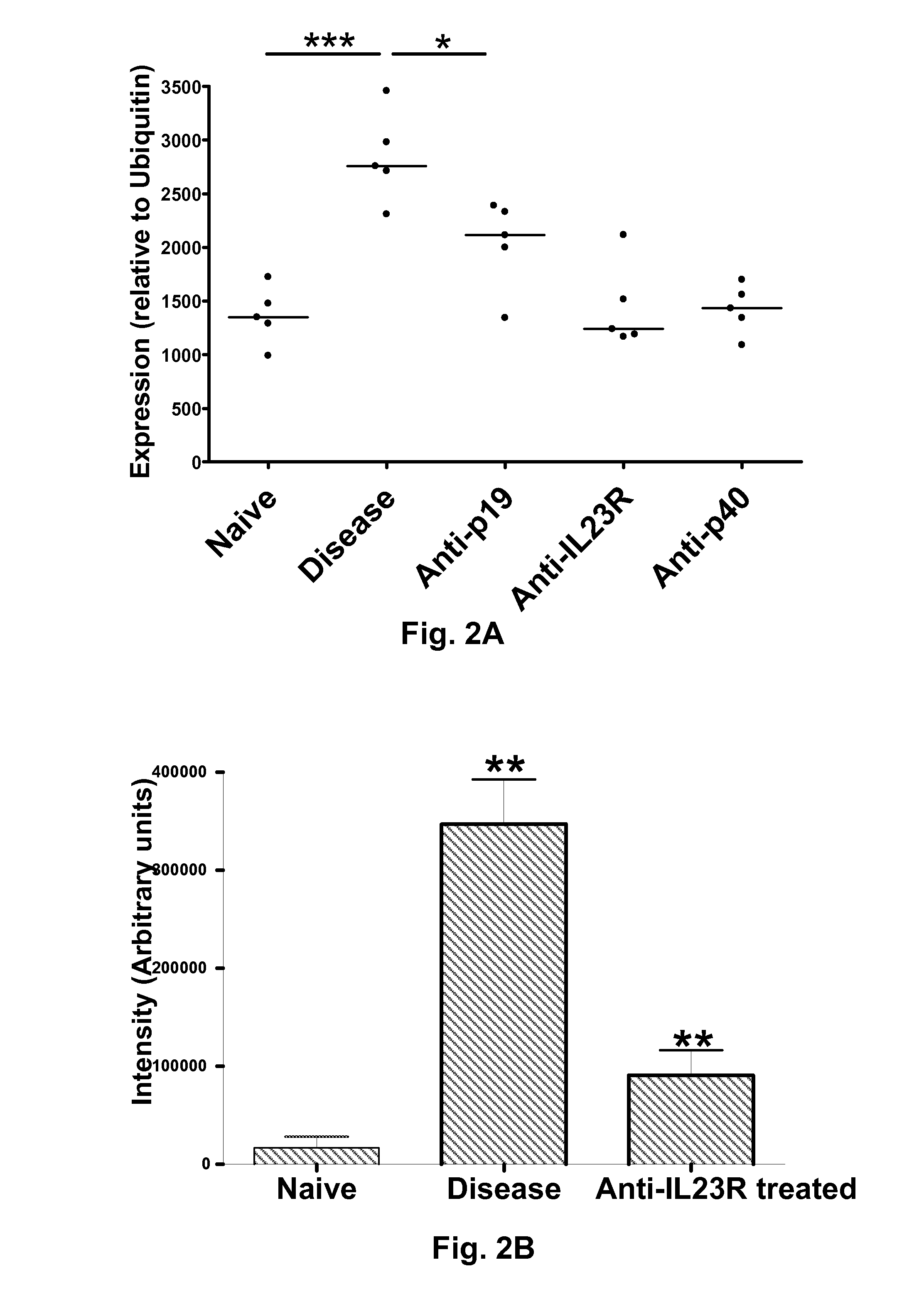
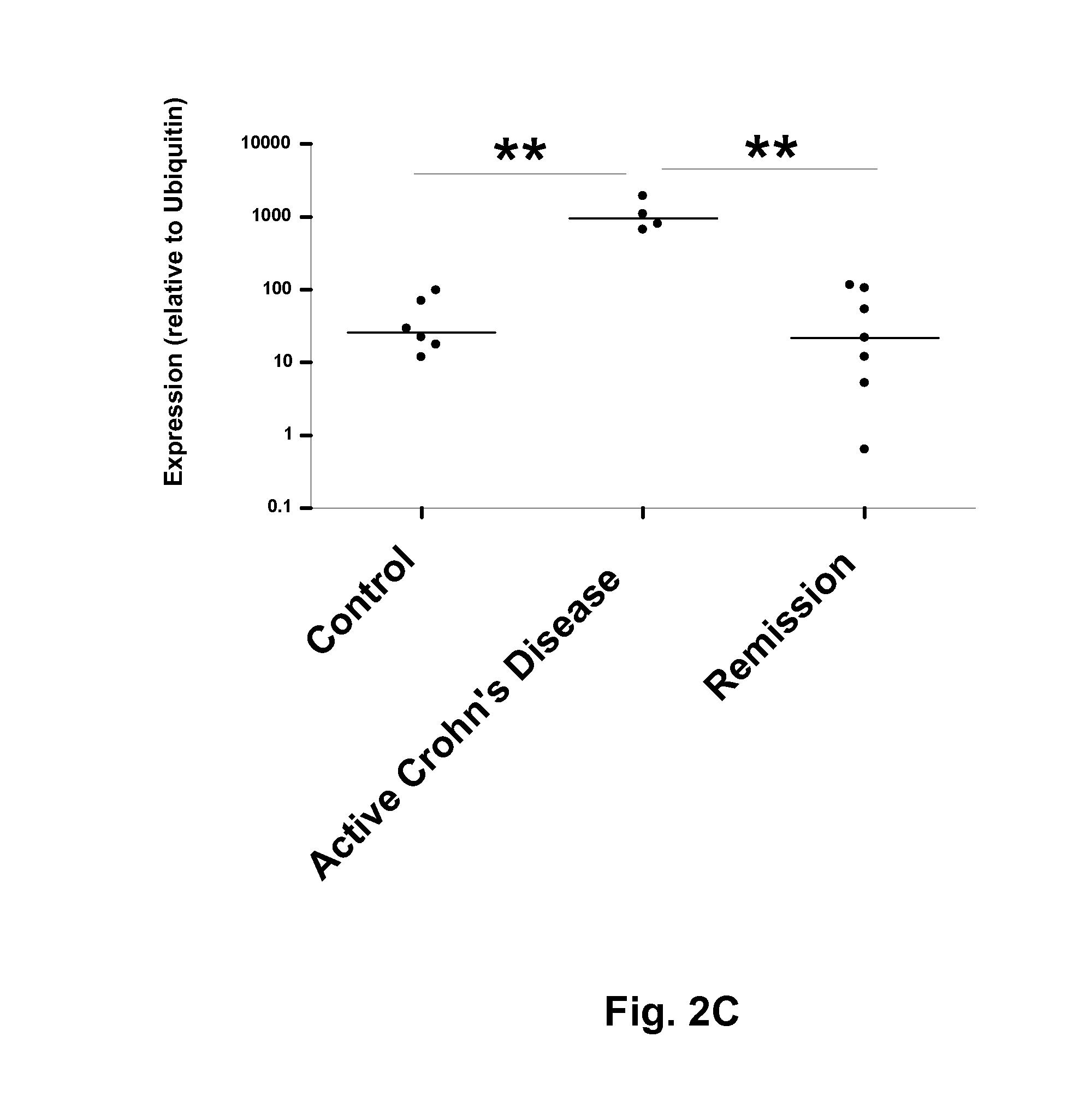
Inflammatory bowel disease biomarkers and related methods of treatment
Biomarkers associated with inflammatory bowel disease (IBD) are provided, as well as methods of using such biomarkers for diagnosing, assessing and monitoring disease progression. The biomarkers may be measured at the protein level or the gene expression level Biomarkers may be tracked individually or in groups of two or more. The disclosed biomarkers may find particular utility in monitoring a course of therapy, such as treatment with an IL-23 antagonist. Changes in biomarker levels can also be used to confirm target engagement and therapeutic efficacy. Changes in biomarkers can also be used inform modification of a therapeutic regimen, for example to increase or decrease dosing of a therapeutic agent, such as an anti-IL-23 or anti-IL-23R anti-body.
Owner:MERCK SHARP & DOHME CORP
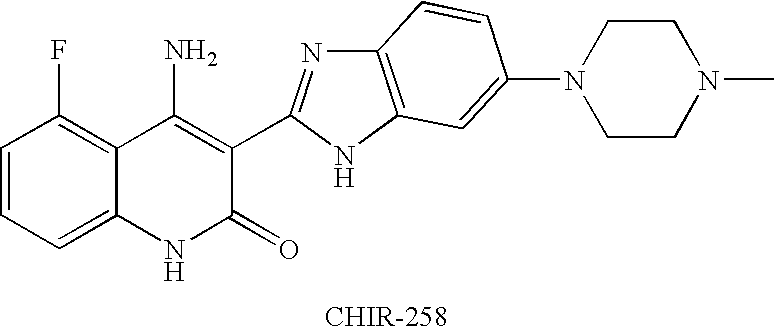
Effects of inhibitors of FGFR3 on gene transcription
InactiveUS8158360B2Improve responseMicrobiological testing/measurementBiological testingMedicineBiomarker identification
Methods of utilizing biomarkers to identify patients for treatment or to monitor response to treatment are taught herein. Alterations in levels of gene expression of the biomarkers, particularly in response to FGFR3 inhibition, are measured and identifications or adjustments may be made accordingly.
Owner:NOVARTIS AG
Expression profiling of tumours
InactiveUS20060265138A1Easy translationImprove throughputMicrobiological testing/measurementBiostatisticsPrimary tumorTissue sample
The present invention relates to methods of profiling tumours and characterisation of the tissue types associated with the tumours. A gene expression profile is obtained from the tissue sample, the genes ranked in order of their relative expression levels and the tissue type identified by comparing the gene ranking obtained with a database of relative gene expression level rankings of different tissue types. This gives a means to identify primary tumours and to determine the identity of a tumour of unknown primary. The invention also provides a method of treatment of a tumour by diagnosis of primary tumours identified by the methods described.
Owner:PETER MACCALLUM CANCER INST
Methods and kits for ascertaining biosafety of an agent
ActiveUS20110287974A1Nucleotide librariesMicrobiological testing/measurementTissue specific geneTissue specific
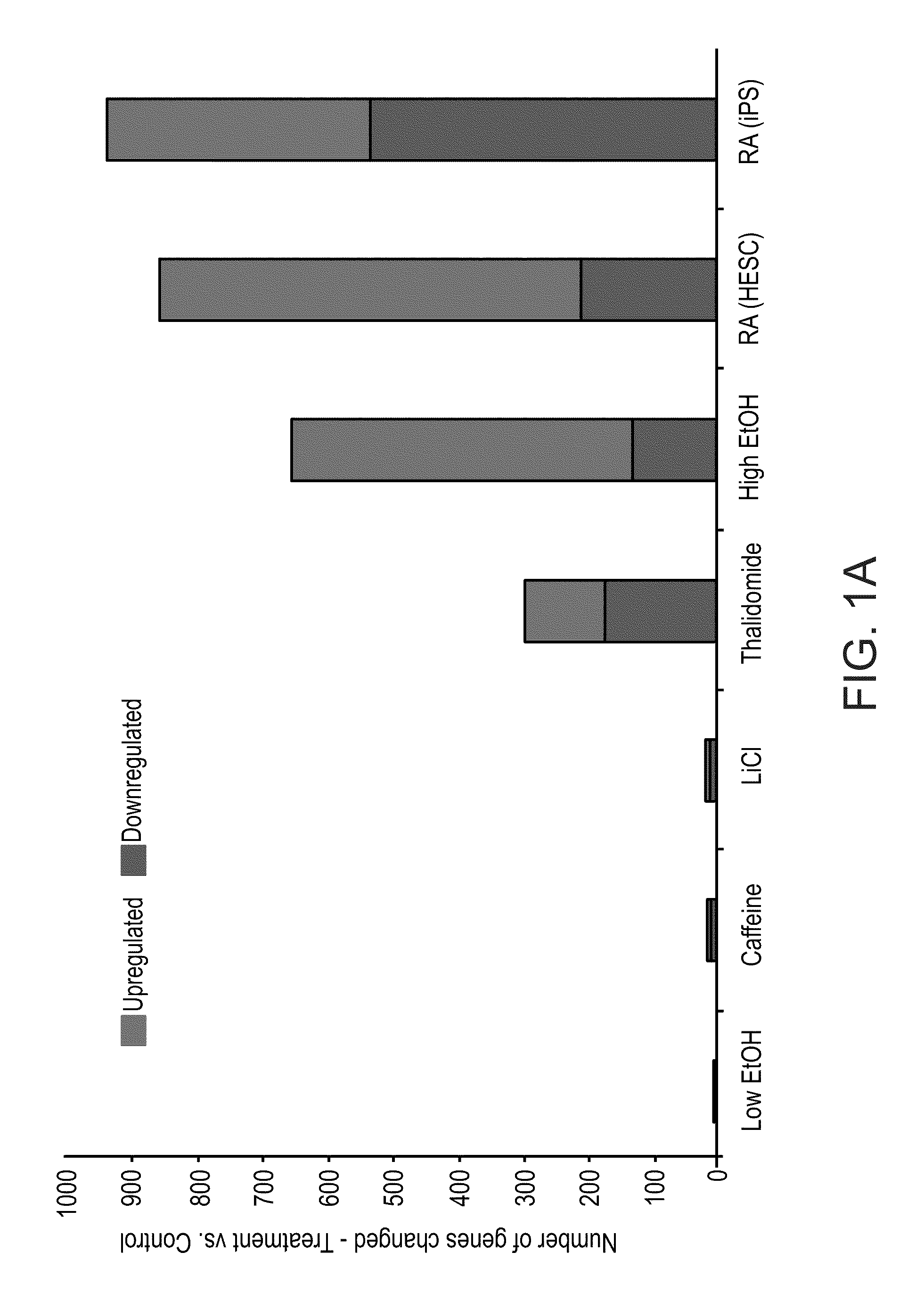
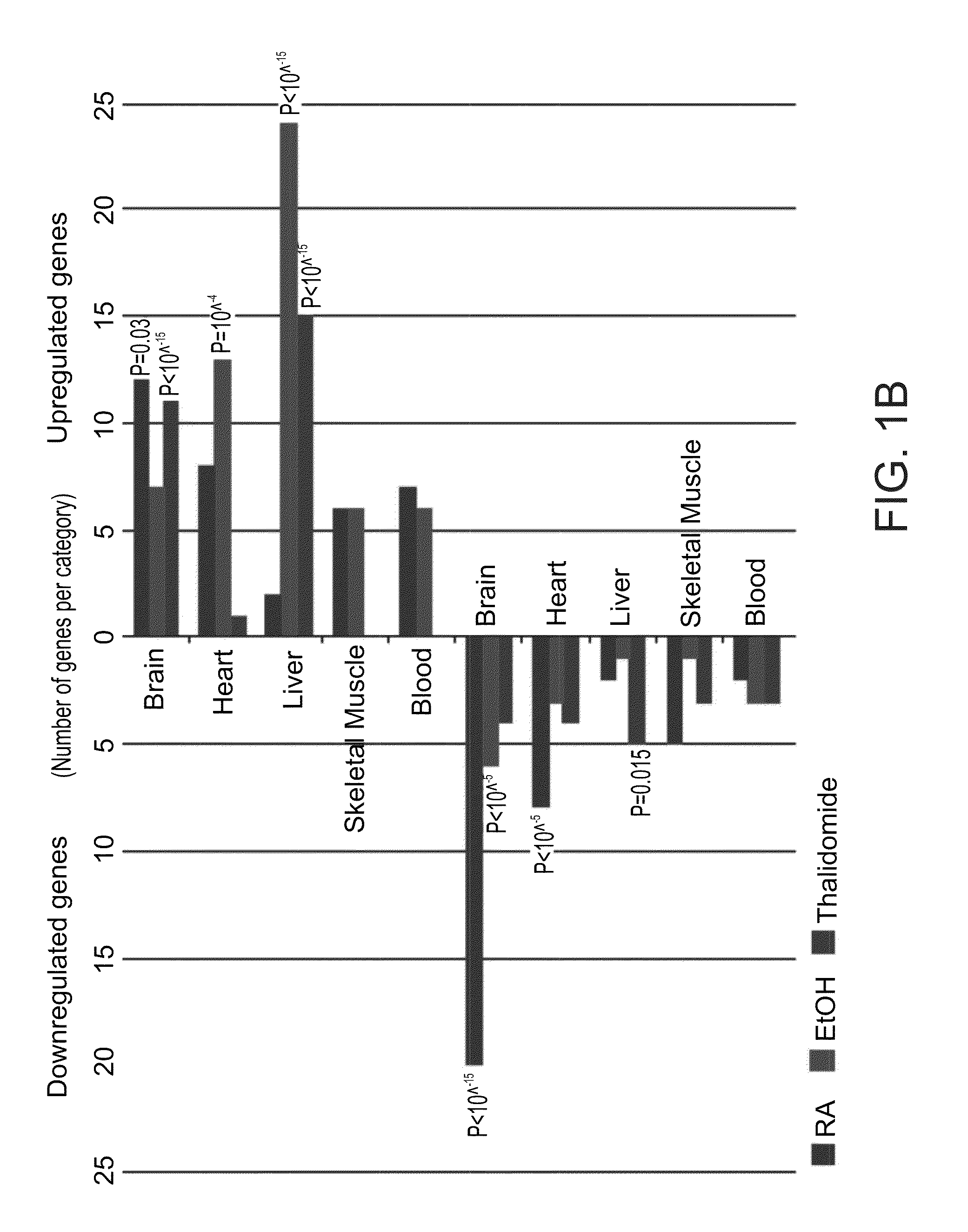
A method of ascertaining the bio-safety of an agent is disclosed. The method comprises:(a) contacting the agent with differentiating human pluripotent stem cells;(b) analyzing a level of gene expression of a plurality of genes in the differentiating human pluripotent stem cells, wherein the agent is qualified as being safe if at least one of the following qualification parameters are fulfilled:(i) the agent causes a difference in the level of gene expression below a predetermined number of genes as compared to control differentiating human pluripotent stem cells that have not been contacted with the agent;(ii) the agent causes a difference in gene expression below a predetermined number of tissue-specific genes of a tissue as compared to control differentiating human pluripotent stem cells that have not been contacted with the agent; or(iii) the agent causes a difference in gene expression below a predetermined number of genes involved in fetal development as compared to control differentiating human pluripotent stem cells that have not been contacted with the agent.
Owner:YISSUM RES DEV CO OF THE HEBREWUNIVERSITY OF JERUSALEM LTD
Fc VARIANTS AND METHODS FOR THEIR PRODUCTION
ActiveUS20140079689A1Reduction in yieldDecreased/elimination of effector functionAnimal cellsSugar derivativesEpitopeHeterologous
Described herein are Fc variants and methods for the efficient production of antibodies and other multimeric protein complexes (collectively referred to herein as heteromultimeric proteins). Heteromultimeric proteins may be capable of specifically binding to more than one target. The targets may be, for example, different epitopes on a single molecule or located on different molecules. The methods combine efficient, high gene expression level, appropriate assembly, and ease of purification for the heteromultimeric proteins. The invention also provides methods of using these heteromultimeric proteins, and compositions, kits and articles of manufacture comprising these antibodies.
Owner:GENENTECH INC
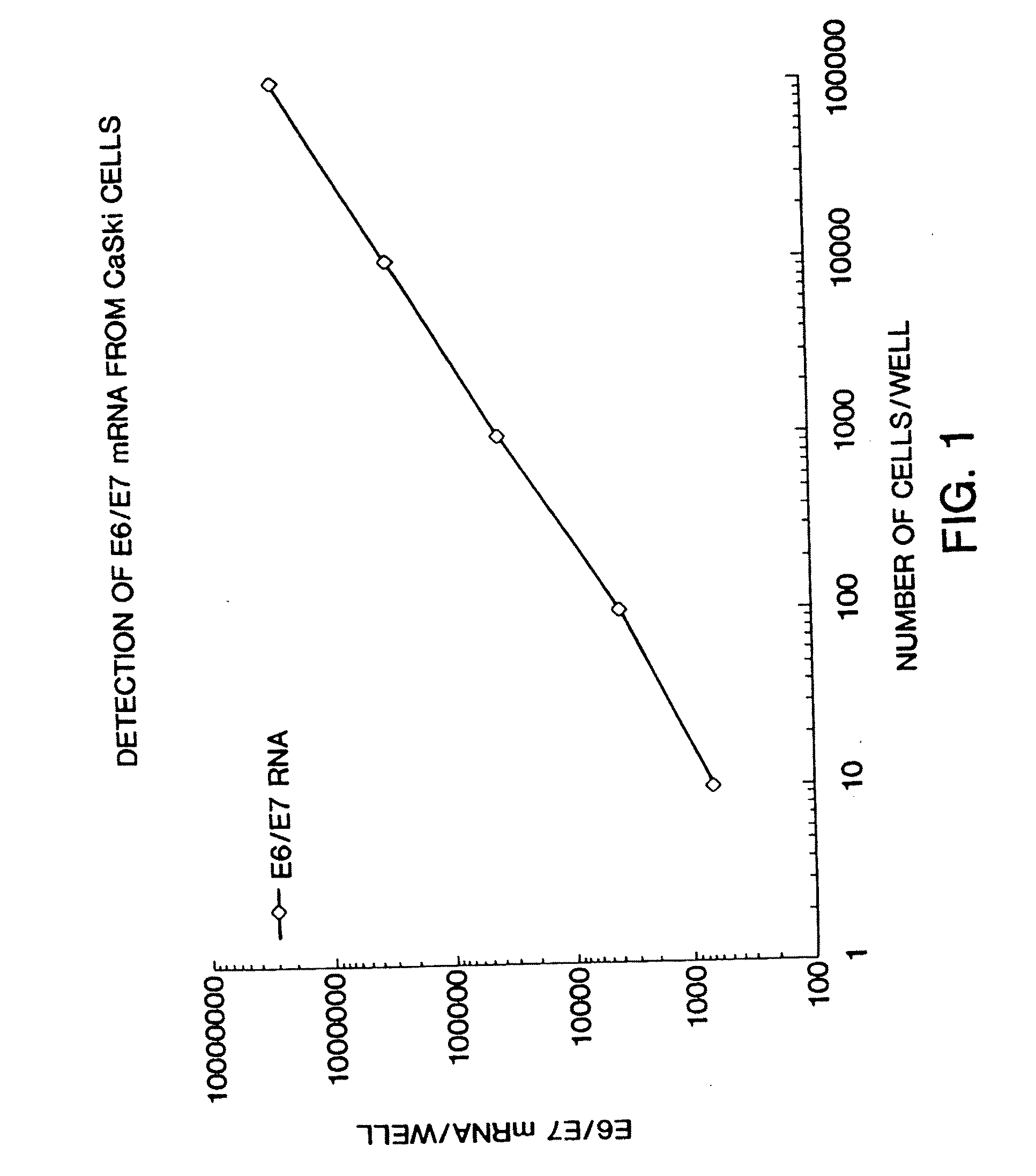
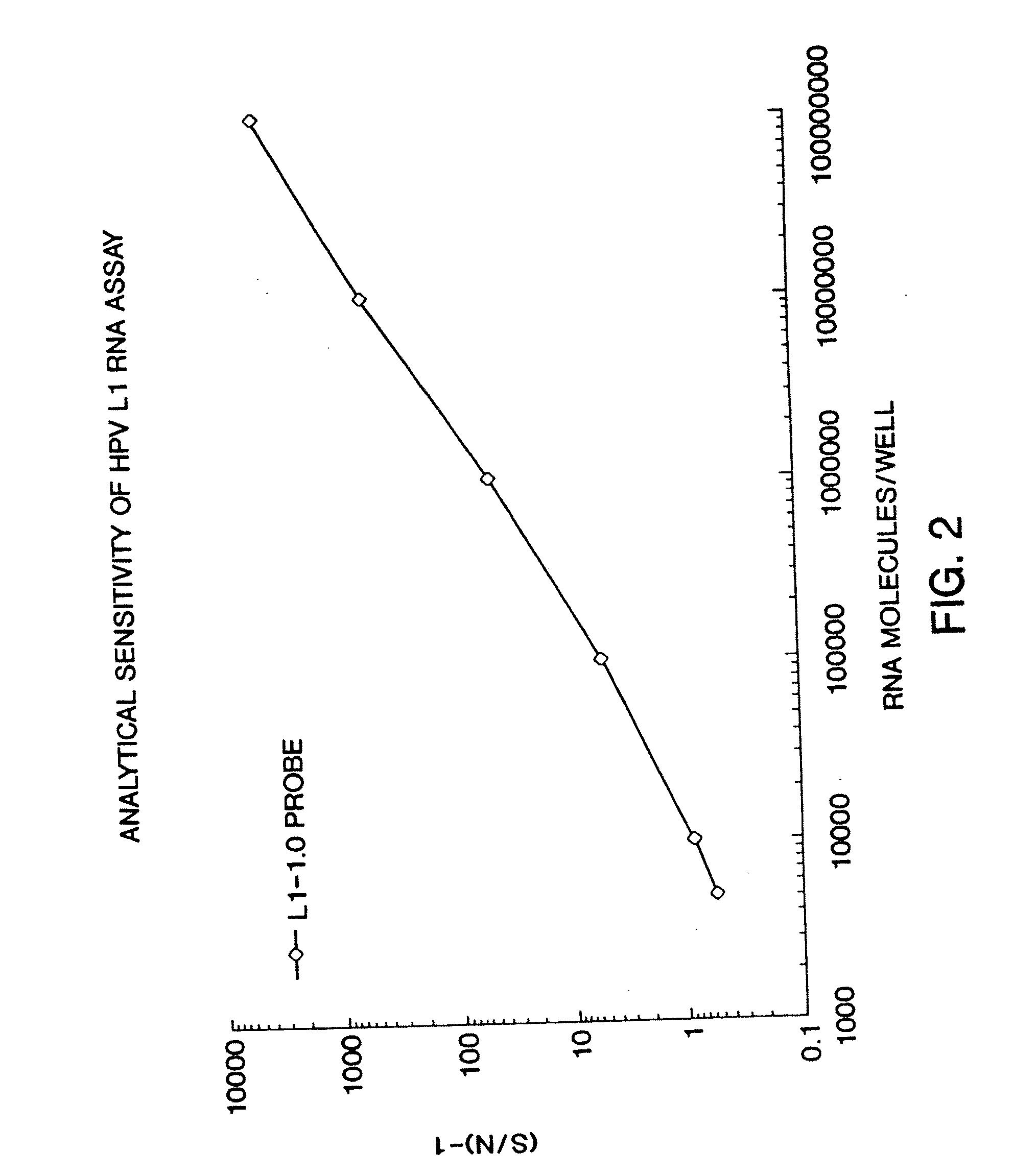
Assessment of human papilloma virus-related disease
InactiveUS20070154884A1Bioreactor/fermenter combinationsBiological substance pretreatmentsMalignant GrowthGene expression level
This invention provides novel methods for assessing HPV infection. Gene expression levels are used to assess the progression of HPV infection from benign to malignant growth. Also provided are kits for carrying out the methods of this invention.
Owner:QIAGEN GAITHERSBURG
Production of heteromultimeric proteins
ActiveUS9637557B2Reduce yieldHigh levelHybrid immunoglobulinsAntipyreticEpitopeAntiendomysial antibodies
Described herein are methods for the efficient production of antibodies and other multimeric protein complexes (collectively referred to herein as heteromultimeric proteins) capable of specifically binding to more than one target. The targets may be, for example, different epitopes on a single molecule or located on different molecules. The methods combine efficient, high gene expression level, appropriate assembly, and ease of purification for the heteromultimeric proteins. The invention also provides methods of using these heteromultimeric proteins, and compositions, kits and articles of manufacture comprising these antibodies.
Owner:F HOFFMANN LA ROCHE & CO AG
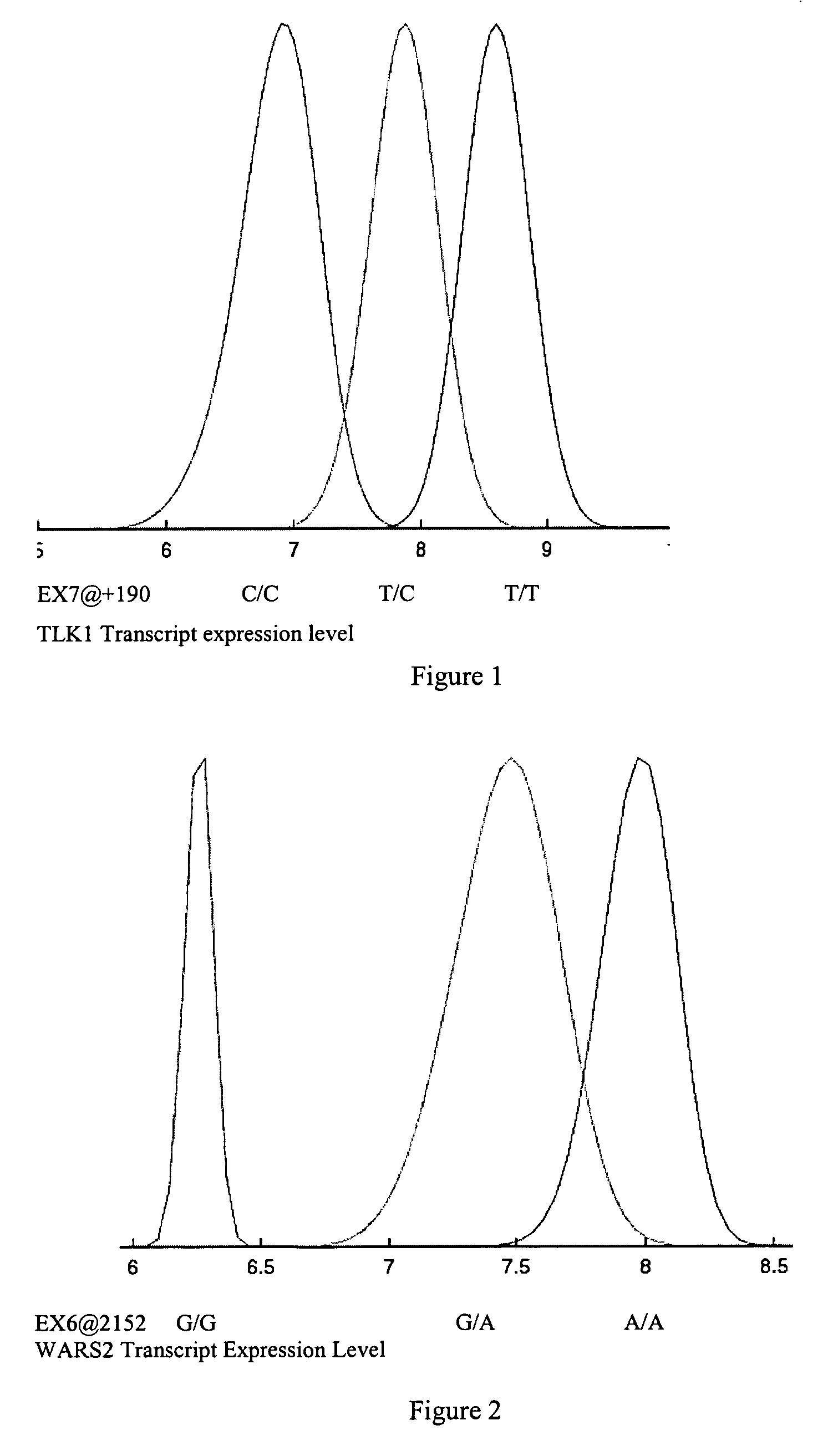
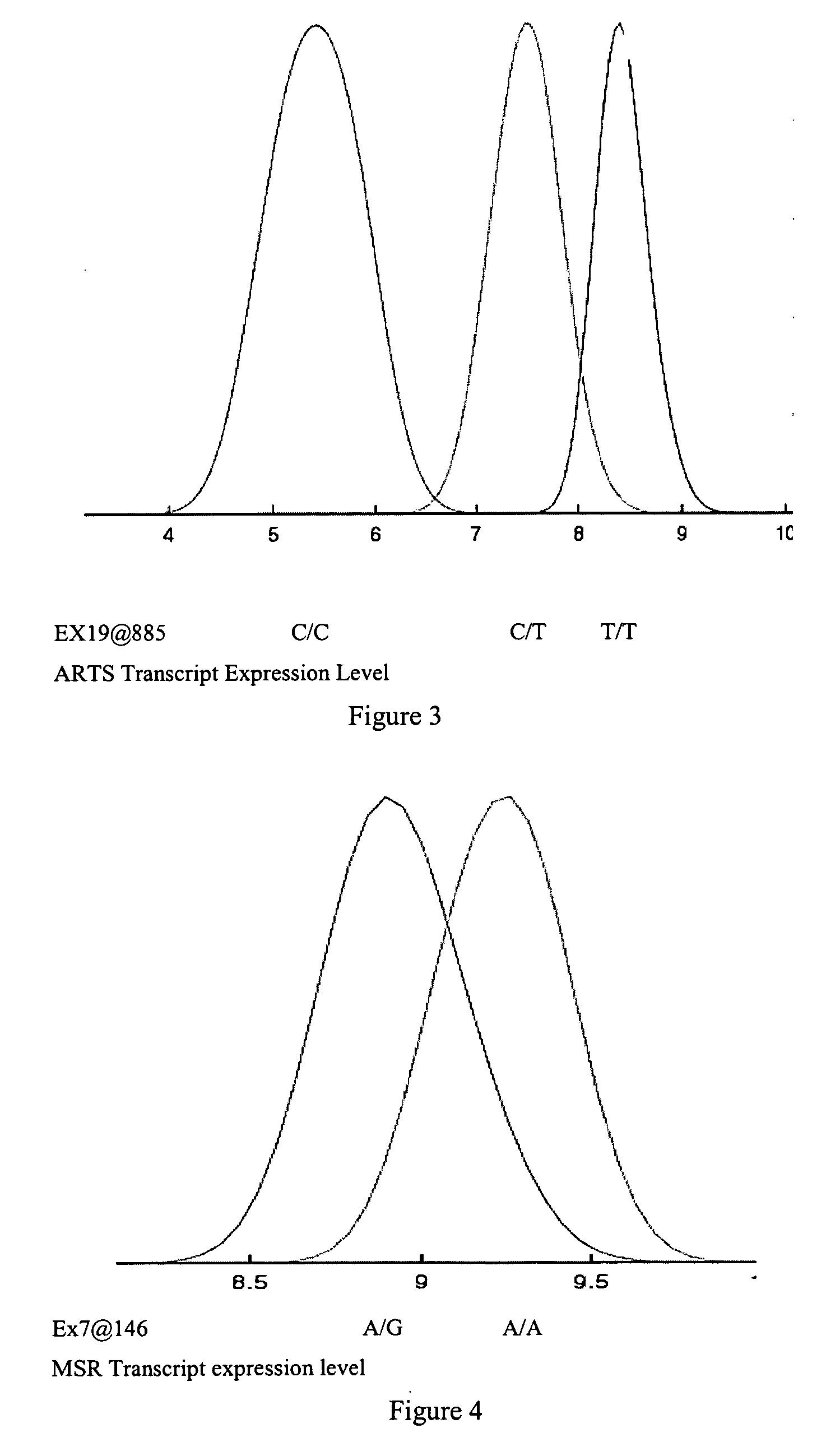
Gene variants and use thereof
InactiveUS20070082347A1Useful predictionBioreactor/fermenter combinationsBiological substance pretreatmentsFKBP1ATAP2
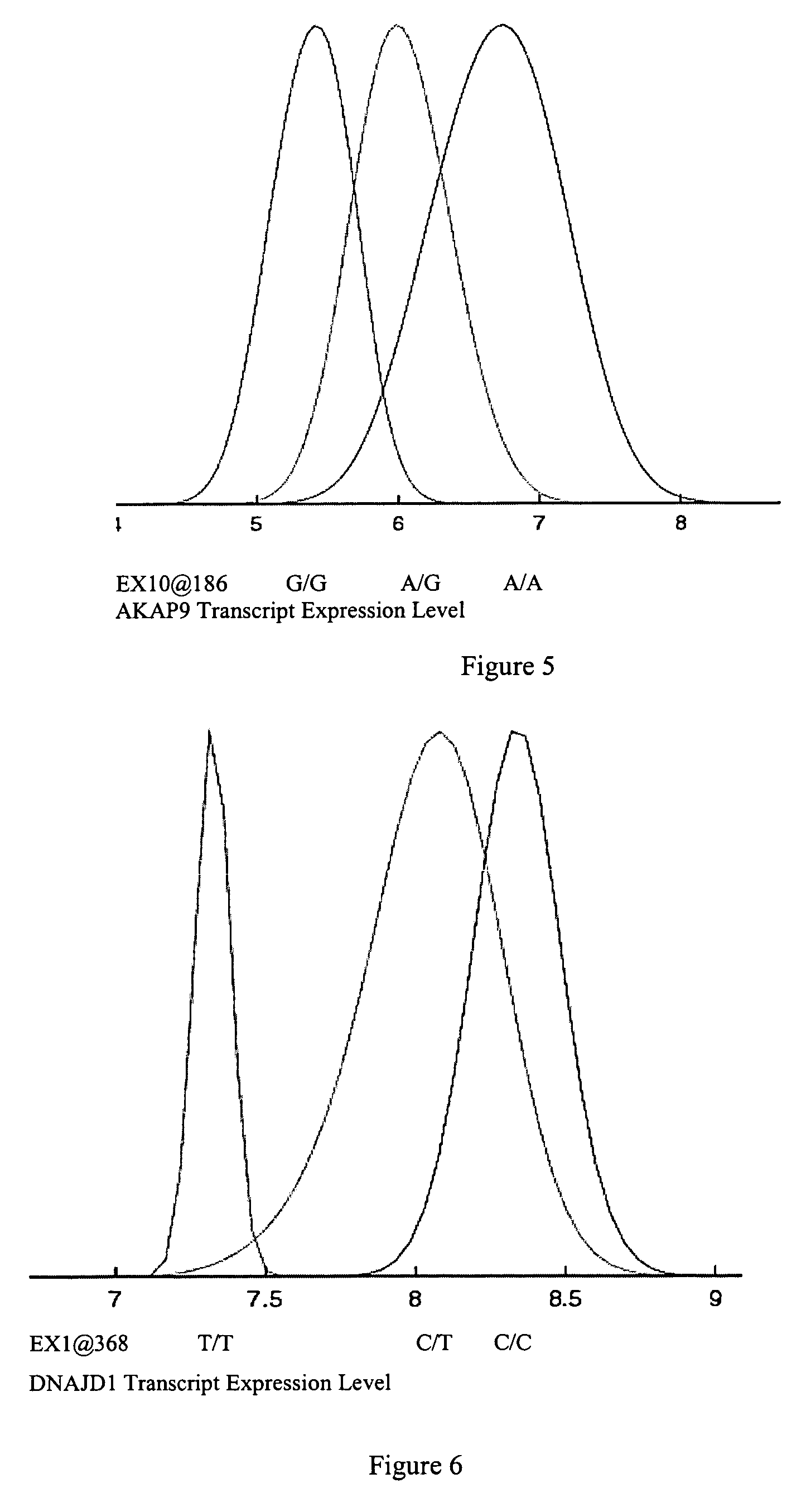
Variants in TLK1, WARS2, ARTS2, MSR, AKAP9, DNAJD1, GOLPH4, RABEP1, TAP2, NARG2, DDX58, CD39, FKBP1a, SRI, XRRA1, IRF5 and AMFR genes are disclosed which are useful as biomarkers for predicting the TLK1, WARS2, ARTS2, MSR, AKAP9, DNAJD1, GOLPH4, RABEP1, TAP2, NARG2, DDX58, CD39, FKBP1a, SRI, XRRA1, IRF5 or AMFR gene expression level and the biological functions associated thereof.
Owner:MYRIAD GENETICS
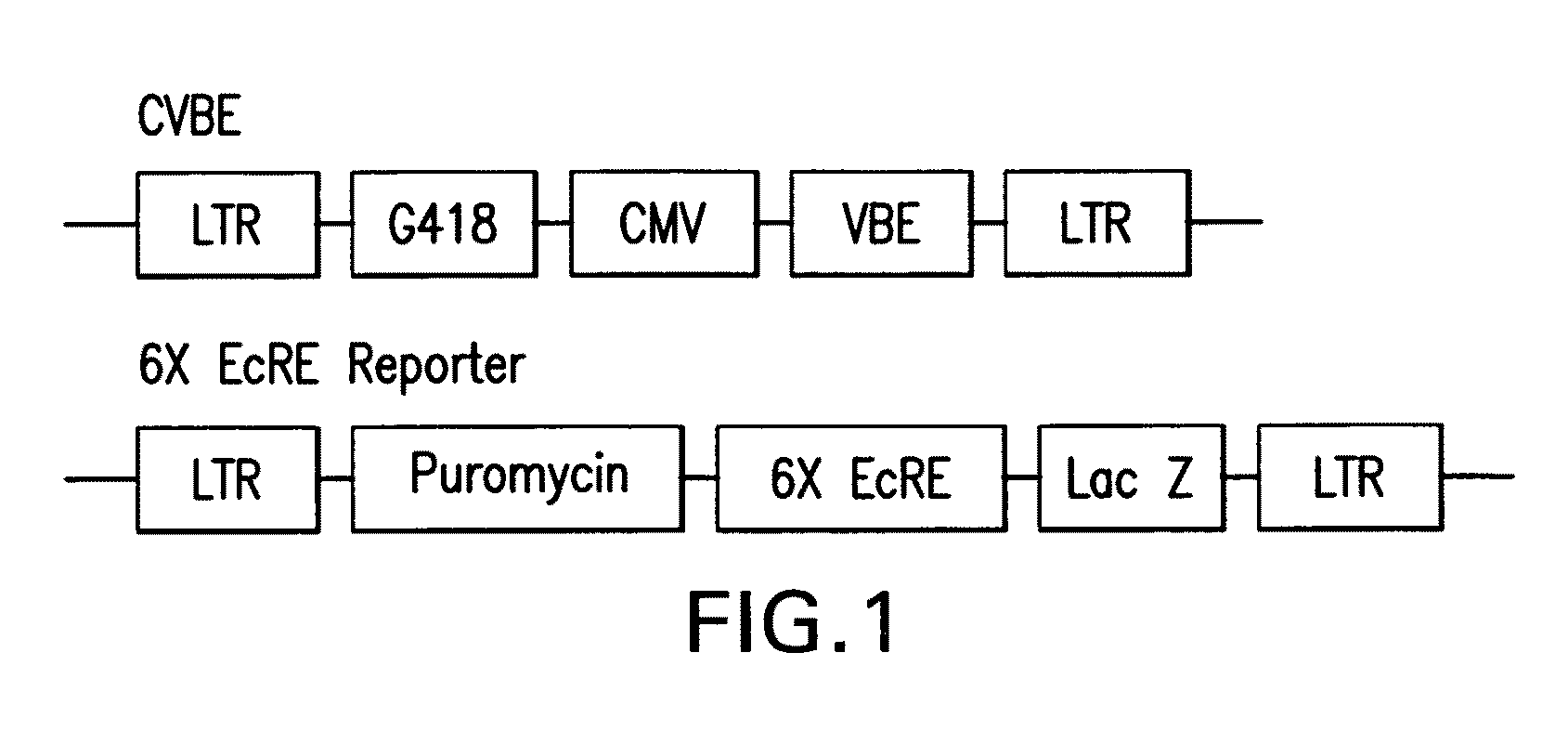
Chiral diacylhydrazine ligands for modulating the expression of exogenous genes via an ecdysone receptor complex
ActiveUS8076517B2Inhibit expressionUrea derivatives preparationOrganic active ingredientsGene expression levelOrganism
The present invention provides diacylhydrazine ligands and chiral diacylhydrazine ligands for use with ecdysone receptor-based inducible gene expression systems. Thus, the present invention is useful for applications such as gene therapy, large scale production of proteins and antibodies, cell-based screening assays, functional genomics, proteomics, metabolomics, and regulation of traits in transgenic organisms, where control of gene expression levels is desirable. An advantage of the present invention is that it provides a means to regulate gene expression and to tailor expression levels to suit the user's requirements.
Owner:PRECIGEN INC
Compositions and Methods for Differential Diagnosis of Chronic Lymphocytic Leukemia
InactiveUS20080280297A1Microbiological testing/measurementImmunoglobulinsReference genesLymphocytic cell
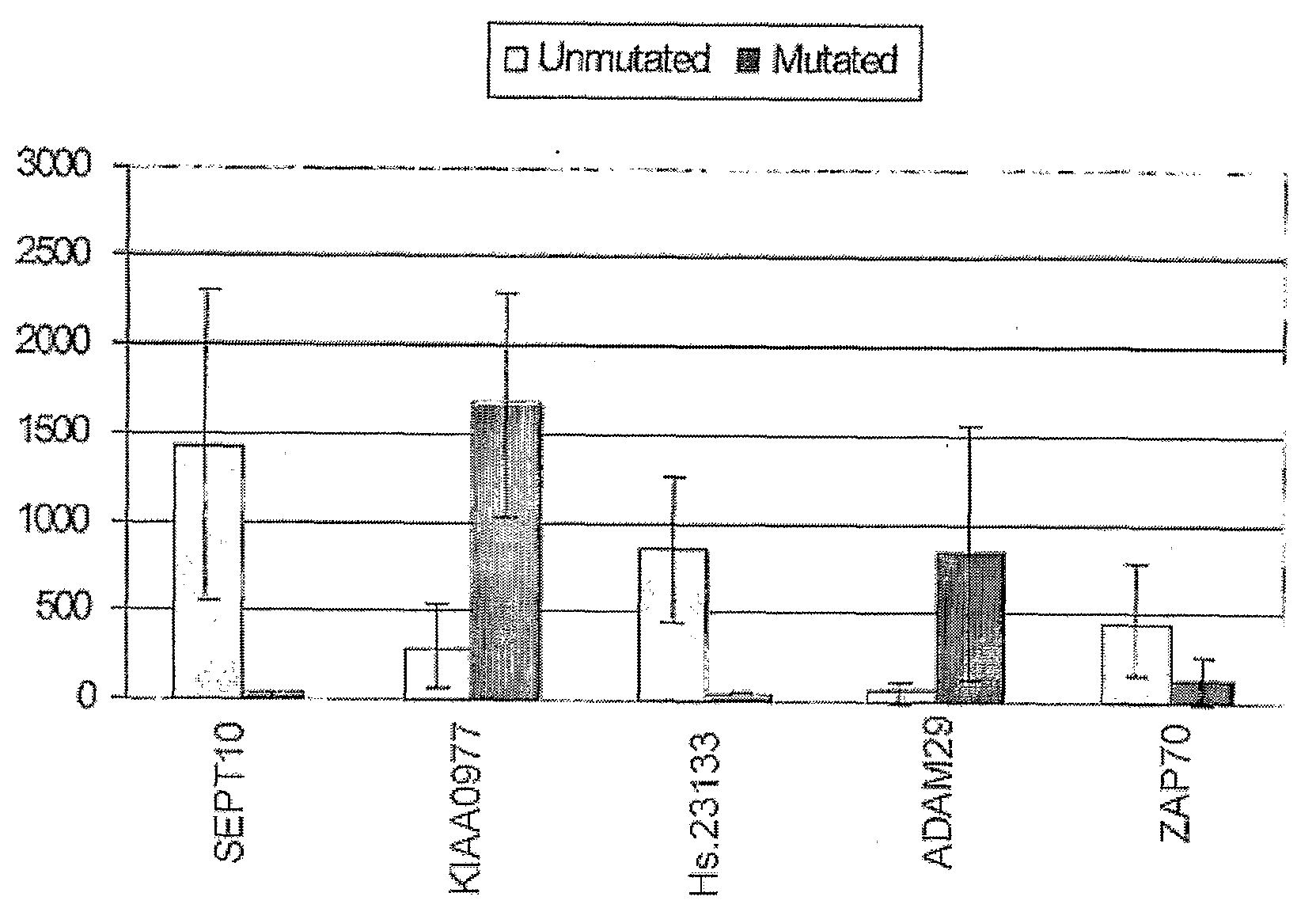
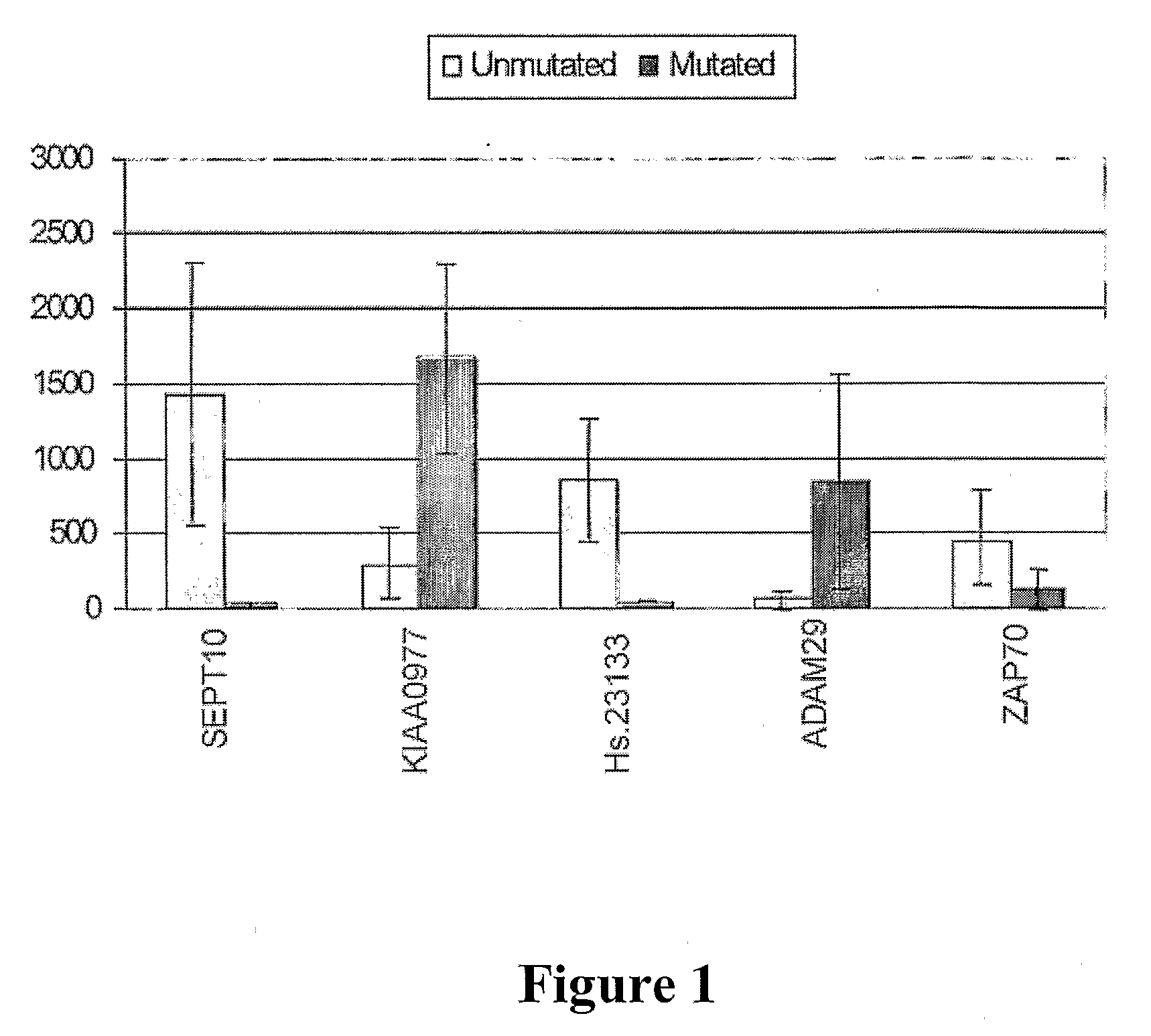
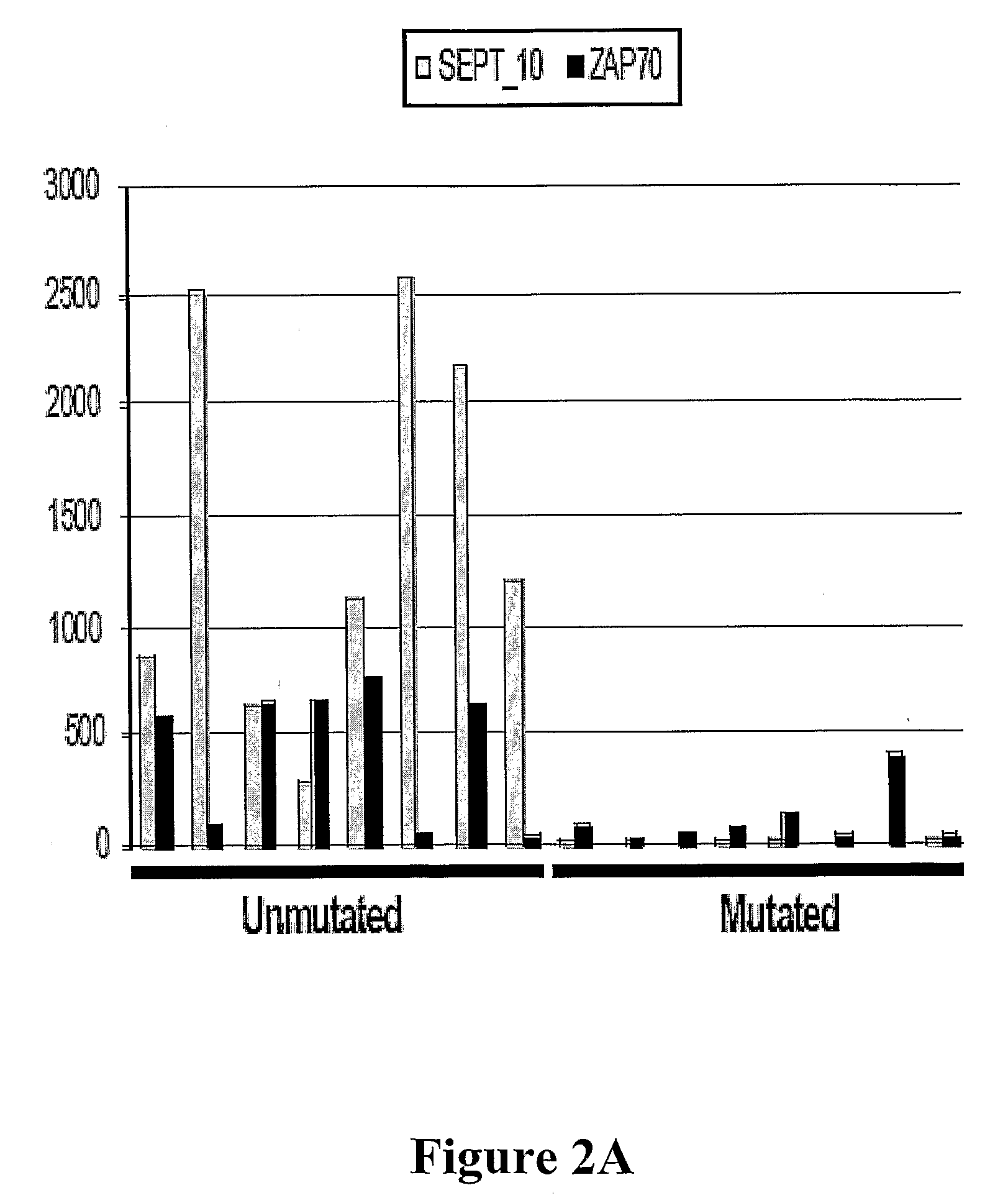
The invention provides compositions and methods for determining a prognosis of a B cell chronic lymphocytic leukemia (CLL) in a subject based on the level of expression of at least one marker gene. Marker genes provided by the invention are SEPTlO, KIAA0799, Hs.23133, and ADAM29. The marker genes can be used to differentially diagnose CLL in a subject based on relative gene expression levels in the subject compared to reference gene expression levels established from a clinically characterized population of patients. The invention also provides diagnostic reagents and compositions and kits based on the marker genes.
Owner:THE TRUSTEES OF COLUMBIA UNIV IN THE CITY OF NEW YORK
Process for the biological production of 1,3-propanediol with high yield
Owner:NUTRITION & BIOSCIENCES USA 4 INC
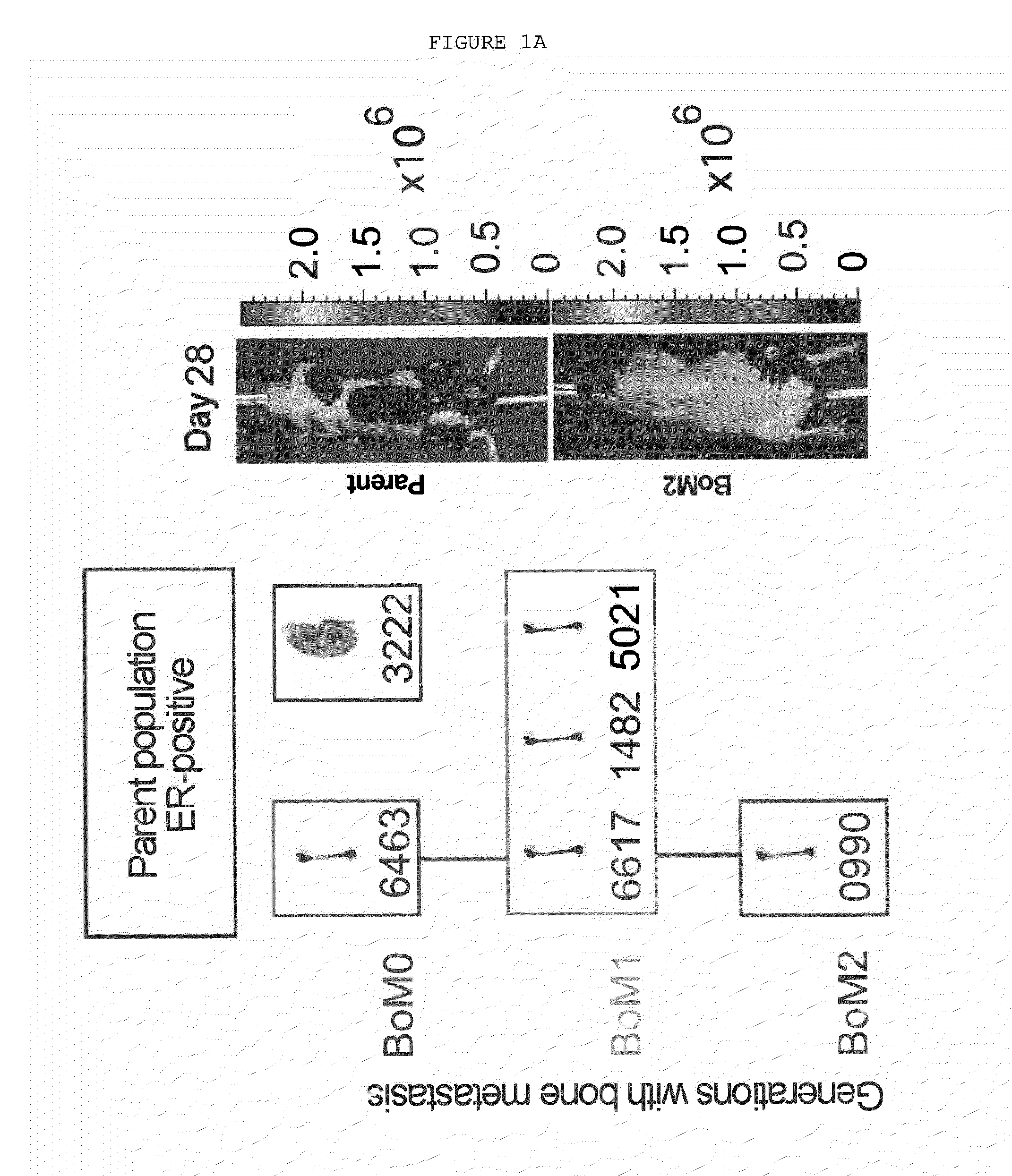
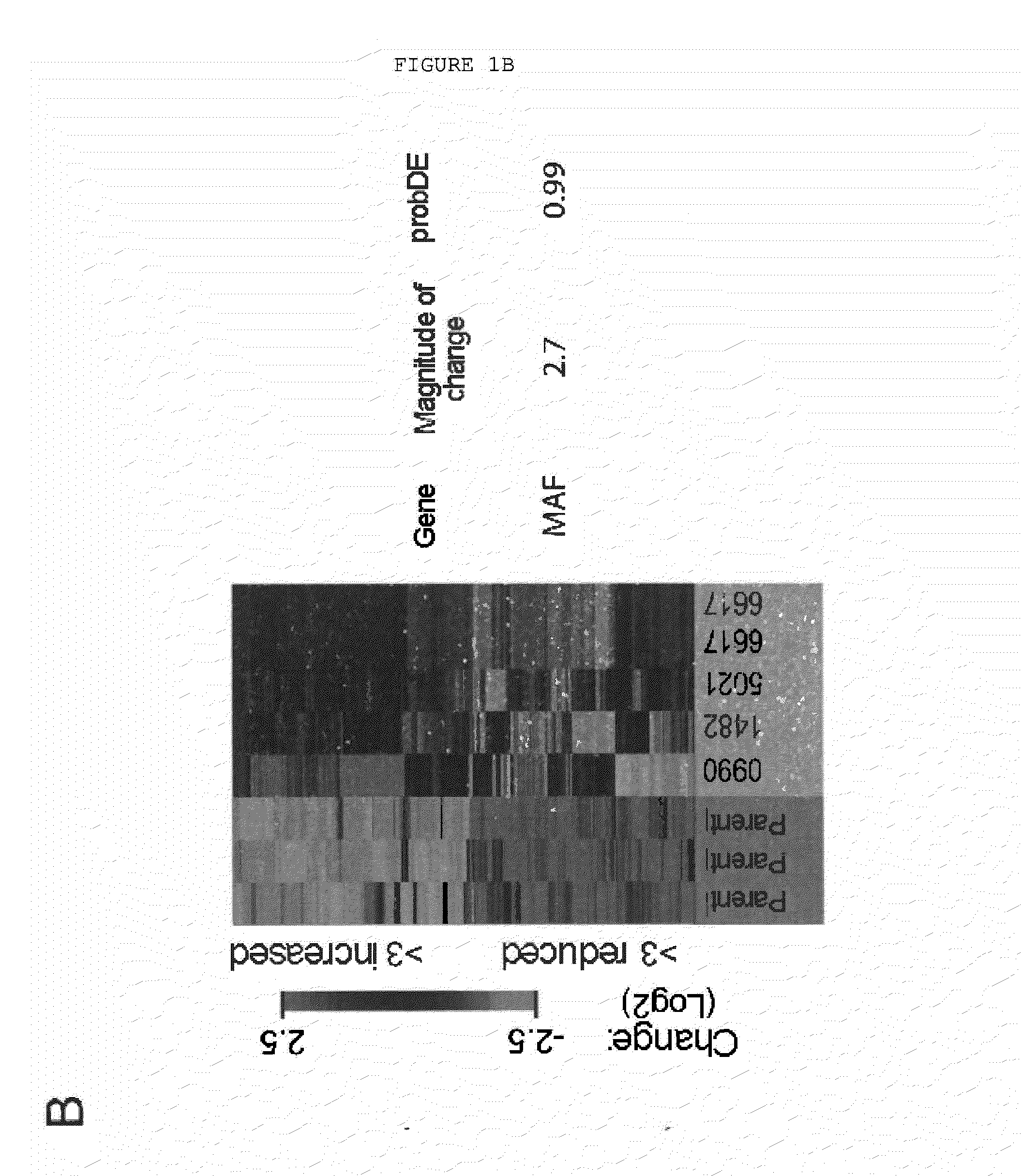
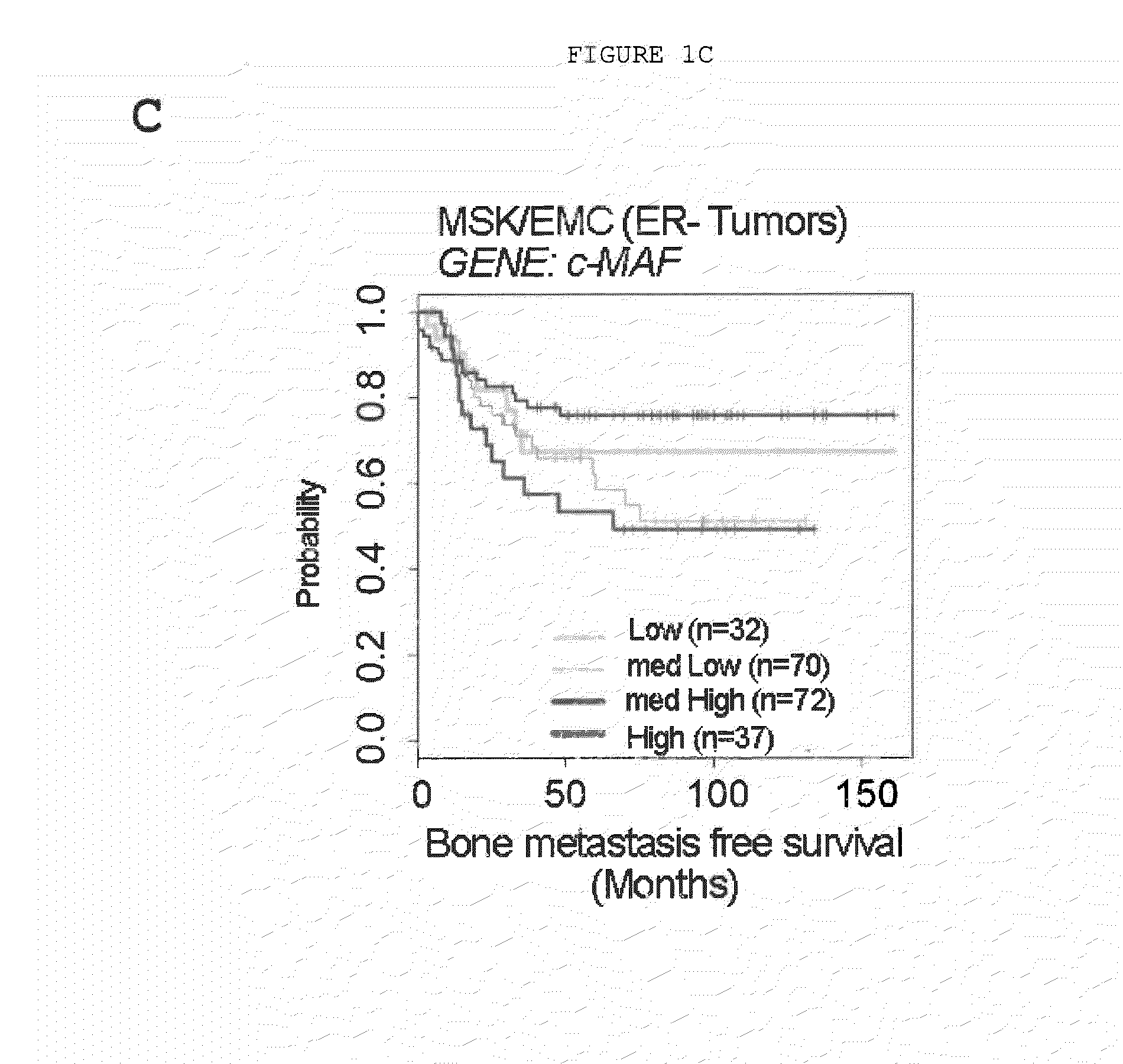
Method for the Diagnosis, Prognosis and Treatment of Breast Cancer Metastasis
ActiveUS20140057796A1Positive diagnosisGreat tendency to develop metastasisSugar derivativesMicrobiological testing/measurementBreast cancer metastasisPrimary tumor
The present invention relates to a method for the diagnosis or the prognosis of metastasis in breast cancer which comprises determining if the c-MAF gene is amplified in a primary tumor sample. Likewise, the invention also relates to a method for the diagnosis or the prognosis of metastasis in ER− breast cancer, as well as to a method for determining the tendency to develop bone metastasis with respect to metastasis in other organs, which comprise determining the c-MAF gene expression level. Finally, the invention relates to the use of a c-MAF inhibitor as therapeutic target for treating the ER− breast cancer metastasis.
Owner:INSTITUCIO CATALANA DE RECERCA I ESTUDIS AVANCATS +1
Method for obtaining gene information and functional genes from species without genome reference sequence
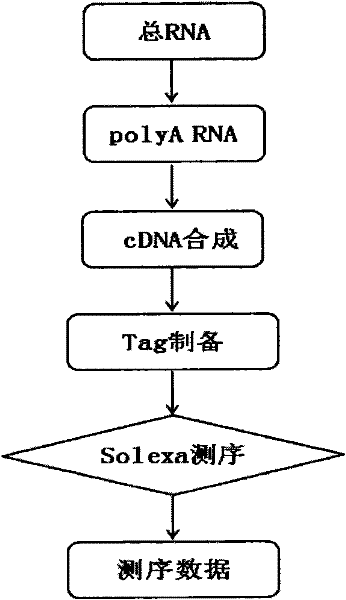
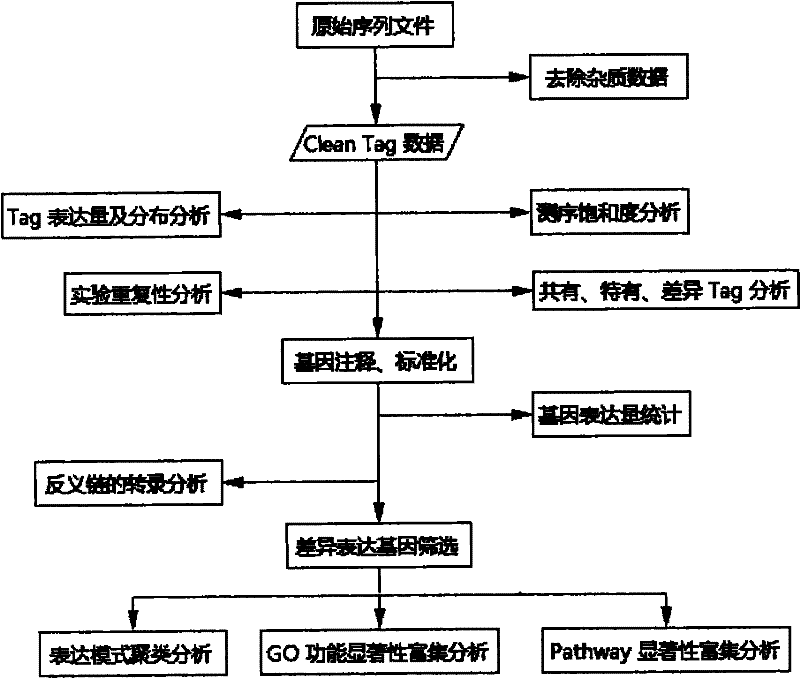
The invention relates to a method for obtaining gene information and functional genes from species without genome reference sequences. The invention also discloses a method for obtaining the gene expression profile of the Asian corn borer. The invention also discloses a method for obtaining transcriptome information and functional genes of the Asian corn borer. The present invention combines DGE-tag technology with RNA-seq technology for the first time to obtain gene expression period, expression level, corresponding metabolic pathway and gene function information from species without genome reference sequence. The method is convenient and fast , accurate and low cost.
Owner:SHANGHAI INST OF BIOLOGICAL SCI CHINESE ACAD OF SCI
Non-invasive detection of fish viruses by real-time PCR
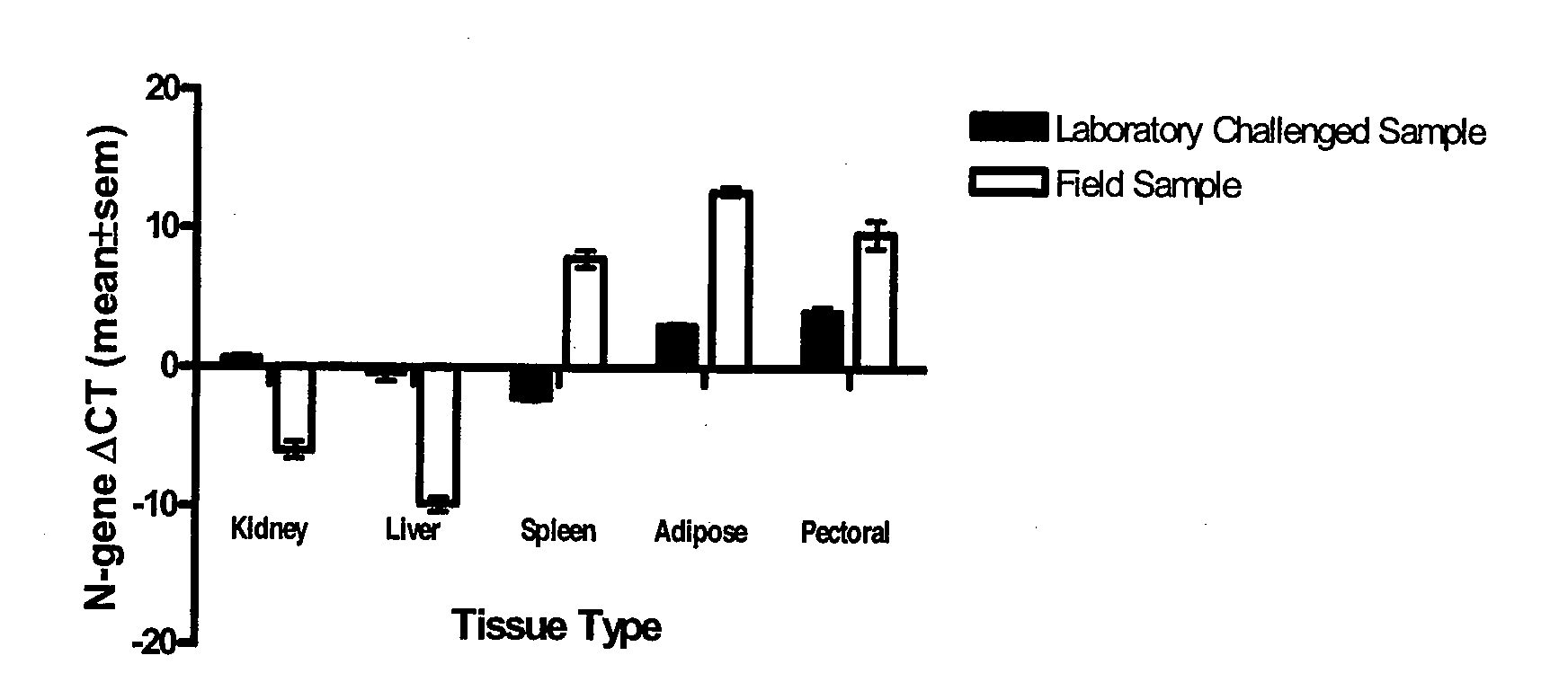
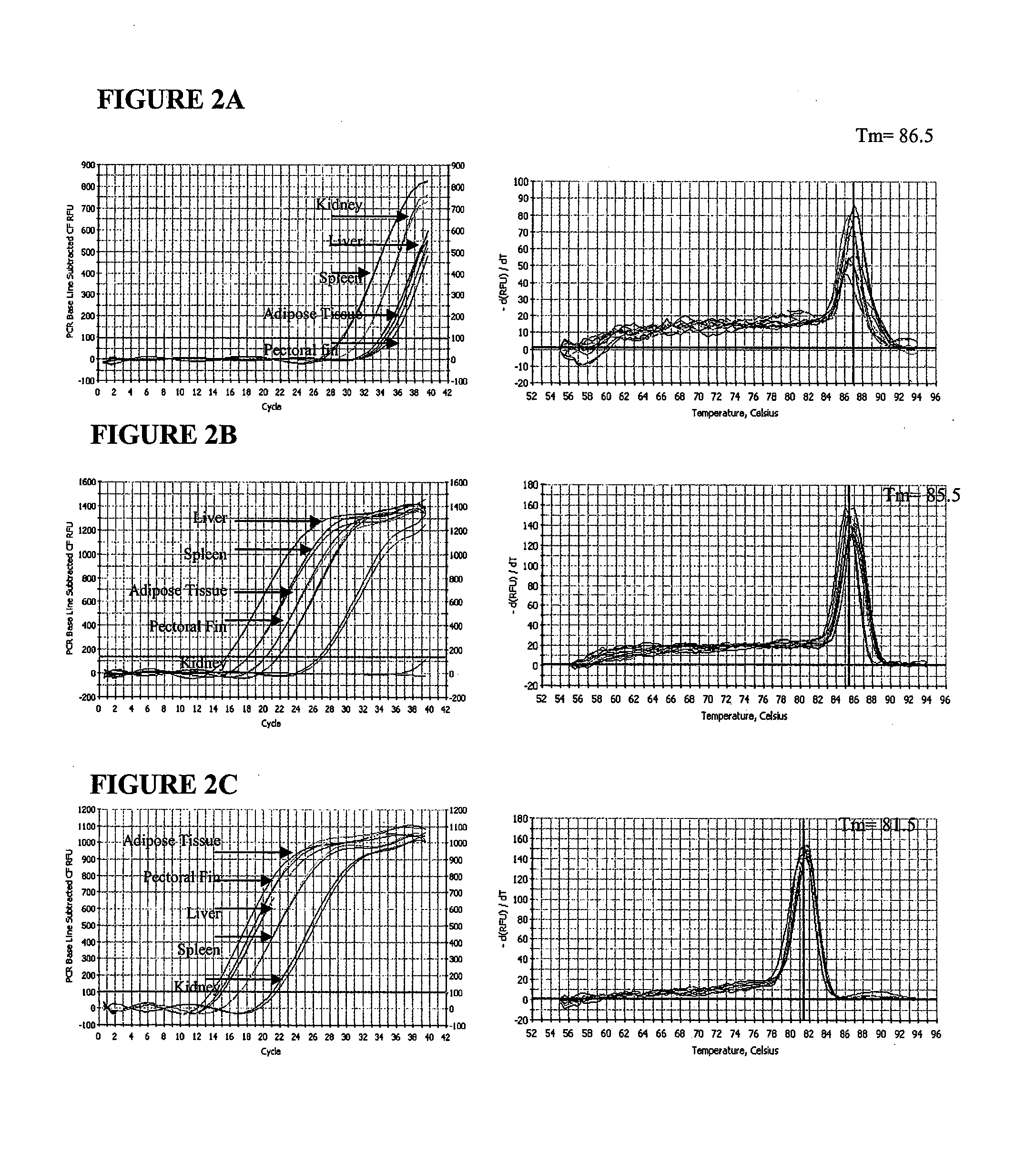
A real-time assay coupled with a non-invasive tissue sampling was developed for the detection and quantification of fish viruses. As a proof of principles, data were presented for the detection and quantification of infectious hypodermal necrosis virus (IHNV) in trout. The primers were designed for IHNV nucleocapsid (N), and surface glycoprotein (G) genes, and trout &bgr;-actin and elongation factor-l&agr; (EF-I &agr;) were used as internal control for the assay. The reaction conditions for the real-time RT-PCR were optimized using cDNA derived from IHNV-infected Epithelioma papulosum cyprinid (EPC) cells. Using both N- and G-gene primers, IHNV was successfully detected in liver, kidney, spleen, adipose tissue and pectoral fin samples of laboratory-challenged and wild samples. The dissociation curves with a single melting peak at expected temperature (85° C. for the N-gene and 86.5° C. for the G-gene) confirmed the specificity of the N- and G-gene amplicons. The IHNV N- and the G-gene expression levels in different tissues of laboratory challenged samples were in the order of spleen, liver, kidney, adipose tissue and pectoral fin, however in the field-collected samples the order of gene expression was liver, kidney, pectoral fin, adipose tissue, and spleen. The N- and G-gene expressions in spleen were found to be dramatically lower in the field-collected samples compared to the laboratory-challenged samples indicating a potential difference in the IHNV replication in the laboratory as opposed to field conditions. The real-time PCR assay was found to be rapid, highly sensitive, and reproducible. Based upon the ability to detect the virus in pectoral fins a non-invasive detection method for IHNV and other fish viruses is developed. Such a non-invasive tissue sampling coupled with real-time PCR assay is very valuable for large-scale virus screening of fish in aquaculture facilities as well as for epidemiological studies.
Owner:ADVANCED BIONUTRITION CORP
Method for producing an L-amino acid using a bacterium with an optimized level of gene expression
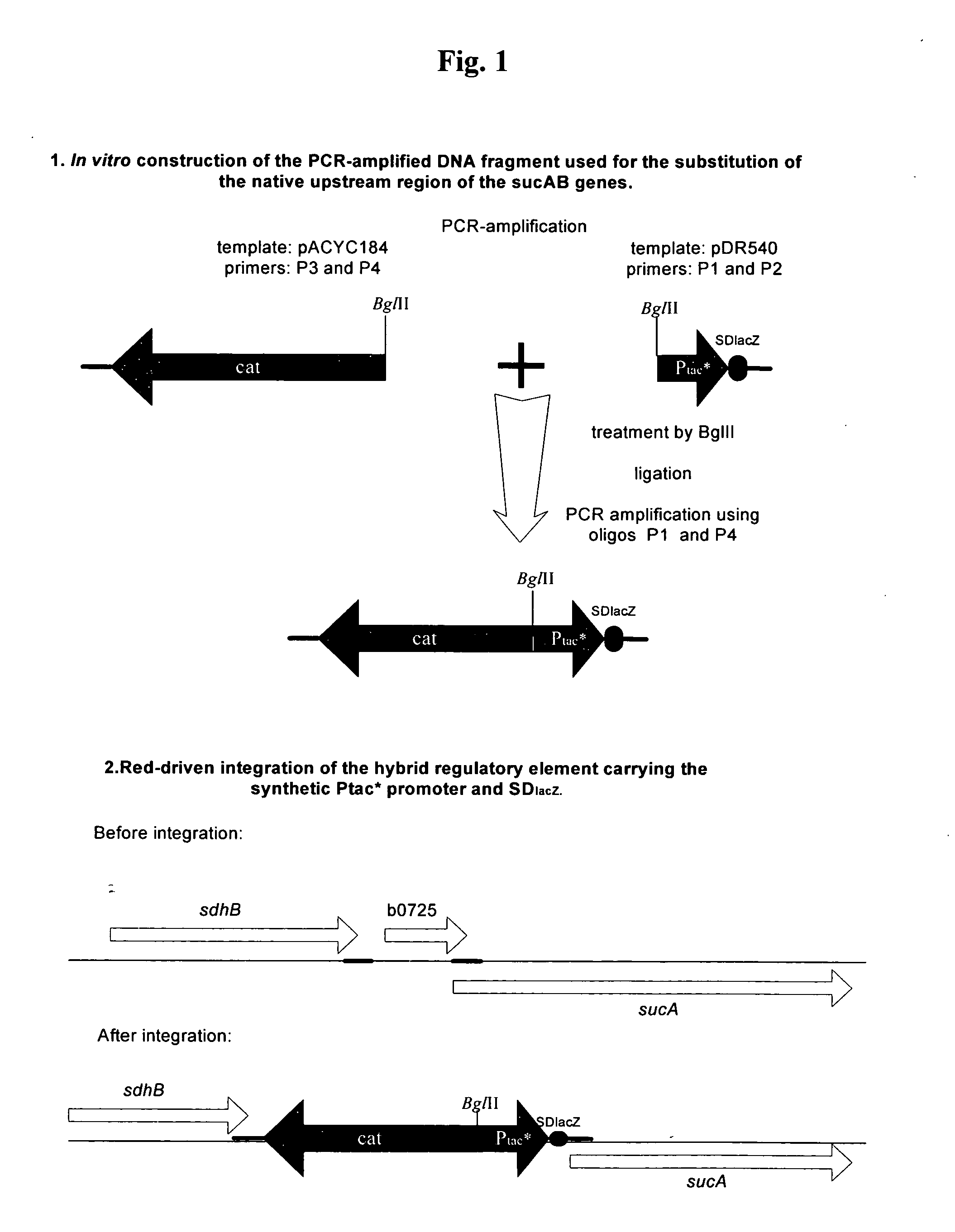
A method is provided for obtaining an L-amino acid or nucleic acid-producing bacterium belonging to the genus Escherichia with an optimized level of expression of the gene which influences the distribution of carbon flow, such as the sucAB genes, comprising introducing into the chromosome of the bacterium a set of in vitro constructed DNA fragments which contain regulatory elements for gene expression instead of the native elements of the regulatory region of the gene, and selecting the colonies with increased L-amino acid productivity. Also, a method is provided for producing an L-amino acid, such as L-glutamic acid, L-proline, L-arginine, L-glutamine, L-leucine, using the bacterium with an optimized level of expression of the sucAB gene.
Owner:AJINOMOTO CO INC
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com