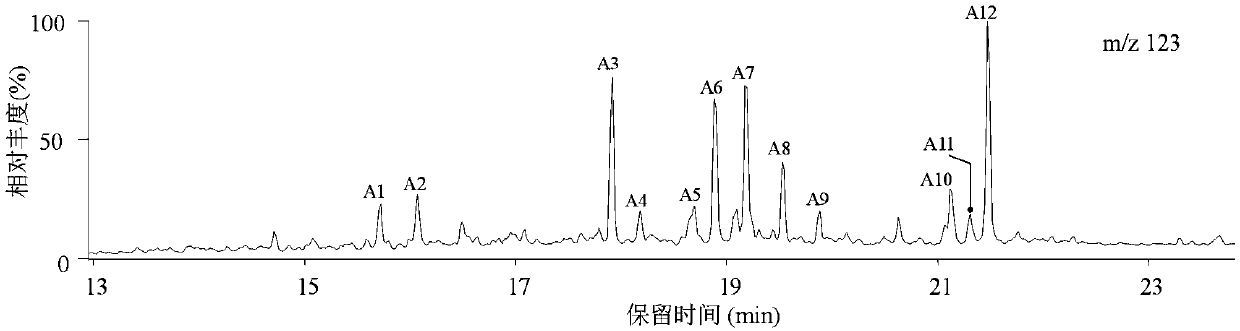
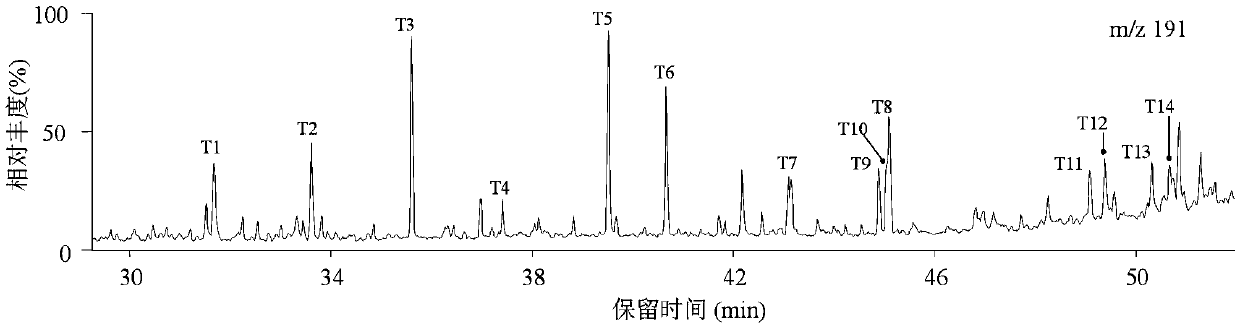
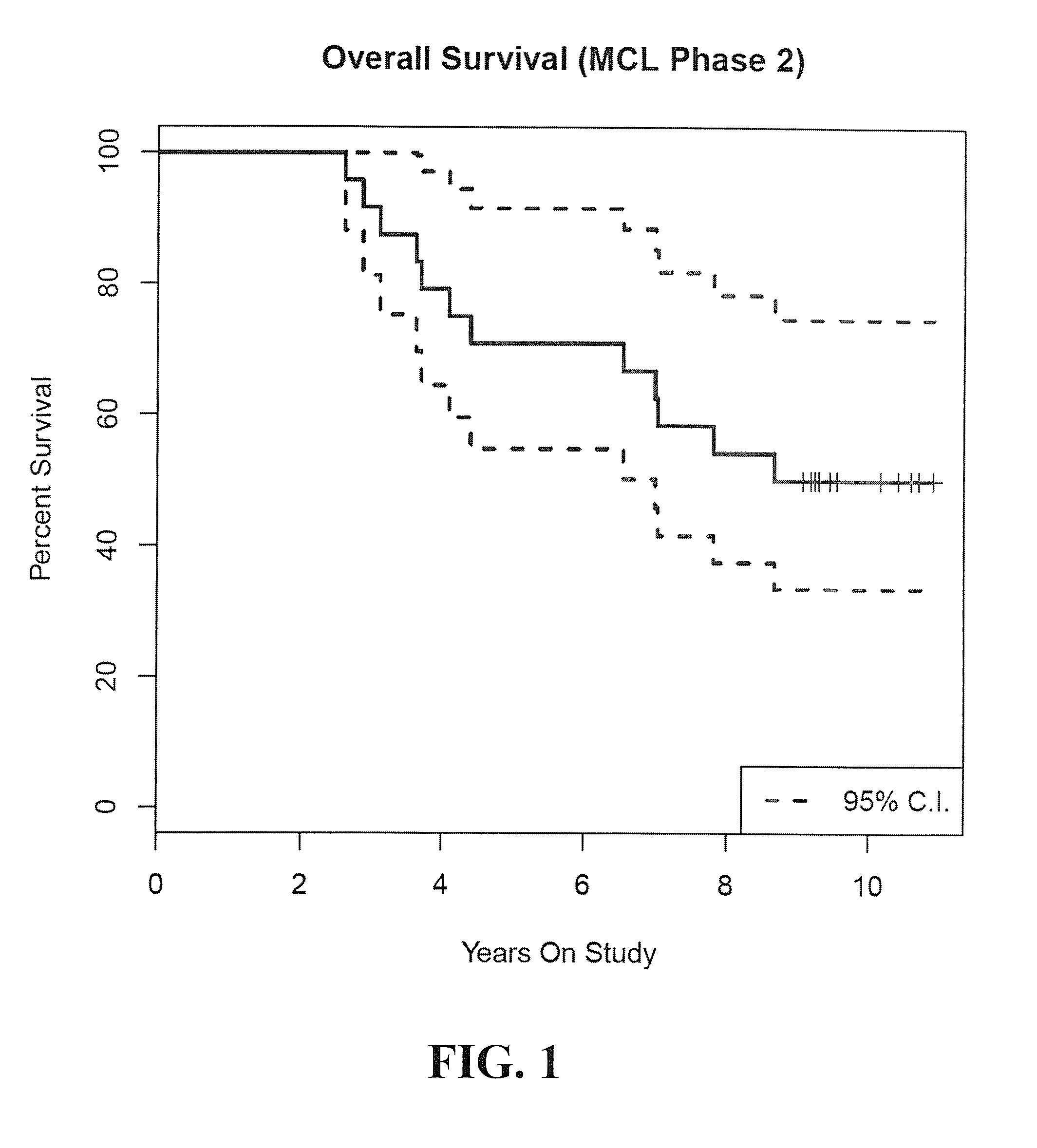
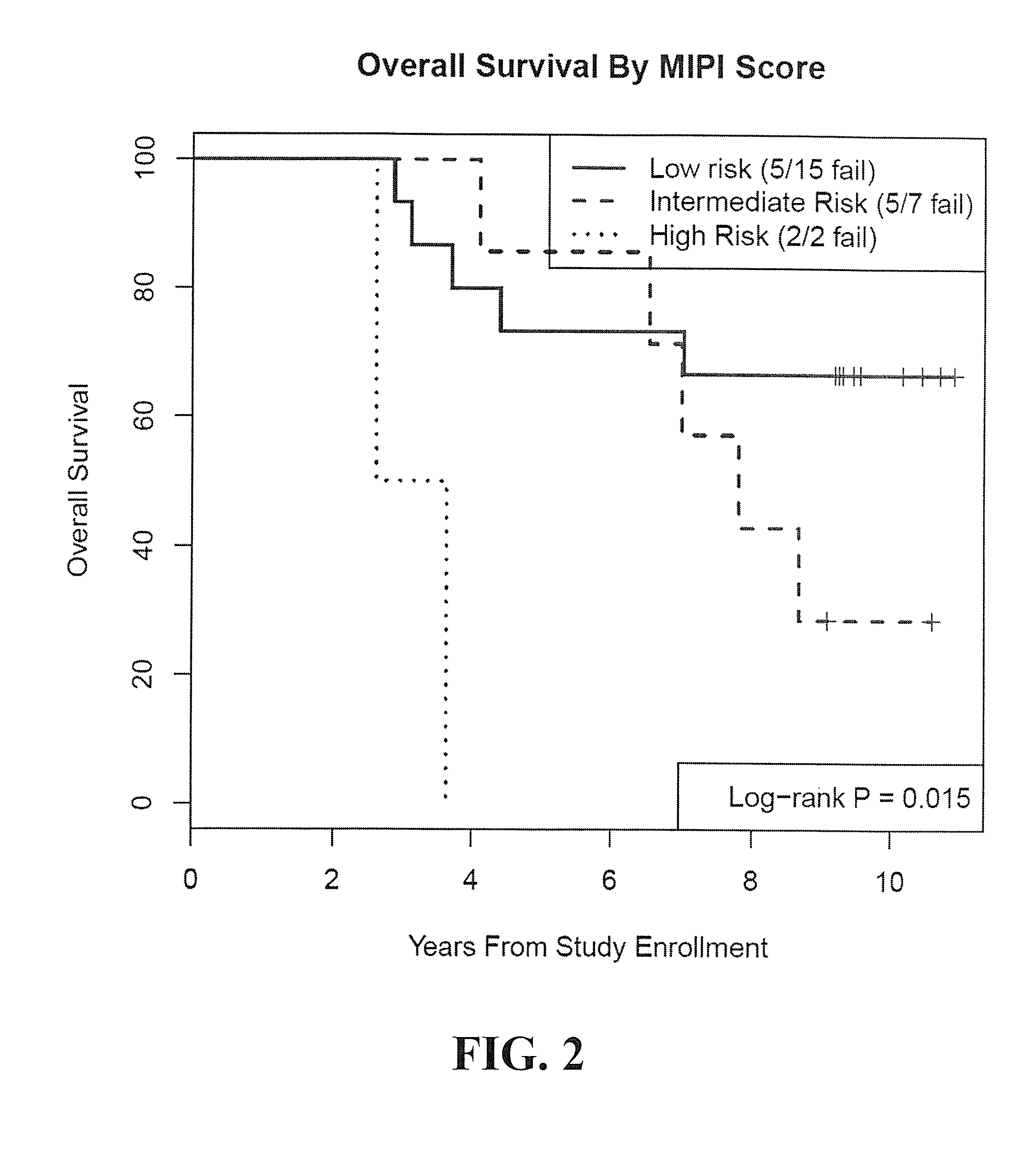
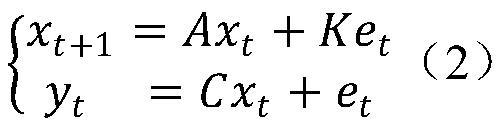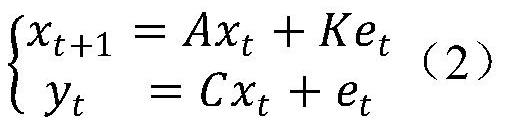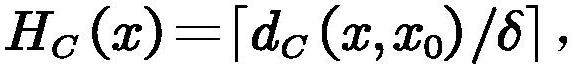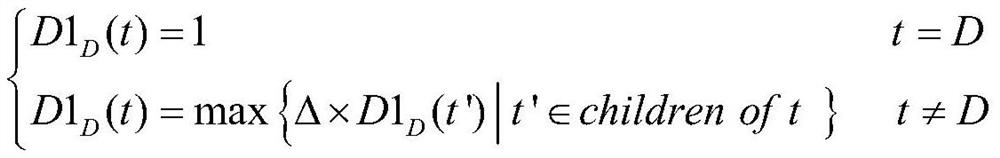Patents
Literature
35 results about "Biomarker identification" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
It is usually identifiable or measurable in the blood, urine or other convenient body fluids by PCR, ELISA, and other conventional immune assays. In this sense, biomarkers can be categorized into genetic, epigenetic, proteomic, glycomic, and imaging biomarkers for cancer diagnosis, prognosis, and epidemiology.
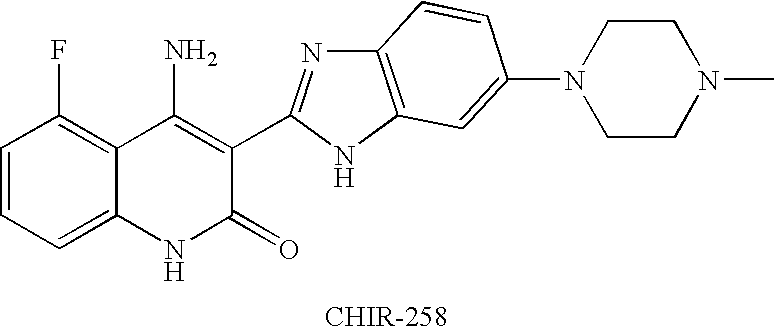
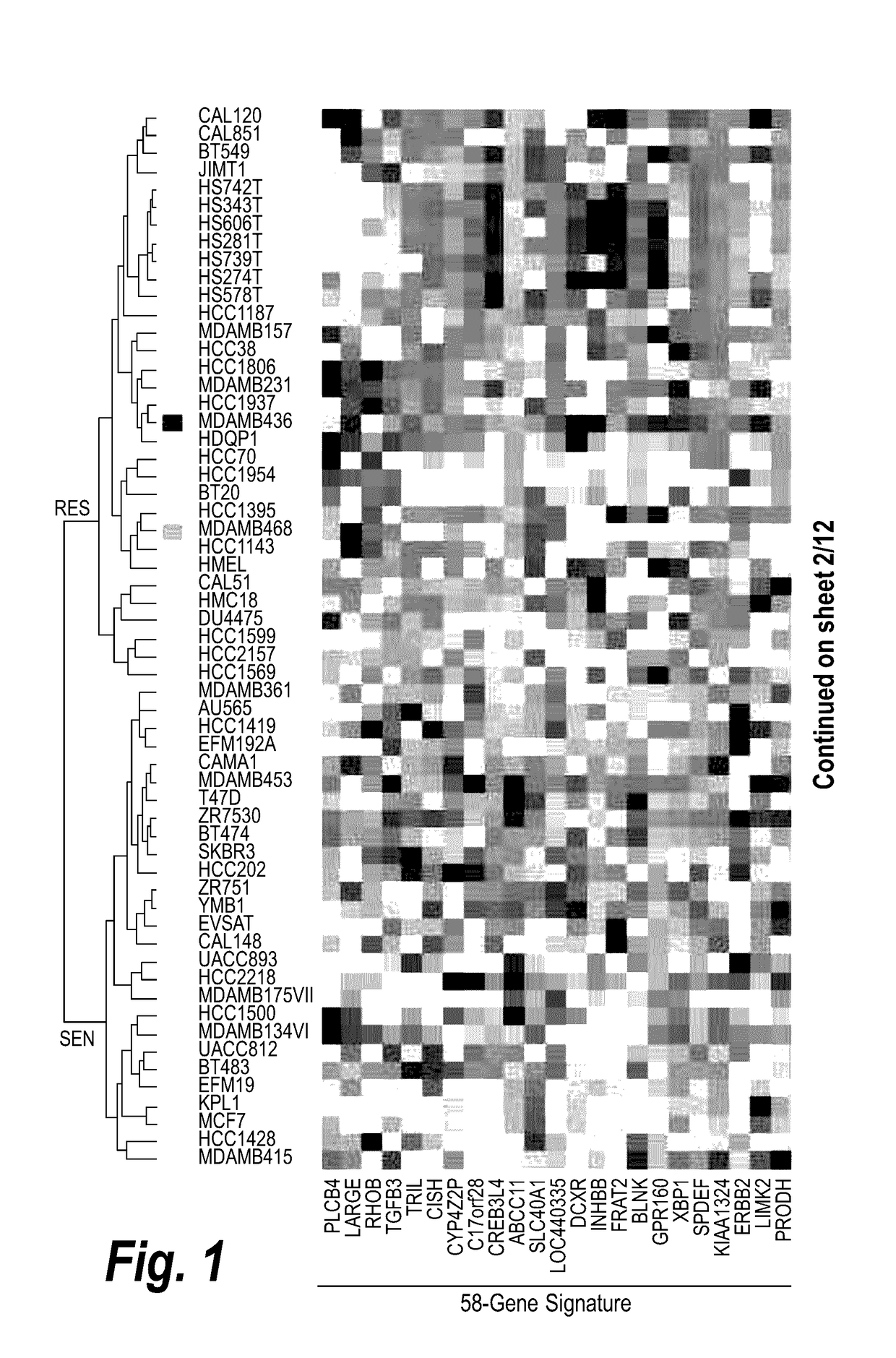
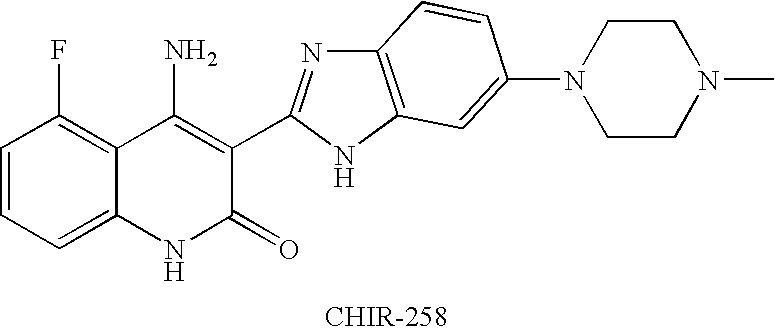
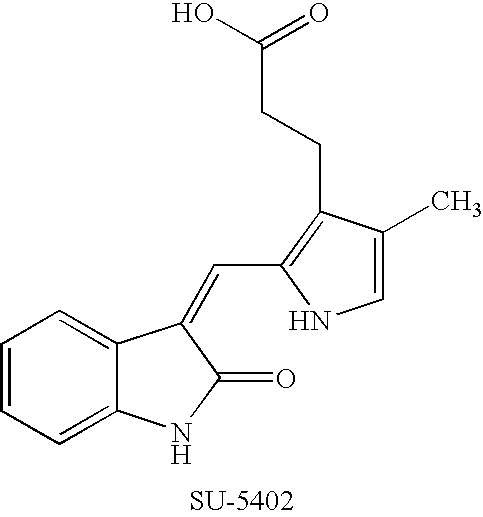
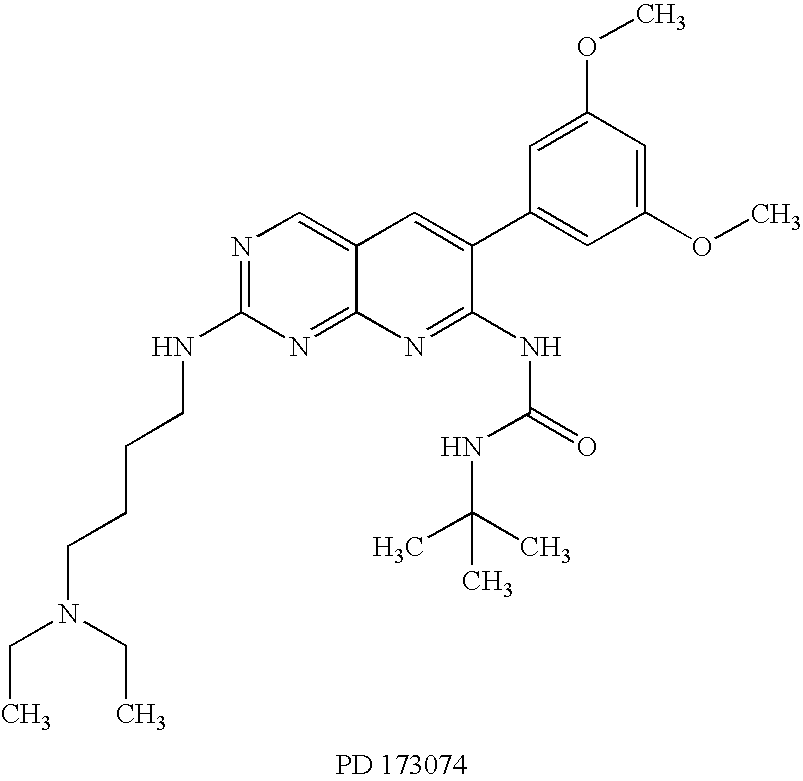
Effects of inhibitors of FGFR3 on gene transcription
InactiveUS8158360B2Improve responseMicrobiological testing/measurementBiological testingMedicineBiomarker identification
Methods of utilizing biomarkers to identify patients for treatment or to monitor response to treatment are taught herein. Alterations in levels of gene expression of the biomarkers, particularly in response to FGFR3 inhibition, are measured and identifications or adjustments may be made accordingly.
Owner:NOVARTIS AG
Thyroid cancer biomarker
InactiveUS20150038376A1Microbiological testing/measurementProtein nucleotide librariesMedicineBiomarker identification
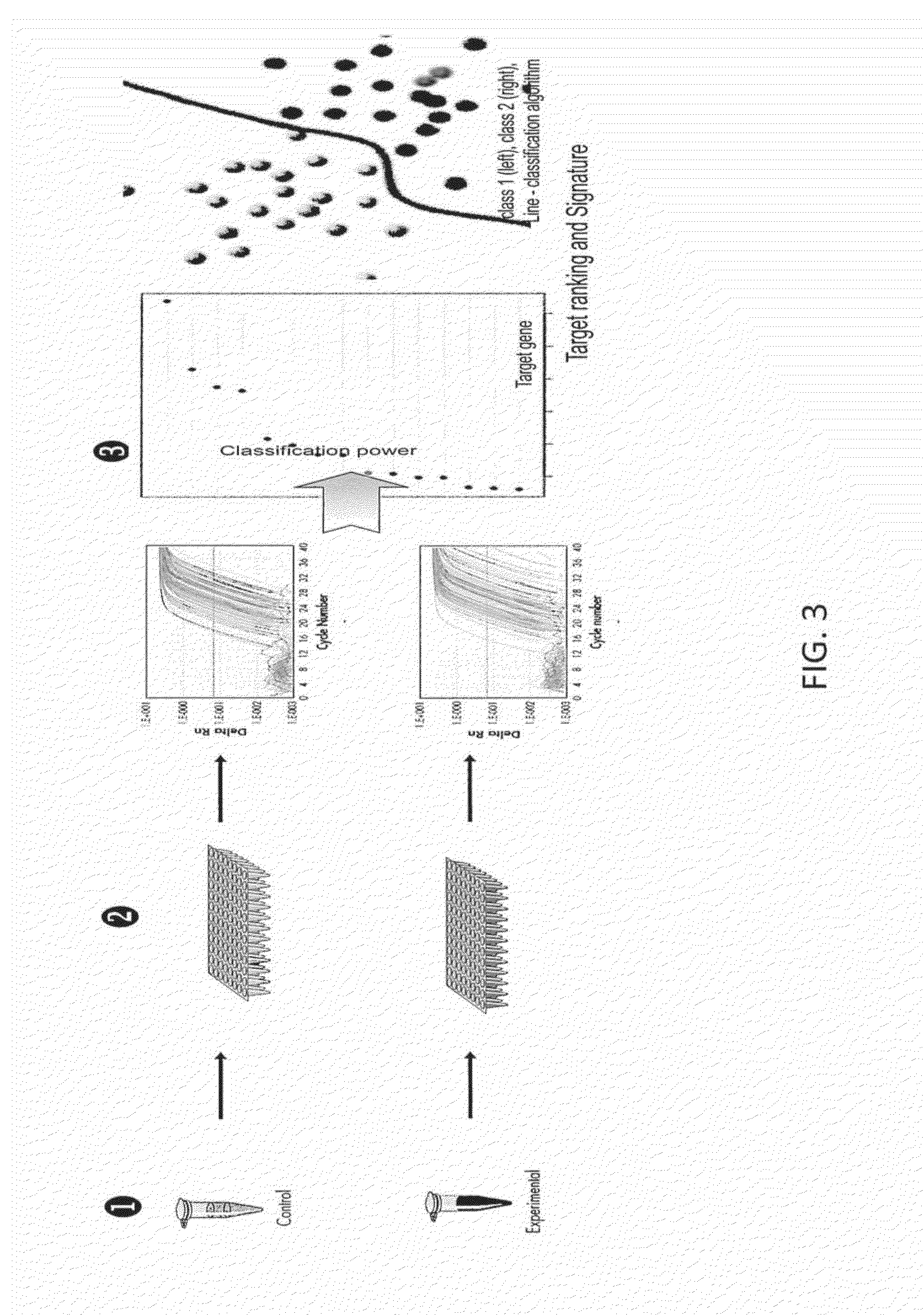
The methods provided herein use microarray data for feature selection and then use selected targets to generate industry standard qPCR arrays with new clinical sample assay data so order to build a classification model. This multi-step method overcomes the disadvantages of traditional biomarker identification.
Owner:CORNELL UNIVERSITY +1
Animal models of pancreatic adenocarcinoma and uses therefor
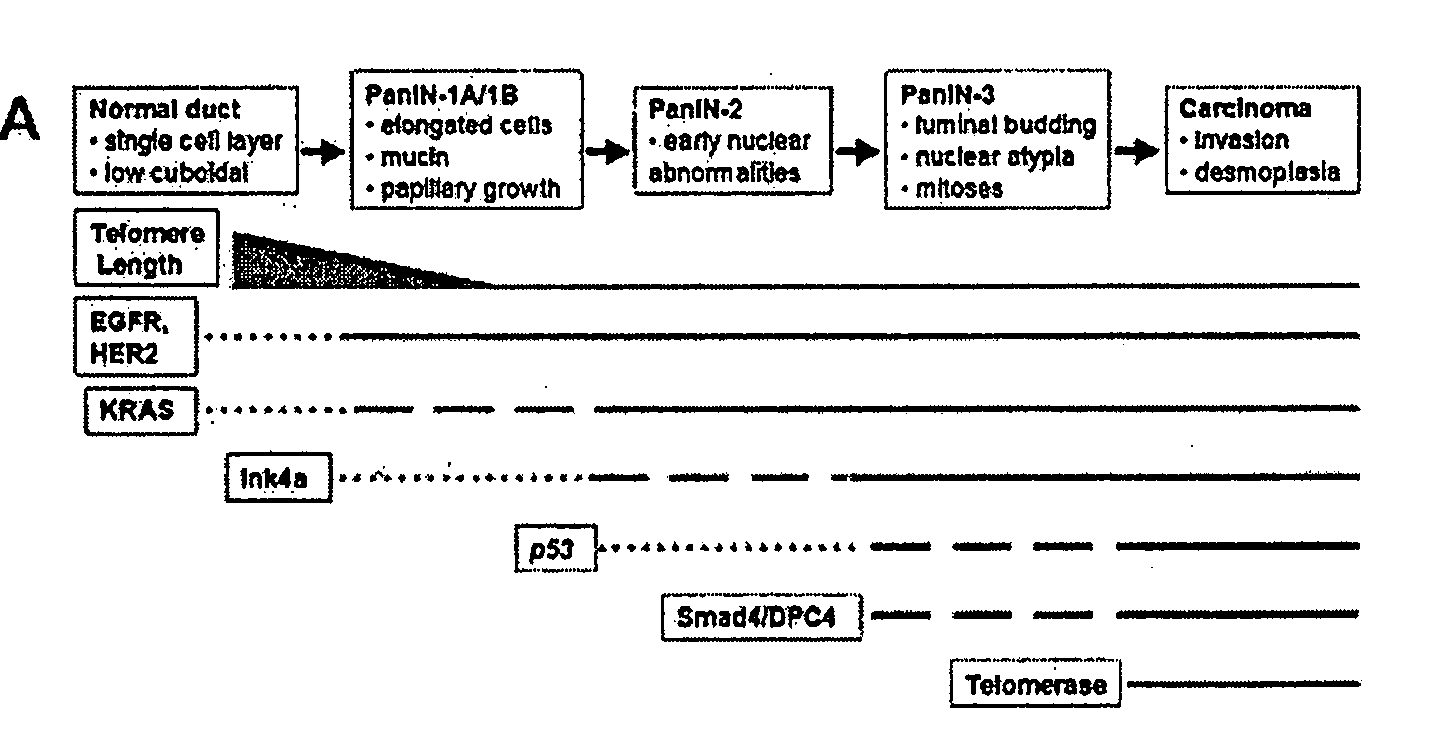
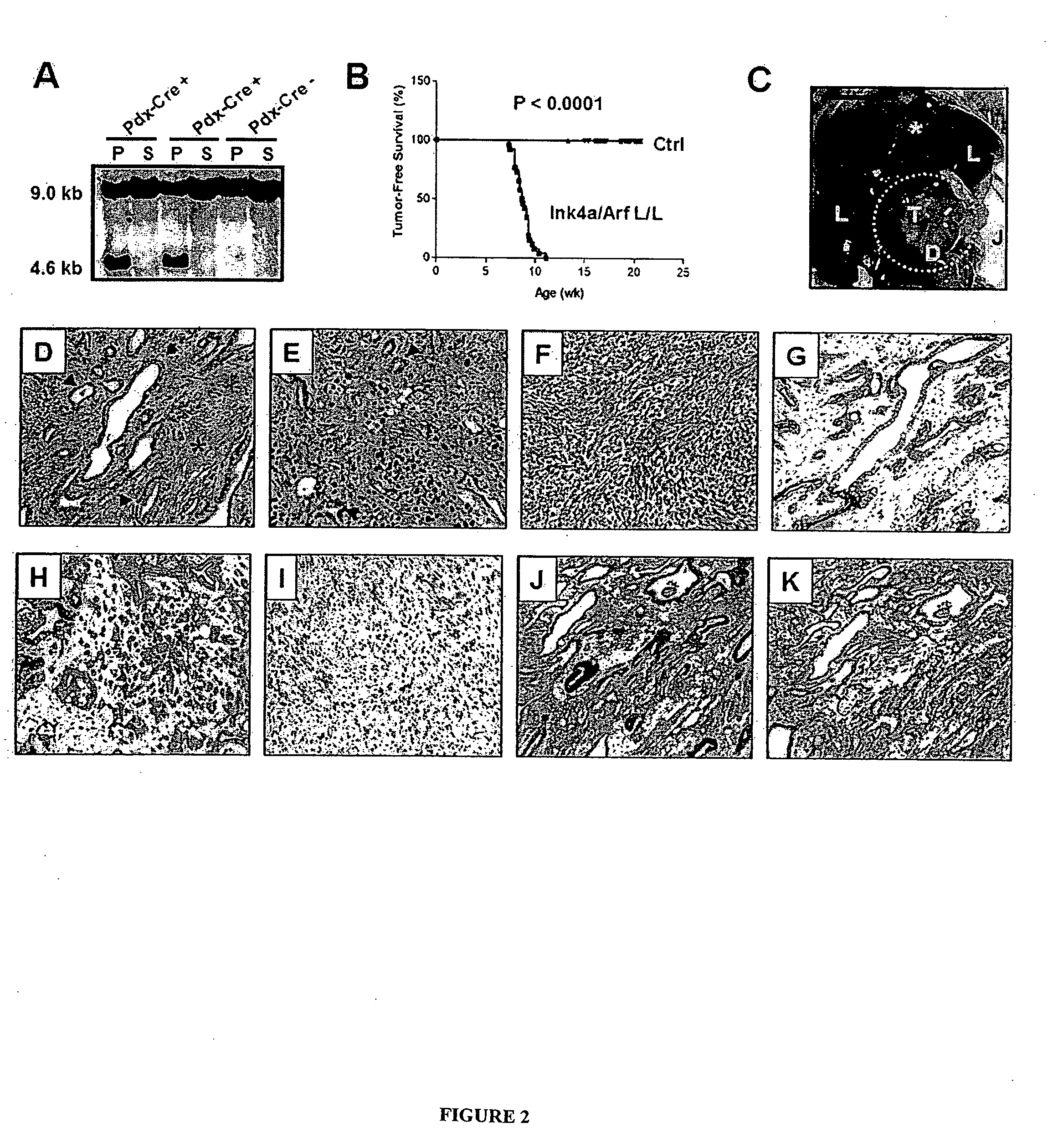
The present invention is based, at least in part, on the generation of an animal model of pancreatic adenocarcinoma which recapitulates the genetic and histological features of human pancreatic adenocarcinoma, including the initiation, maintenance, and progression of the disease. Accordingly, the present invention provides animal models of cancer, e.g., pancreatic adenocarcinoma, wherein an activating mutation of Kras has been introduced, and any one or more known or unknown tumor suppressor genes or loci, e.g., Ink4a / Arf, Ink4a, Arf, p53, Smad4 / Dpc, Lkb1, Brca2, or Mlh1, have been misexpressed, e.g., have been misexpressed leading to decreased expression or non-expression. The animal models of the invention may be used, for example, to identify biomarkers of pancreatic cancer, to identify agents for the treatment or prevention of pancreatic cancer, and to evaluate the effectiveness of potential therapeutic agents.
Owner:THE TRUSTEES OF THE UNIV OF PENNSYLVANIA +1
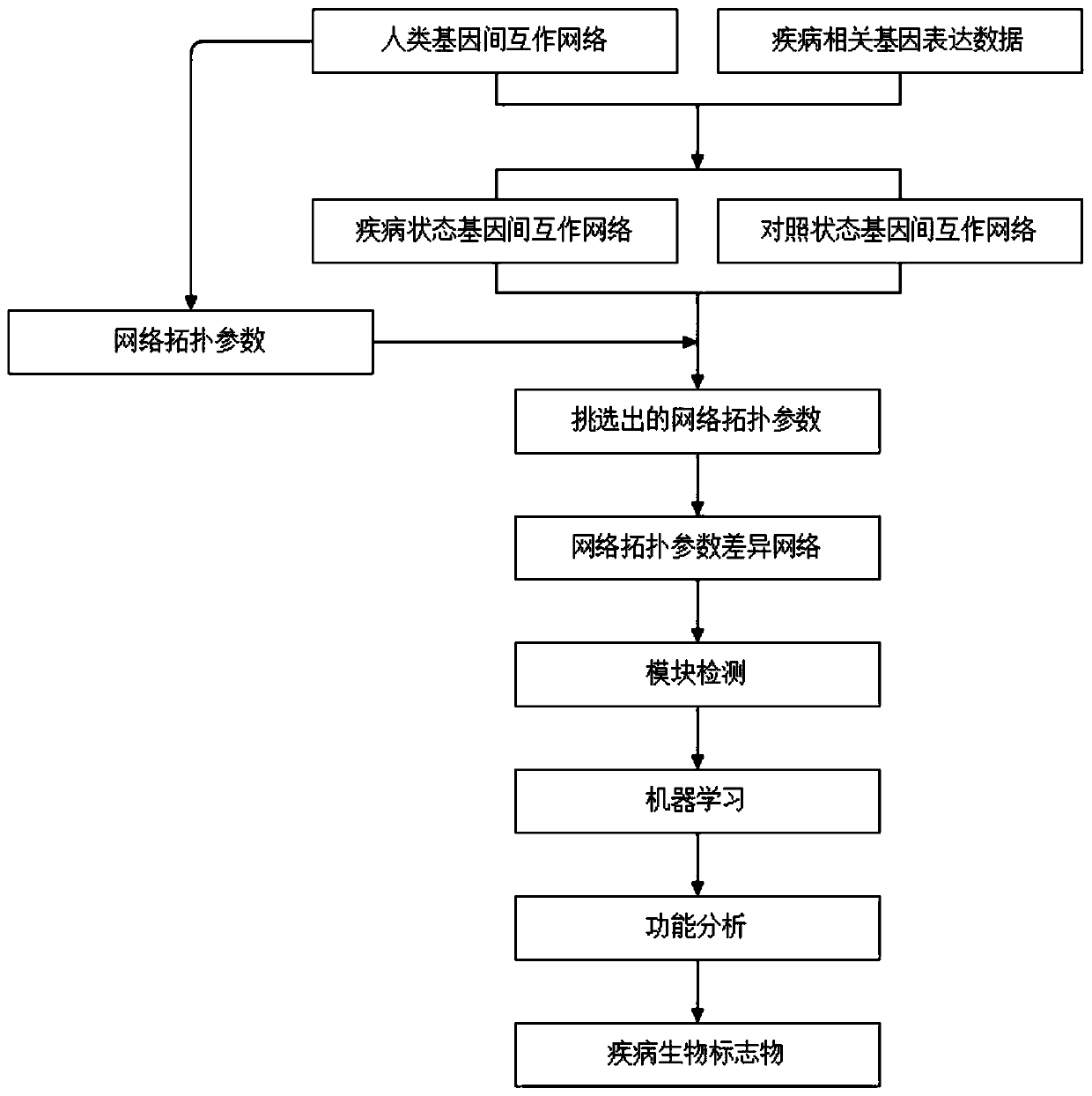
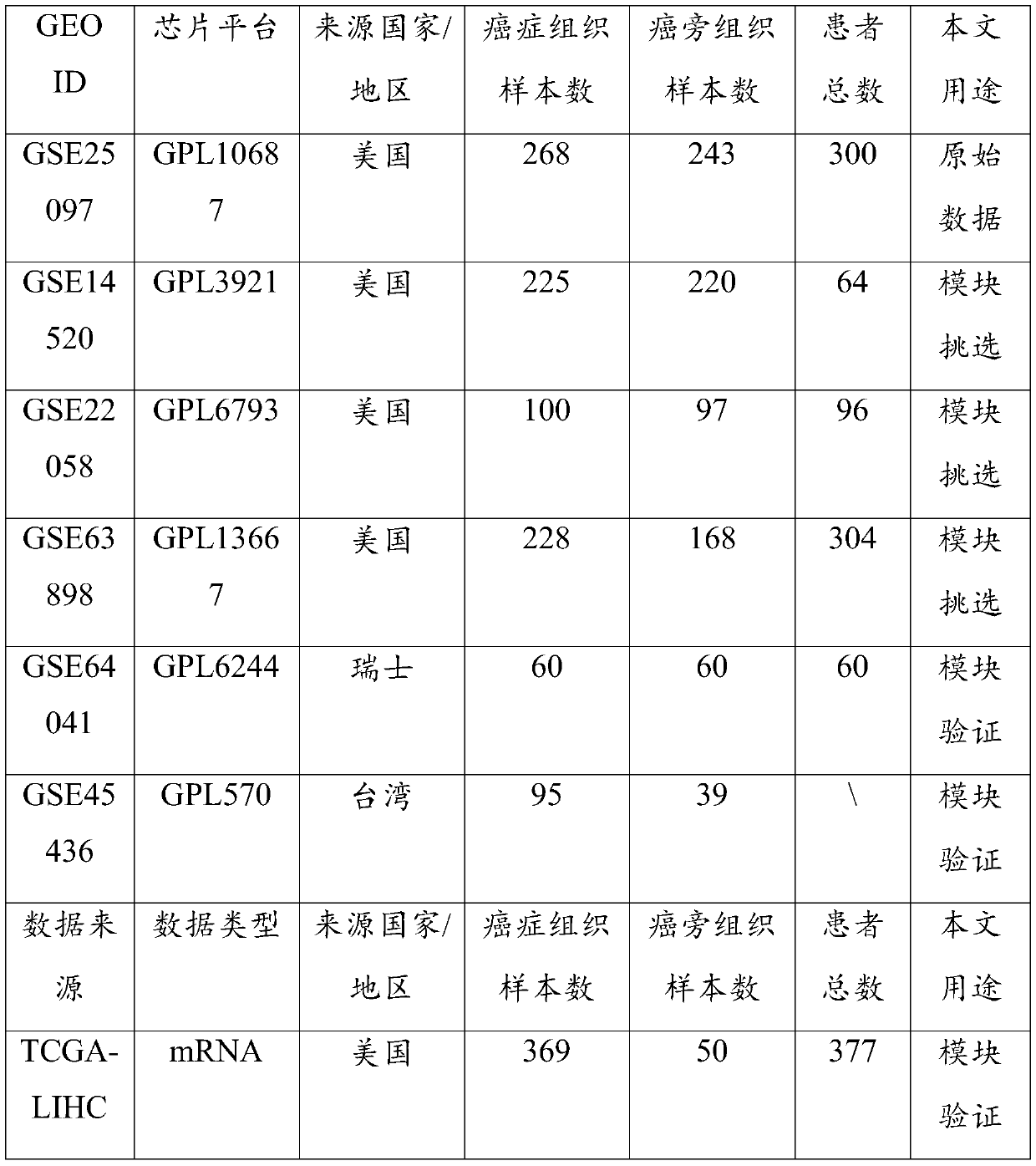
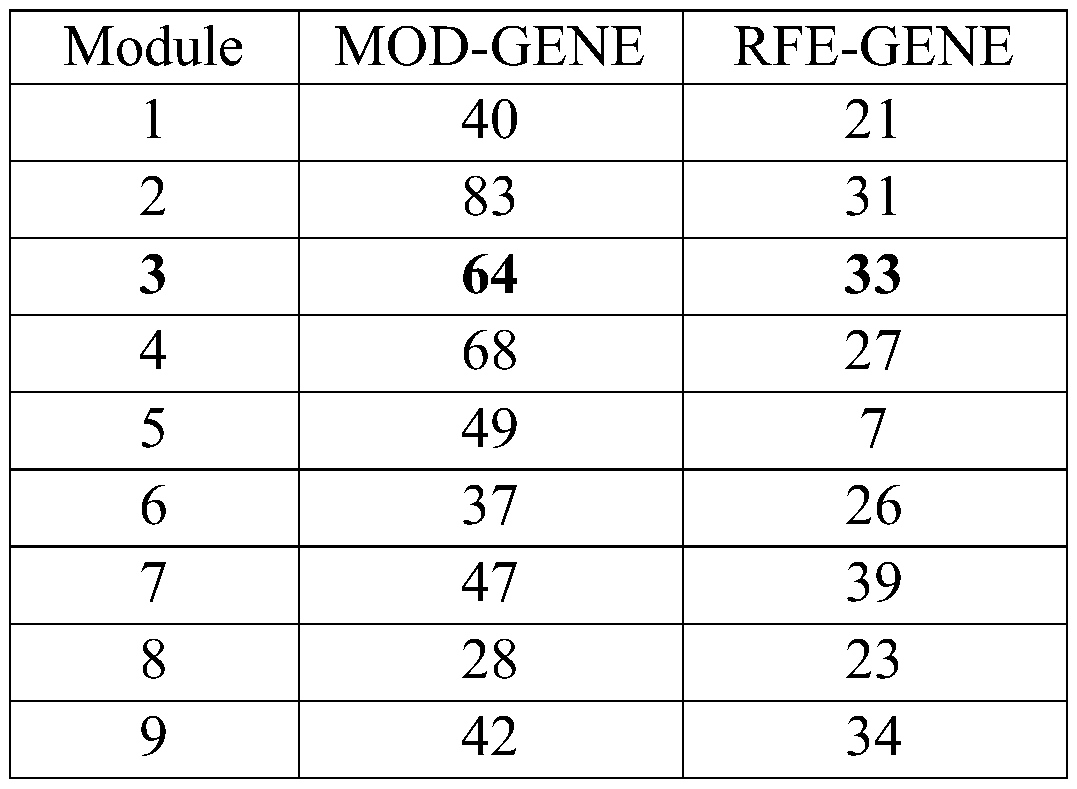
Cancer biomolecule marker screening method and system based on network topology parameters
ActiveCN110444248ACharacter and pattern recognitionProteomicsBiomarker identificationScreening method
The invention discloses a cancer biomolecule marker screening method and system based on network topology parameters. The method comprises the steps that a human gene interaction network and gene chipexpression data are acquired and integrated to obtain a gene interaction network based on gene expression data; a disease state and control state gene interaction network is constructed; network topology parameter difference genes of the disease state and control state gene interaction network are calculated, and a network topology parameter difference change network is obtained based on the network parameter difference genes; network module mining is performed on the network topology parameter difference network; feature selection is performed on an obtained difference network module to obtain genes capable of distinguishing normality and diseases in all modules; the classification effect of the genes selected from the modules on the diseases is detected, and the difference network module is screened according to the classification effect to serve as a biomolecule marker candidate. The invention provides a novel complex disease biomarker identification method based on omics data, andexperiments prove that the method has certain accuracy and effectiveness.
Owner:SHANDONG UNIV
Biomarkers of target modulation, efficacy, diagnosis and/or prognosis for raf inhibitors
InactiveUS20100004253A1Improving patient careBiocideMicrobiological testing/measurementBiomarker identificationGene expression level
Methods of utilizing biomarkers to identify patients for treatment or to monitor response to treatment are taught herein. Alterations in levels of gene expression of the biomarkers, particularly in response to Raf kinase inhibition, are measured and identifications or adjustments may be made accordingly.
Owner:NOVARTIS AG
Methods and apparatus for identifying disease status using biomarkers
InactiveUS20070255113A1Facilitate ascertaining the disease status of an individualBiostatisticsMedical automated diagnosisAssociated organismBiomarker identification
Methods and apparatus for identifying disease status according to various aspects of the present invention include analyzing the levels of one or more biomarkers. The methods and apparatus may use biomarker data for a condition-positive cohort and a condition-negative cohort and select multiple relevant biomarkers from the plurality of biomarkers. The system may generate a statistical model for determining the disease status according to differences between the biomarker data for the relevant biomarkers of the respective cohorts. The methods and apparatus may also facilitate ascertaining the disease status of an individual by producing a composite score for an individual patient and comparing the patient's composite score to one or more threshold for identifying potential disease status.
Owner:BIOMARKER TECH +1
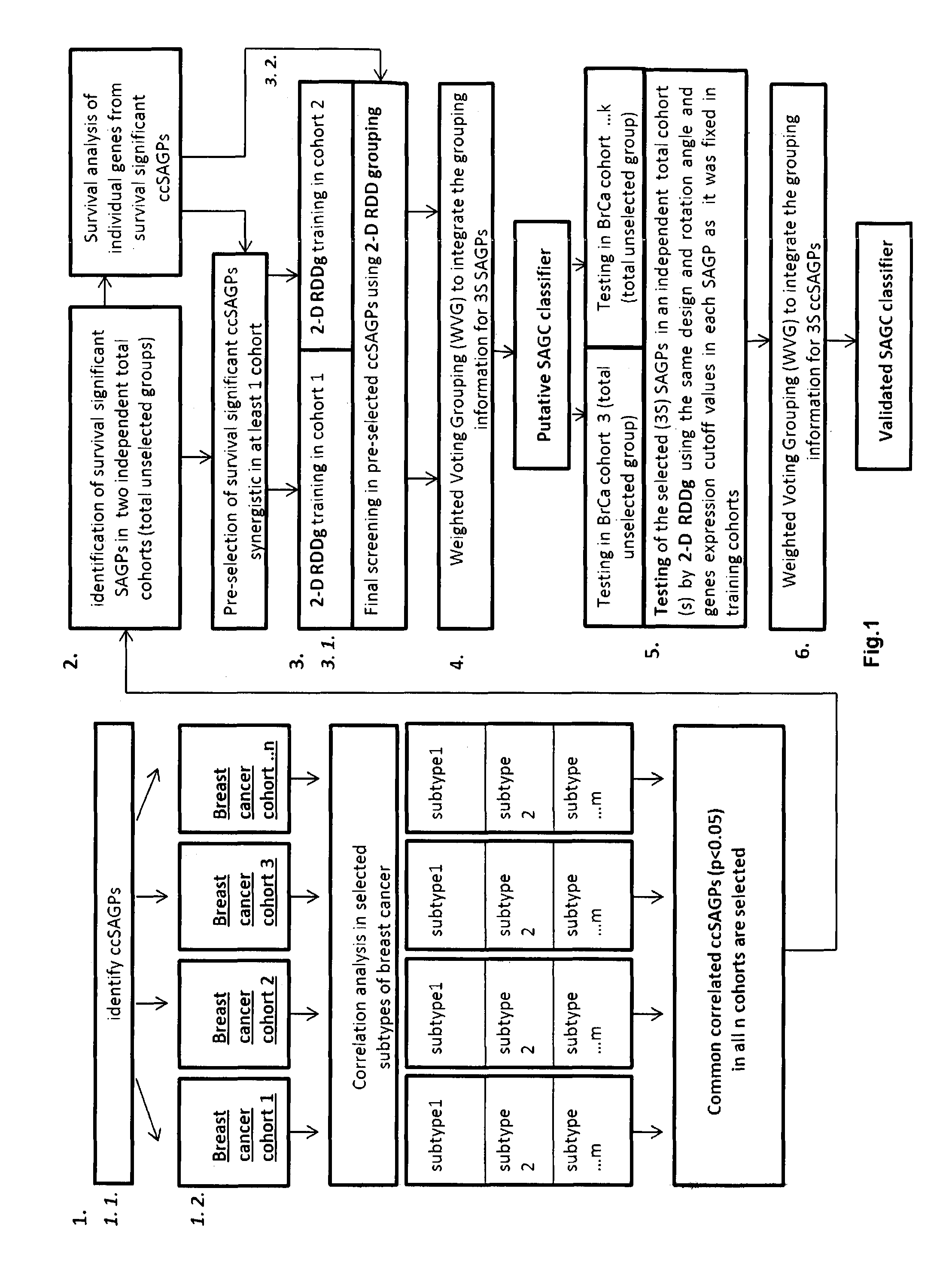
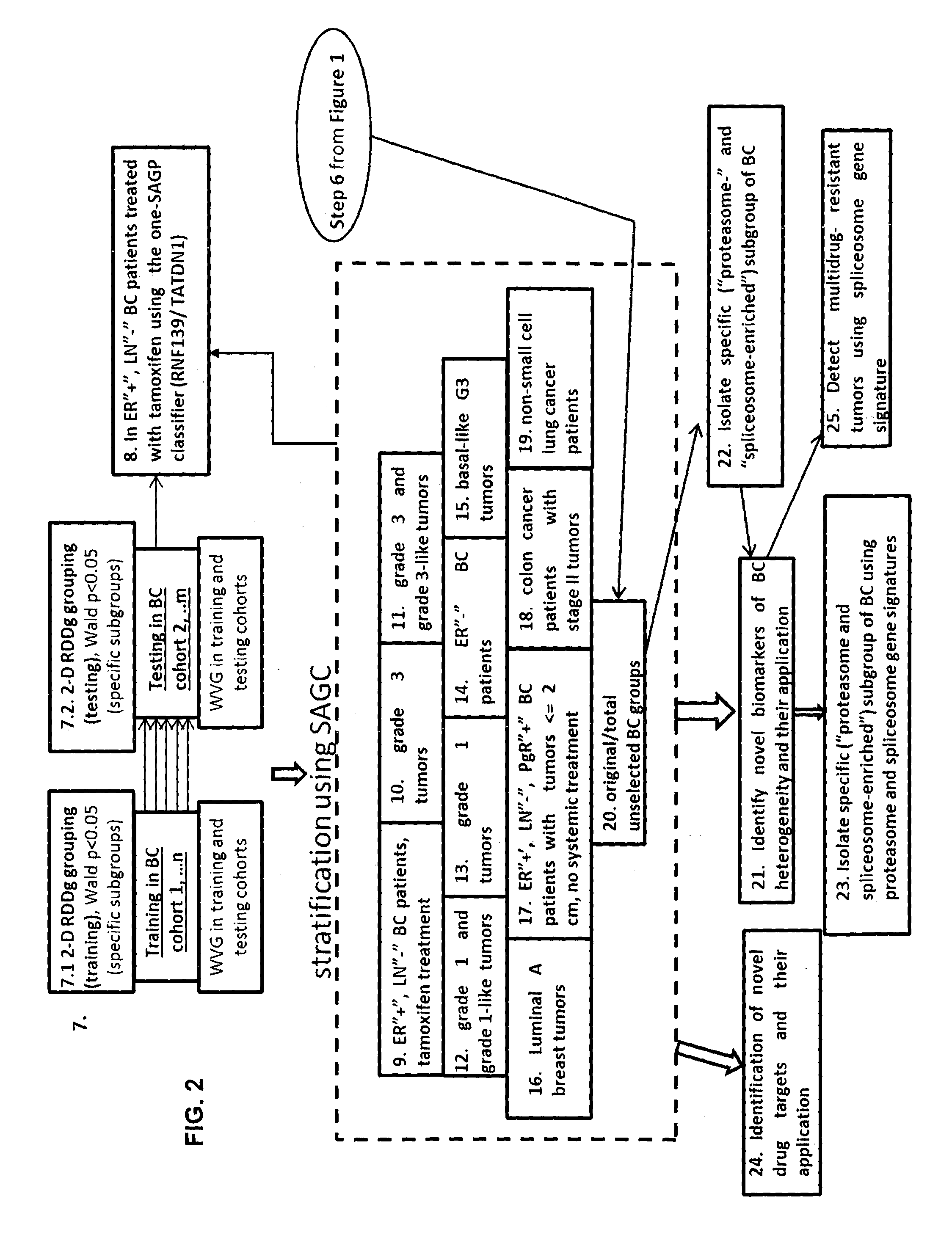
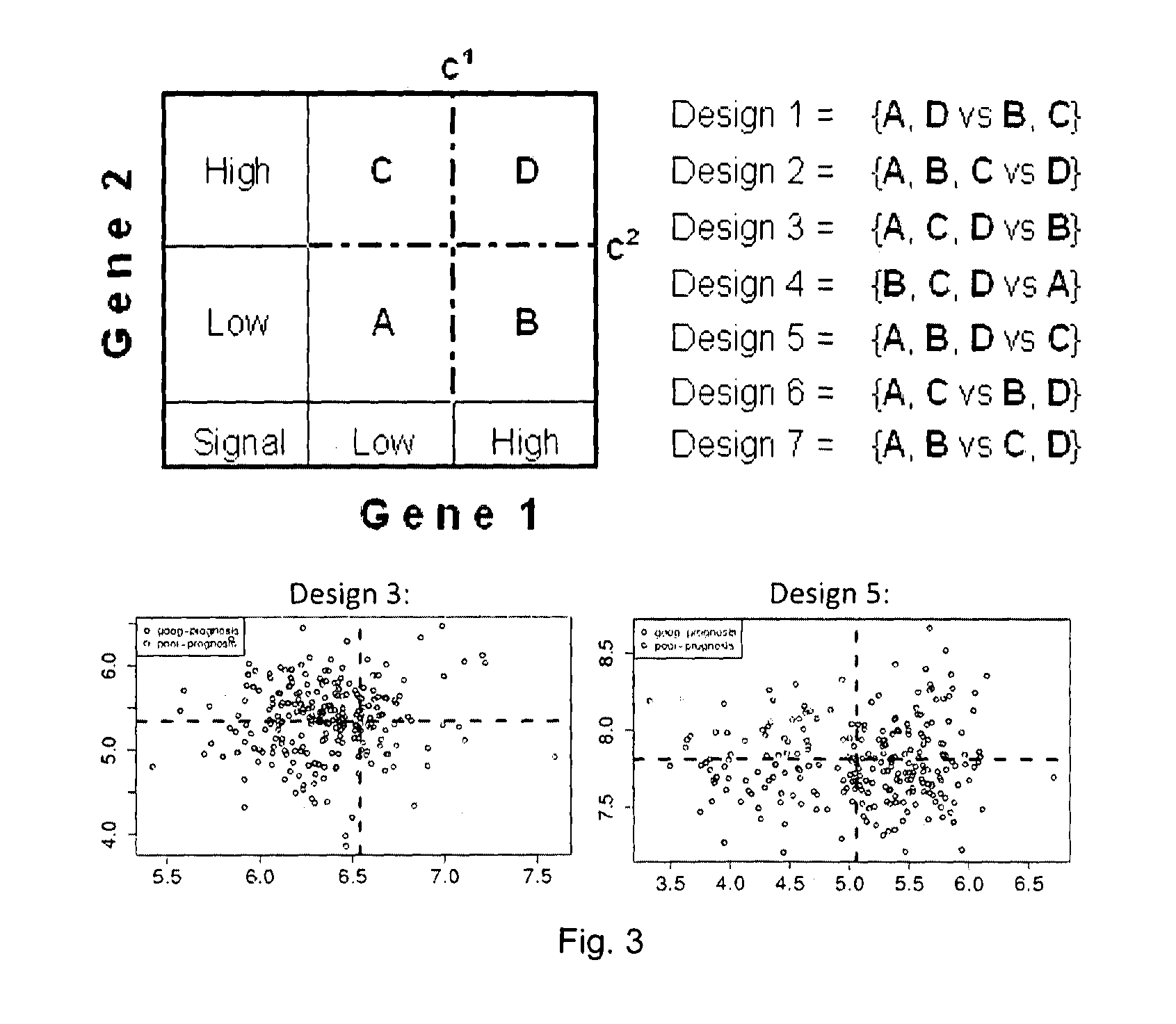
Sense-antisense gene pairs for patient stratification, prognosis, and therapeutic biomarkers identification
InactiveUS20160259883A1Quality improvementHighly prognostically significantMicrobiological testing/measurementLibrary screeningPrognostic signaturePatient stratification
The present invention relates to a method of identification of clinically and genetically distinct sub-groups of patients subject to a medical condition, particularly breast, lung, and colon cancer patients using a composition of respective gene expression values for certain gene pairs. Sense-antisense gene pairs (SAGPs) which are relevant for a medical condition and the disease prognosis are used by the method to generate statistical models based on the expression values of the SAGPs. SAGPs for which the statistical models are found to have high value in prognosis of the variation of medical condition and the diseases are selected and integrated in the prognostic signature including specified parameters (e.g. cut-off values) of the prognostic model. It further relates to using respective gene expression values for these genes to predict patient′ risk groups (in context of patient's survival or / and disease progression) and to using the predicted groups for identification of patient risk, and specific and robust prognostic biomarkers with mechanistic interpretations of biological changes (associated with the gene signatures) appropriating for an implementation of therapeutic targeting.
Owner:AGENCY FOR SCI TECH & RES
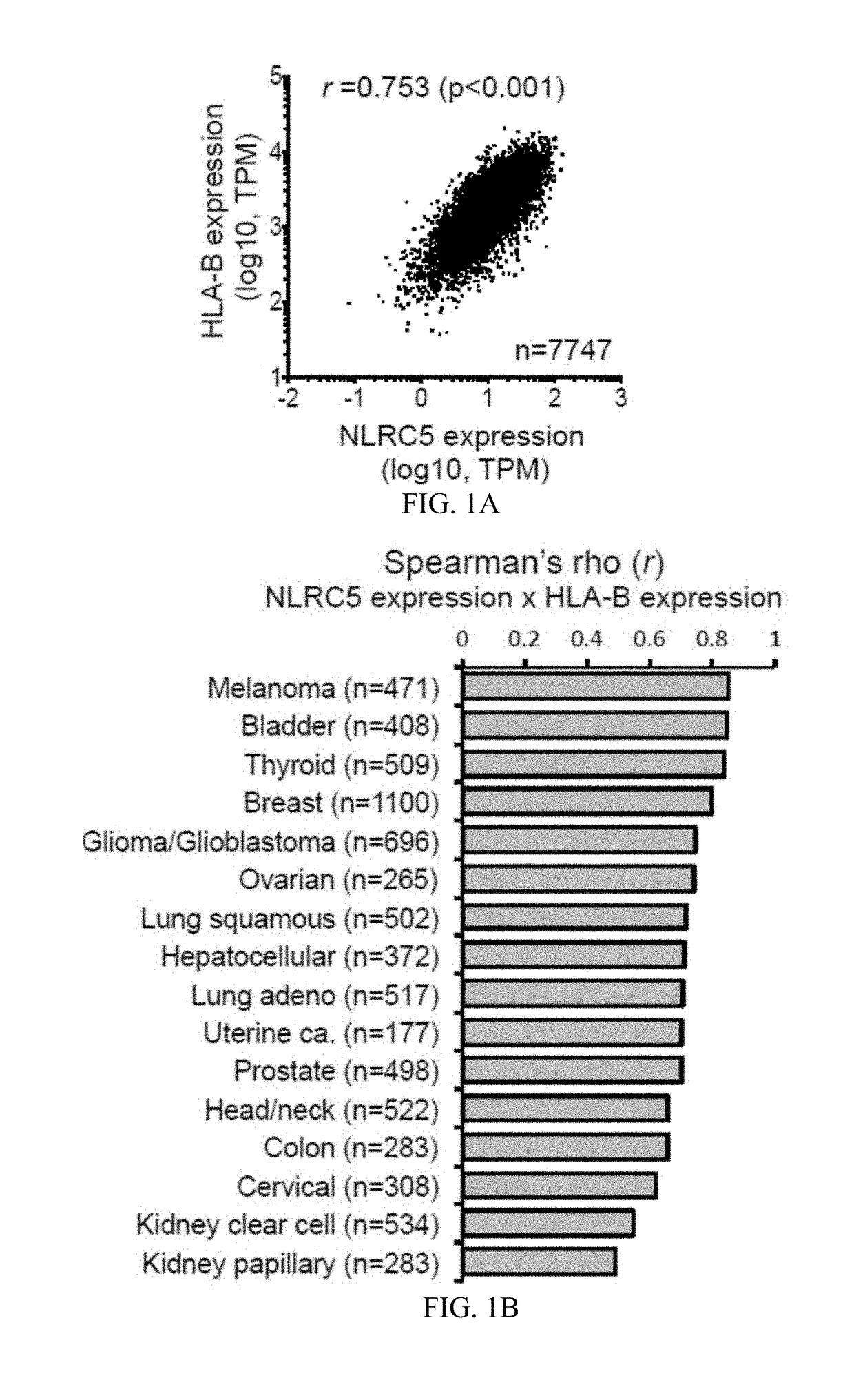
Nlrc5 as a biomarker for cancer patients and a target for cancer therapy
InactiveUS20170321285A1Reduce capacityHigh expressionMicrobiological testing/measurementImmunoglobulins against cell receptors/antigens/surface-determinantsCancer cellBiomarker identification
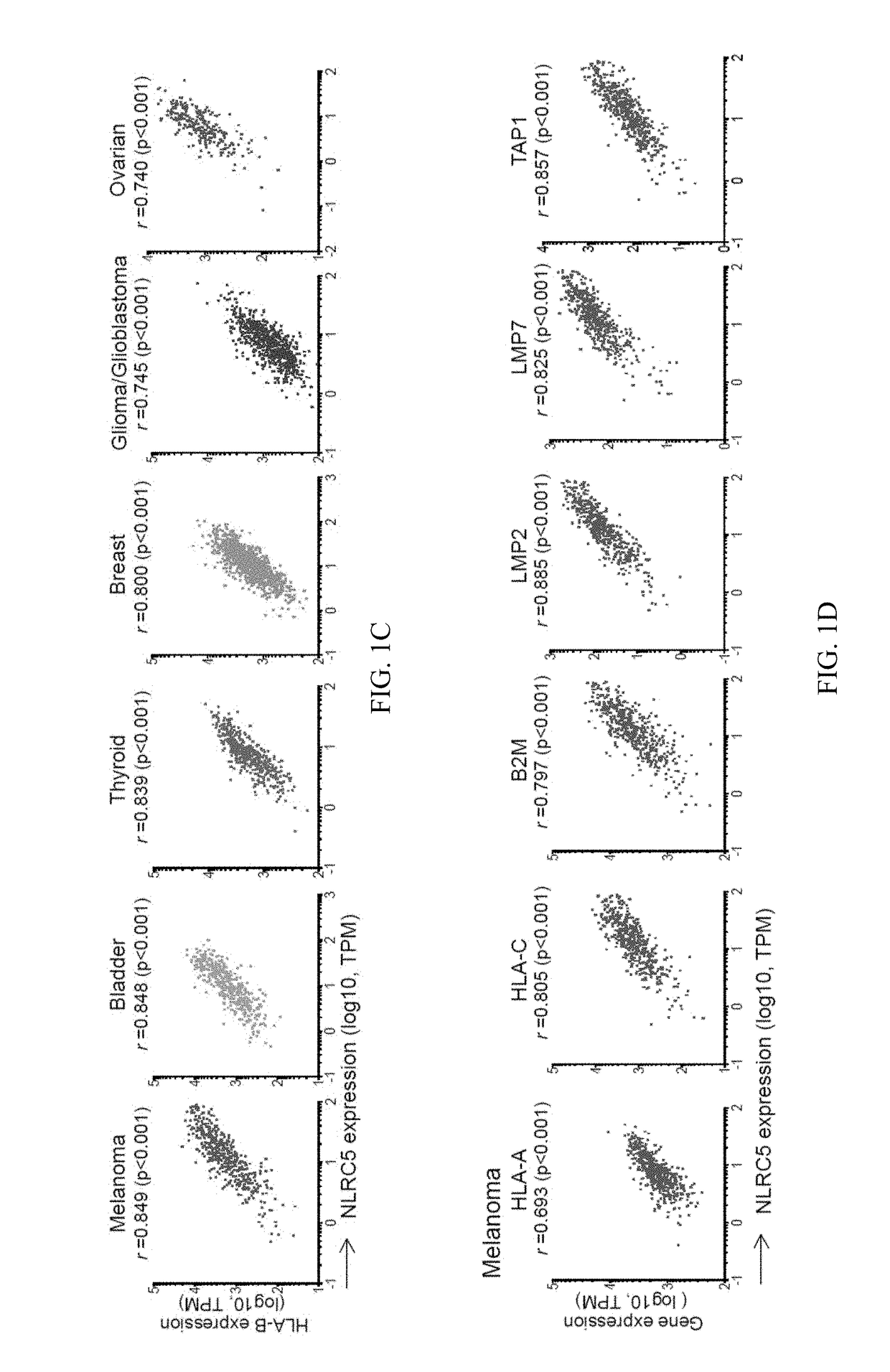
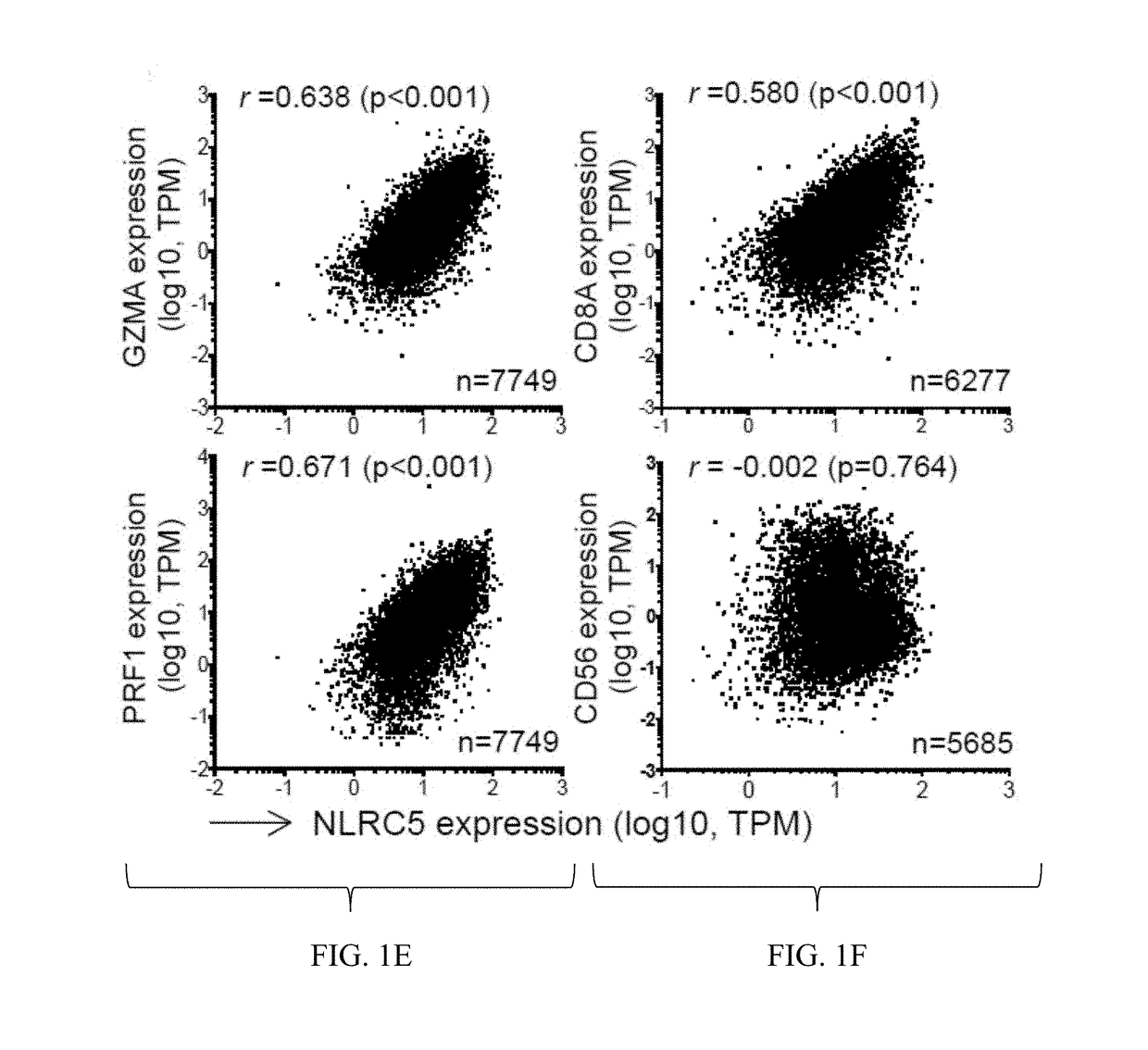
The invention pertains to biomarkers for identifying a cancer that is likely or not likely to evade the immune system of a subject, thus, is likely or not likely to show better prognosis (prognostic biomarker) and / or better responses to cancer therapies (predictive biomarker). The invention provides a method of identifying a subject as having a cancer that is likely to evade the immune system of the subject based on one or more of the following biomarkers in the cancer cells of the subject: a) reduced amount of NLRC5 mRNA or protein; b) reduced activity of NLRC5 protein; c) a mutation that reduces the activity of NLRC5 protein; d) increased methylation of nlrc5 or a portion thereof; and e) reduced copy number of nlrc5. These variables are useful to predict both patient survival (prognostic biomarker) and patient responses to immunotherapies (predictive biomarker). Furthermore, this invention provides a method of identifying a subject as having a cancer that is likely to evade the immune system of the subject with greater prediction power by utilizing multiple variables, in addition to above a)-e) variables, including neoantigen load, mutation number, or expression of genes involved in immune responses, including but not limited to CTLA4, PD1, PD-L1 and PD-L2. The invention also pertains to a method of treating a cancer likely to evade the immune system of the subject by administering an immunotherapy and a therapy designed to activate the MHC class I antigen presentation pathway by activating the expression and / or activity of NLRC5 protein.
Owner:TEXAS A&M UNIVERSITY
Systems and methods for predicting patient outcome to cancer therapy
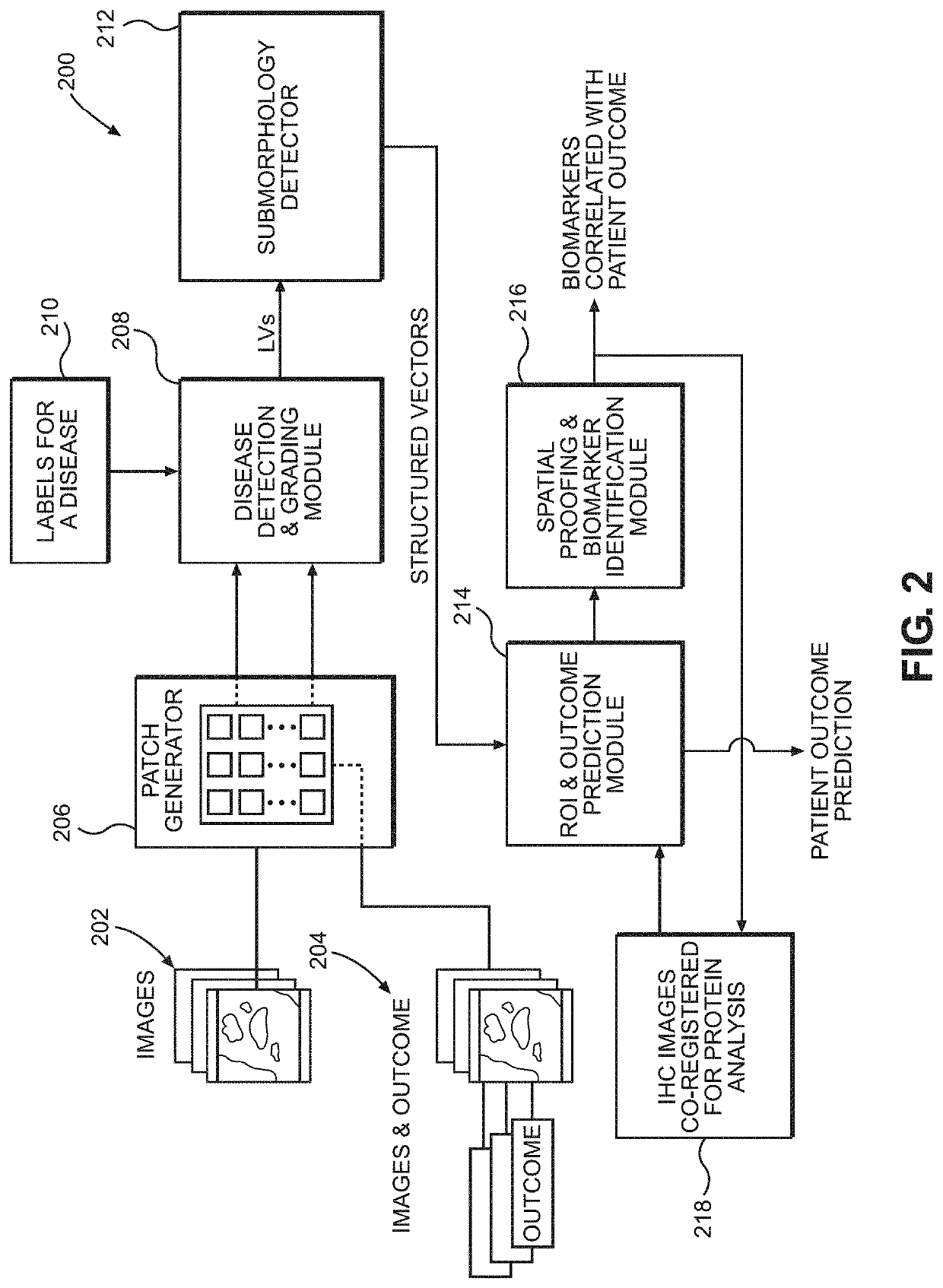
ActiveUS20210193323A1Lower confidenceMedical data miningHealth-index calculationDisease outcomeMorphological pattern
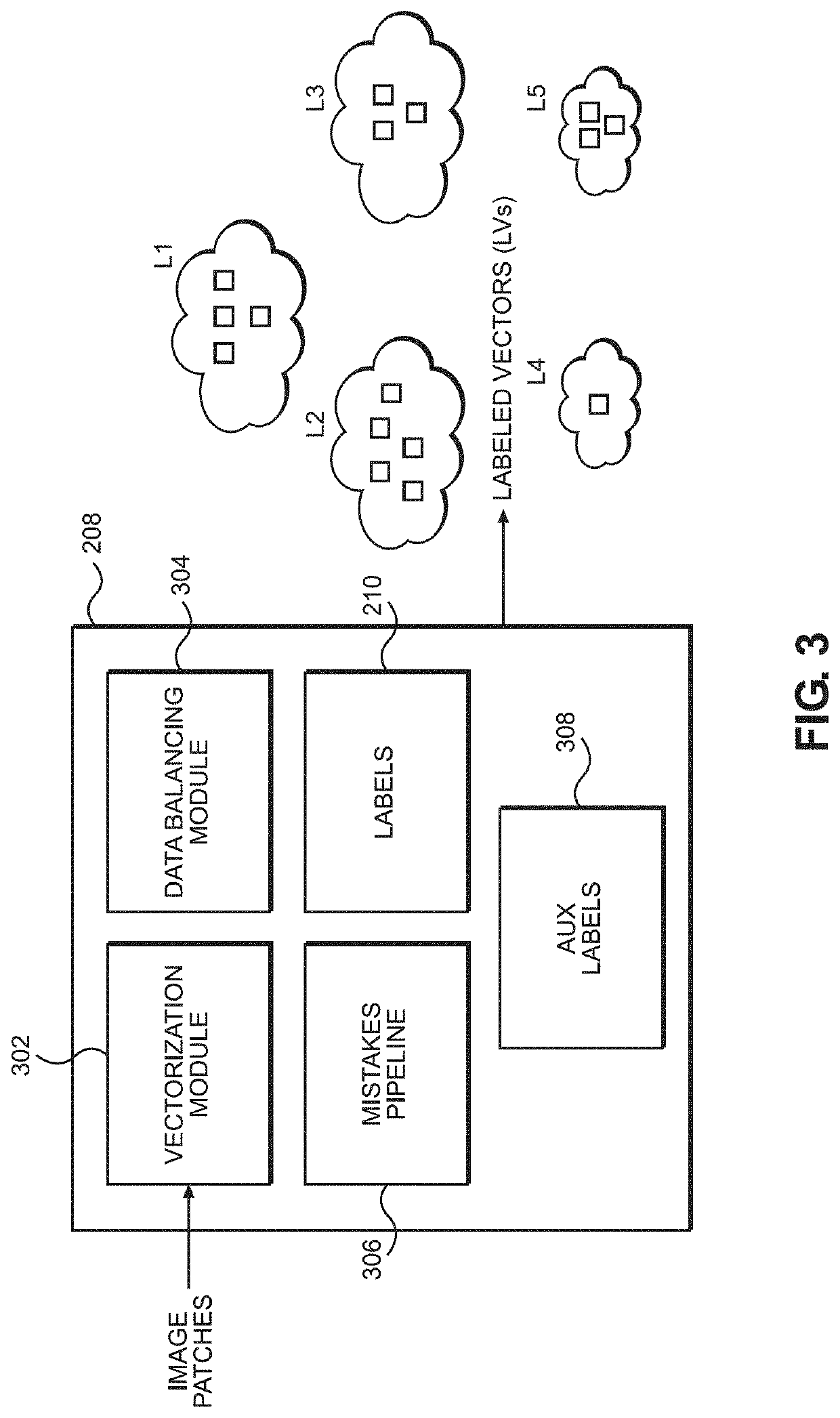
Disclosed are systems and methods for predicting patient response to a treatment option. In one embodiment, the image slides from patient tissue samples are divided into patches and morphological patterns correlated with a disease outcome are labeled and given a patch-level score, based on whether the morphological patterns occur only in patients with good outcomes or patients with poor outcomes. A patient-level score can be generated based, at least partly, on the patch-level scores. Patch-level scores can identify regions of interest for targeted biomarker identification.
Owner:PATHOMIQ INC
Methods and apparatus for identifying disease status using biomarkers
InactiveUS20110077931A1Facilitate ascertaining the disease status of an individualAnalogue computers for chemical processesBiostatisticsBiomarker identificationBiomarker (petroleum)
Methods and apparatus for identifying disease status according to various aspects of the present invention include analyzing the levels of one or more biomarkers. The methods and apparatus may use biomarker data for a condition-positive cohort and a condition-negative cohort and select multiple relevant biomarkers from the plurality of biomarkers. The system may generate a statistical model for determining the disease status according to differences between the biomarker data for the relevant biomarkers of the respective cohorts. The methods and apparatus may also facilitate ascertaining the disease status of an individual by producing a composite score for an individual patient and comparing the patient's composite score to one or more thresholds for identifying potential disease status.
Owner:PROVISTA DIAGNOSTICS INC
Biomarkers to identify patients that will respond to treatment and treating such patients
ActiveUS9663825B2Microbiological testing/measurementBiological material analysisGenes mutationCancer type
The invention relates to methods for using biomarkers to identify cancer patients that will or are likely to respond to treatment. Specifically, the invention relates to the use of one or more of three association studies of cancer types, gene mutations, or gene expression levels in order to identify cancer patients that will or are likely to respond to treatment with a histone deacetylase (HDAC) inhibitor, alone or in combination with another cancer treatment. The methods may, optionally, further include treating such patient with an HDAC inhibitor, alone or in combination with another cancer treatment.
Owner:ACETYLON PHARMA
Effects of Inhibitors of Fgfr3 on Gene Transcription
InactiveUS20090048266A1Improve responseOrganic active ingredientsMicrobiological testing/measurementMedicineBiomarker identification
Methods of utilizing biomarkers to identify patients for treatment or to monitor response to treatment are taught herein. Alterations in levels of gene expression of the biomarkers, particularly in response to FGFR3 inhibition, are measured and identifications or adjustments may be made accordingly.
Owner:NOVARTIS AG
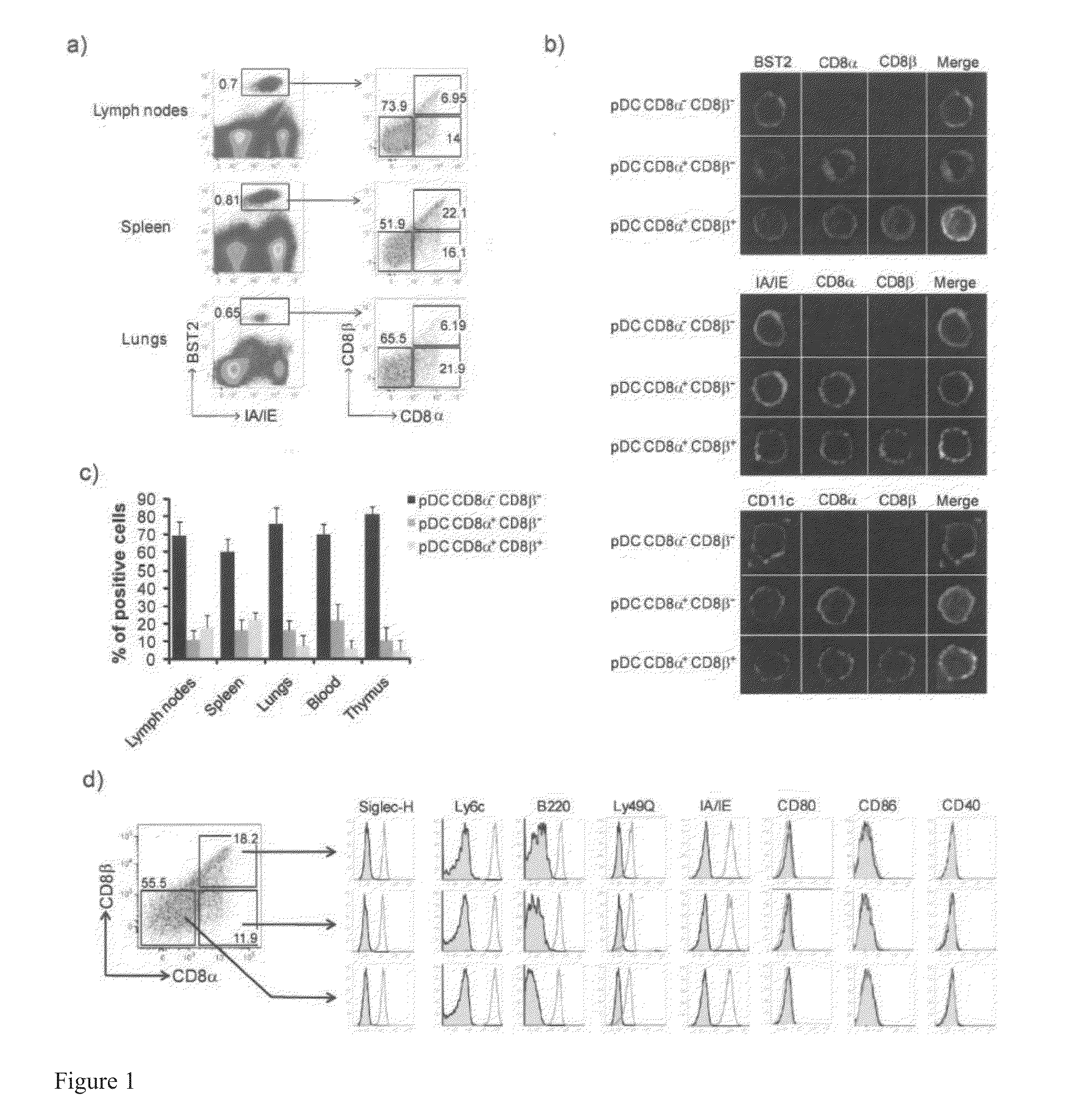
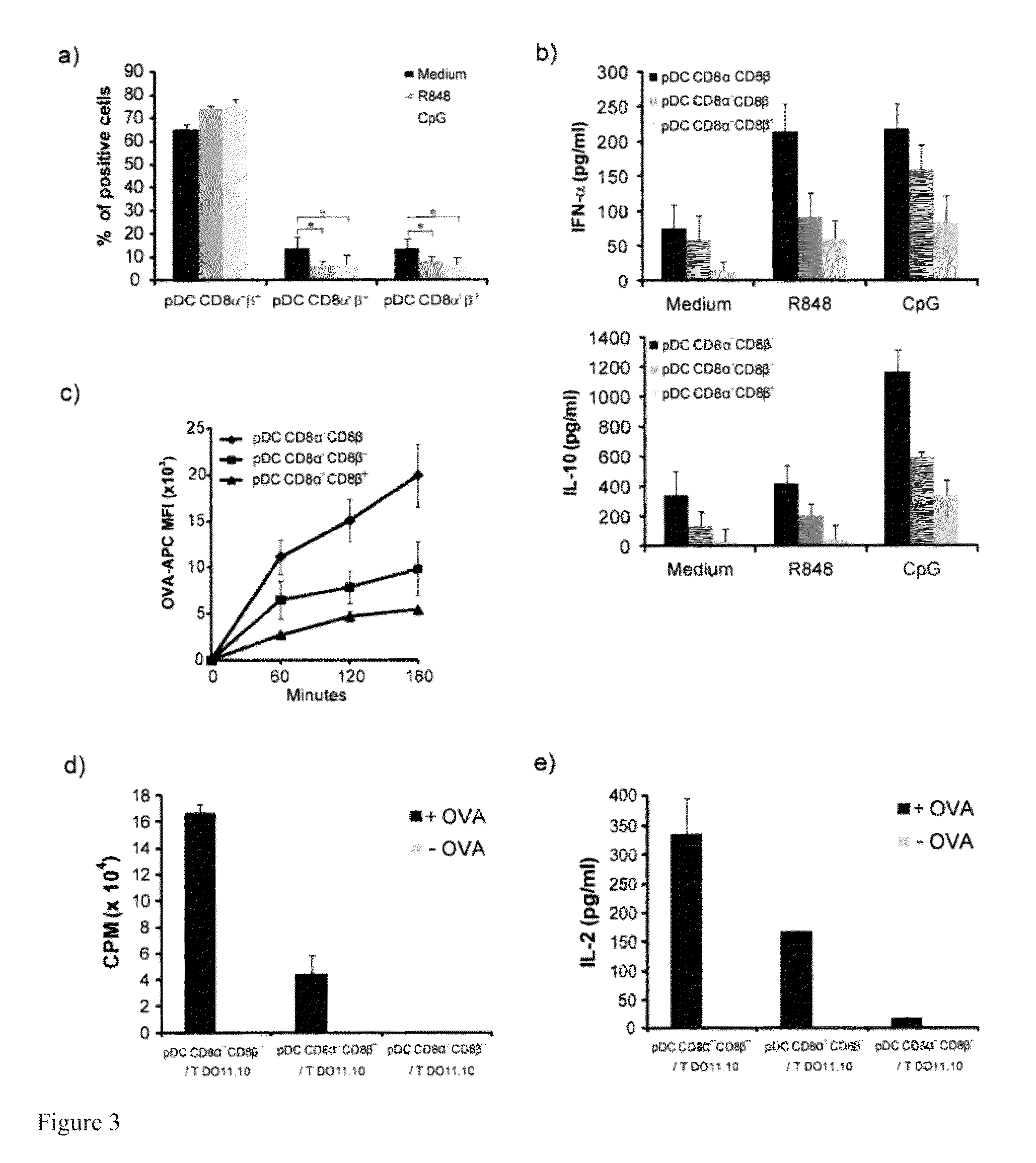
Tolerogenic Plasmacytoid Dendritic Cells Co-Expressing Cd8-Alpha And Cd8-Beta And Methods Of Inducing The Differentiation Of Regulatory T Cells Using Same
InactiveUS20130142830A1Avoid developmentSpeed up the conversion processMicrobiological testing/measurementSnake antigen ingredientsImmunologic disordersPlasmacytoid dendritic cell
This invention discloses an unexpected discovery that plasmacytoid dendritic cells (pDCs) may be segregated into immunogenic or tolerogenic species based on novel biomarkers discovered herein. Exemplary biomarkers include CD8α+β+, CD8α+β−, CD8α−β−, C1q, and IL-9R. For example, pDCs with CD8α+β+, CD8α+β− are tolerogenic and CD8α−β− is immunogenic. Also disclosed are isolated pDCs, compositions comprising the pDCs, methods for isolating the pDCs, methods for treating immune-hyper-reactivity, such as airway hyper-reactivity, food allergy, asthma, and autoimmune disorders, by using compositions containing tolerogenic antigen presenting cells, preferably pDCs disclosed herein. Also disclosed are methods for identifying tolerogenic antigen presenting cells by using one or more novel biomarkers disclosed herein, including RALDH expression, CD8α, CD8βC1qa, C1qc, and IL-9R. Also disclosed are methods for inducing Treg cells by using the pDCs disclosed herein.
Owner:UNIV OF SOUTHERN CALIFORNIA
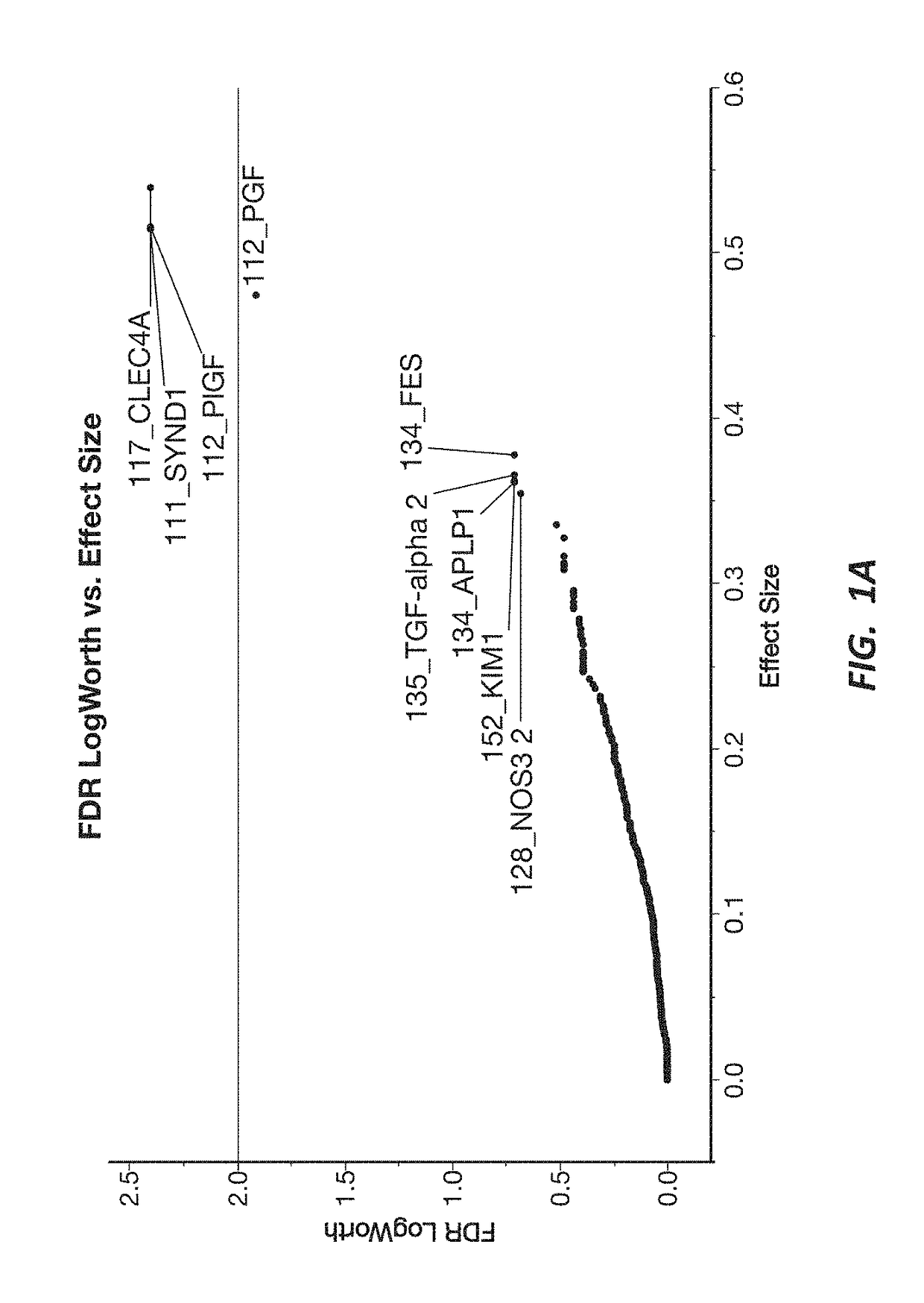
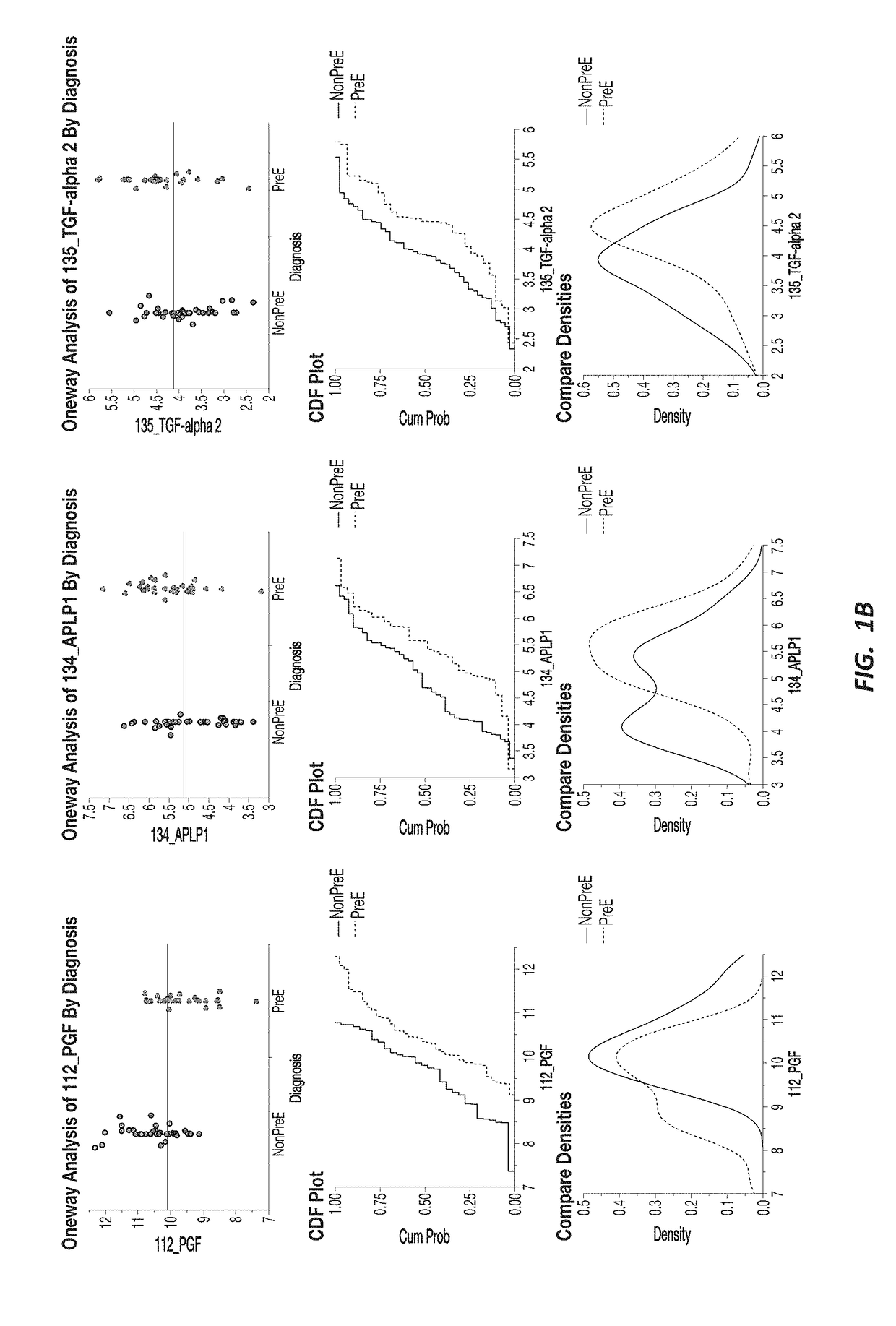
Preeclampsia biomarkers and related systems and methods
Disclosed herein are methods, kits, tests, and systems for detecting, predicting, monitoring, or ruling out preeclampsia in pregnant women. Also provided herein are novel diagnostic markers, methods of data analysis, assay formats, and kits employing such markers to improve one or more characteristics of a test for identifying or ruling out preeclampsia based on biomarkers from patient samples.
Owner:BIORA THERAPEUTICS INC
Methods for biomarker identification and biomarker for non-small cell lung cancer
InactiveUS20120004116A1Health-index calculationMicrobiological testing/measurementBiomarker identificationBiomarker (petroleum)
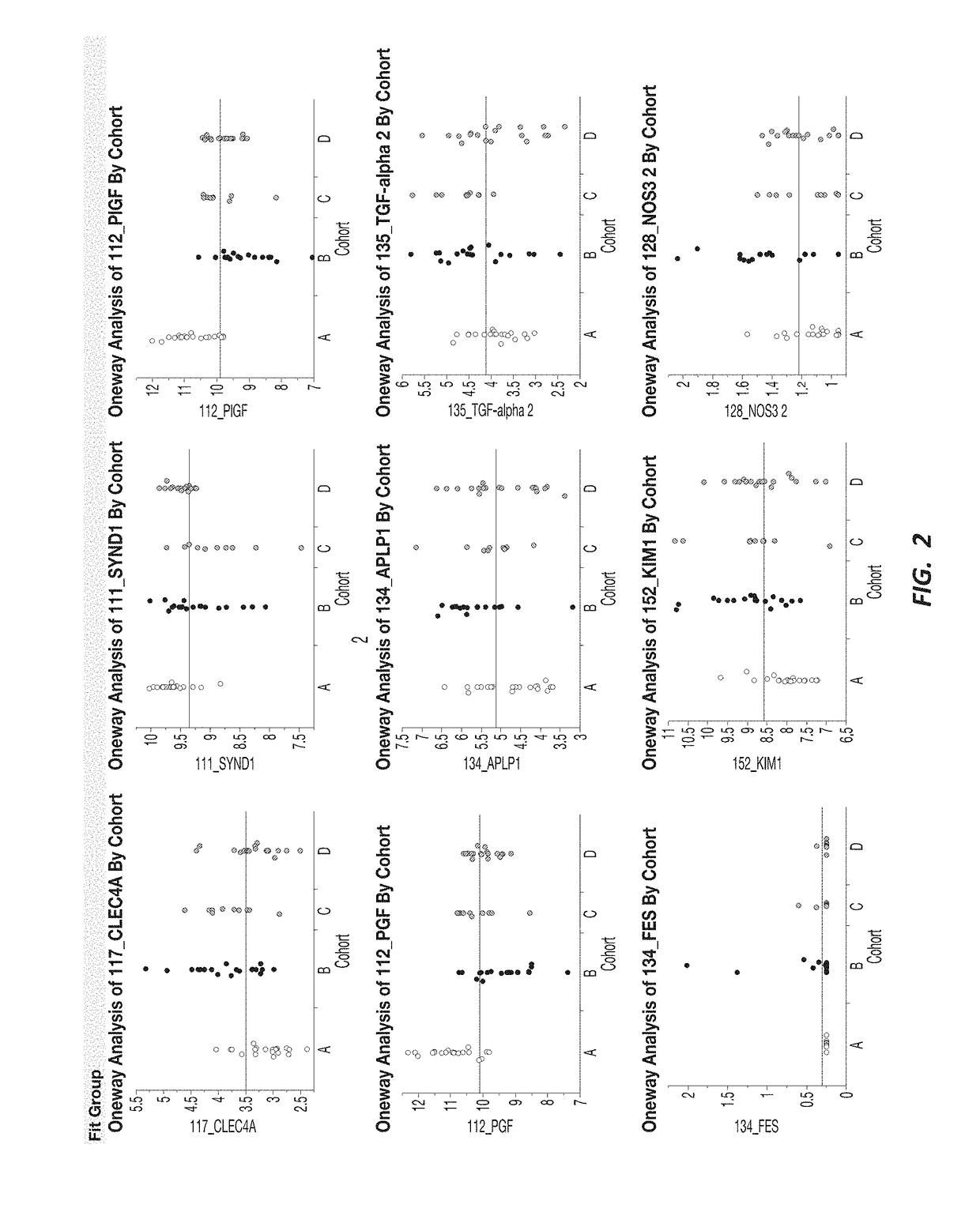
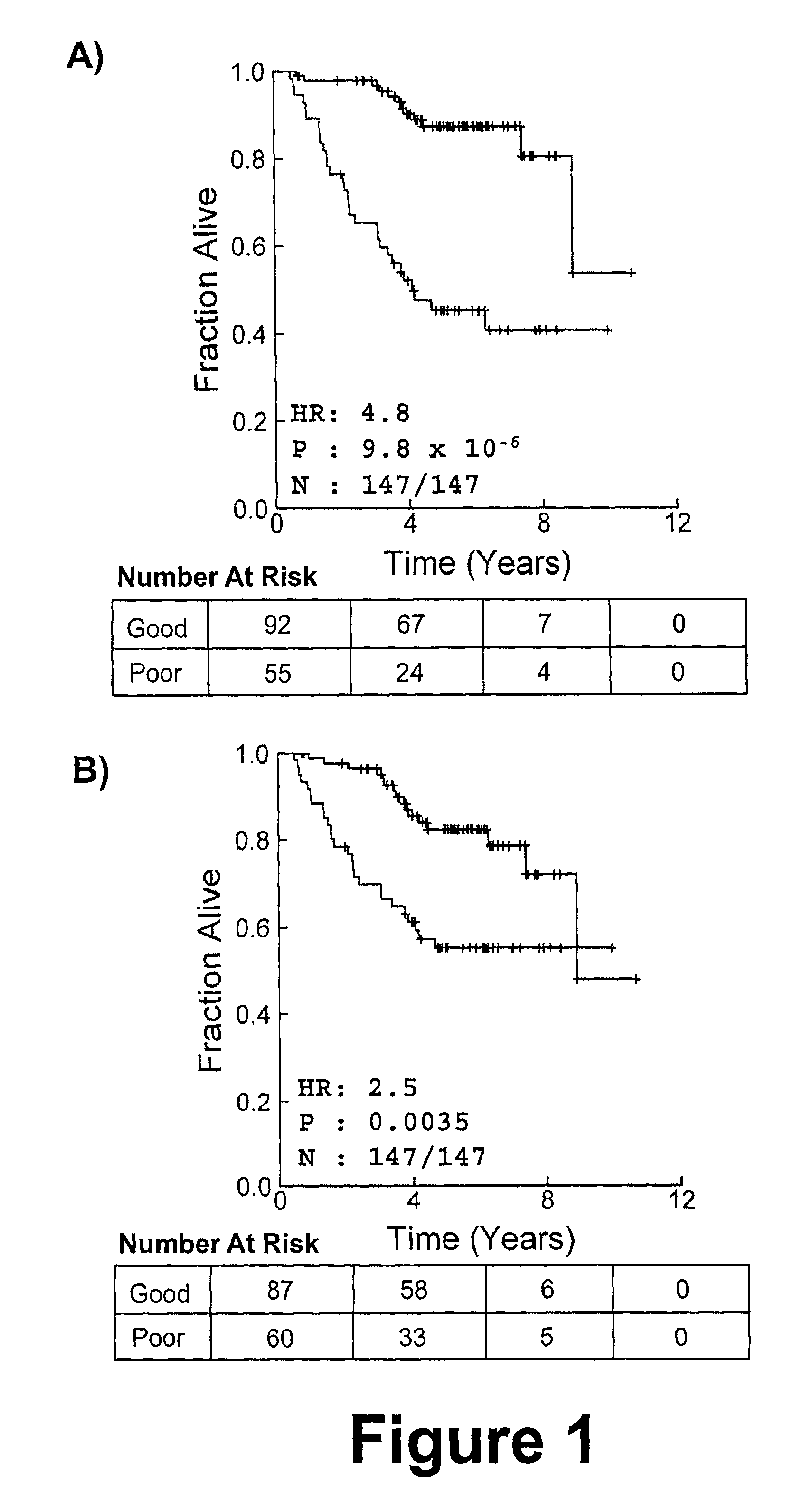
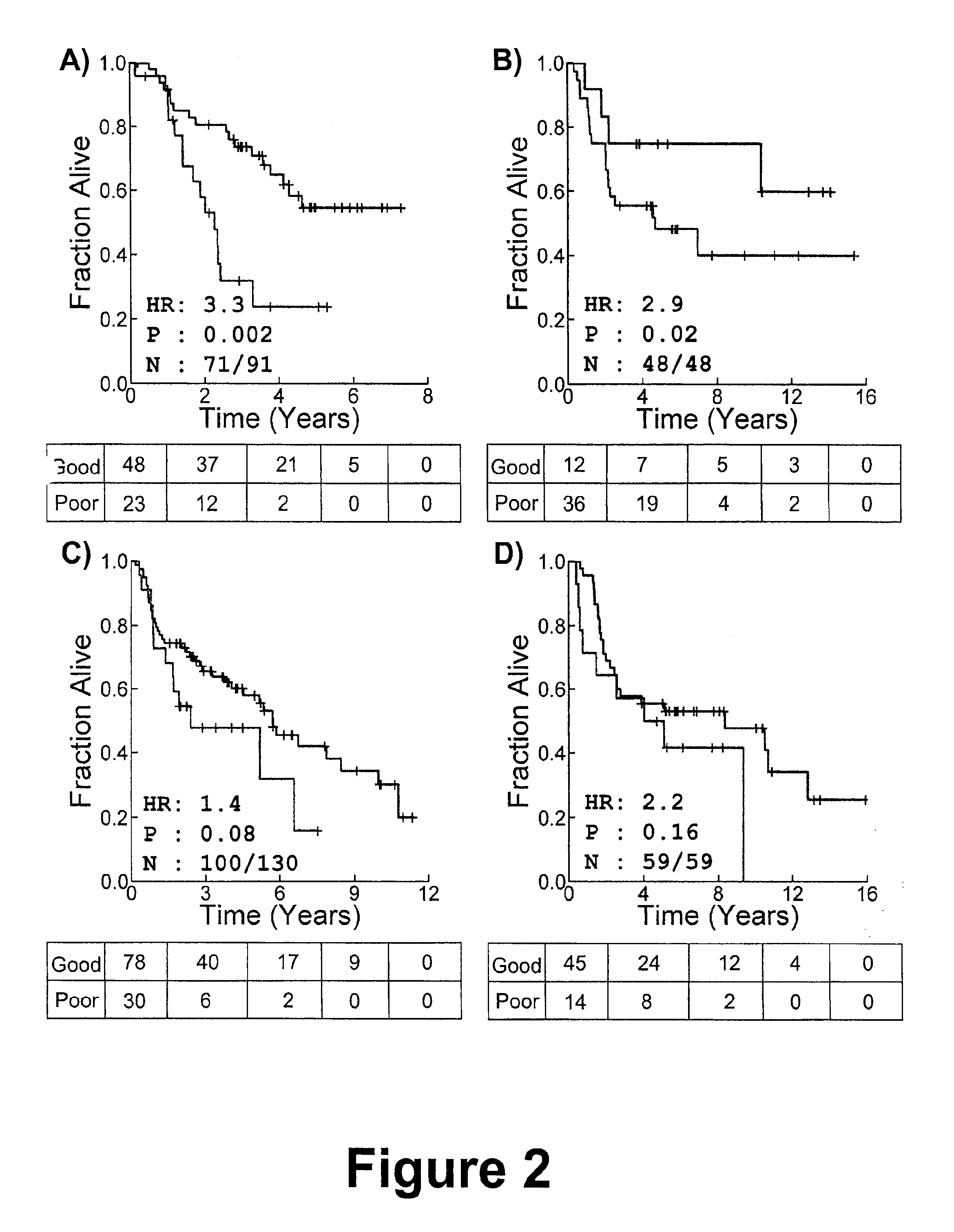
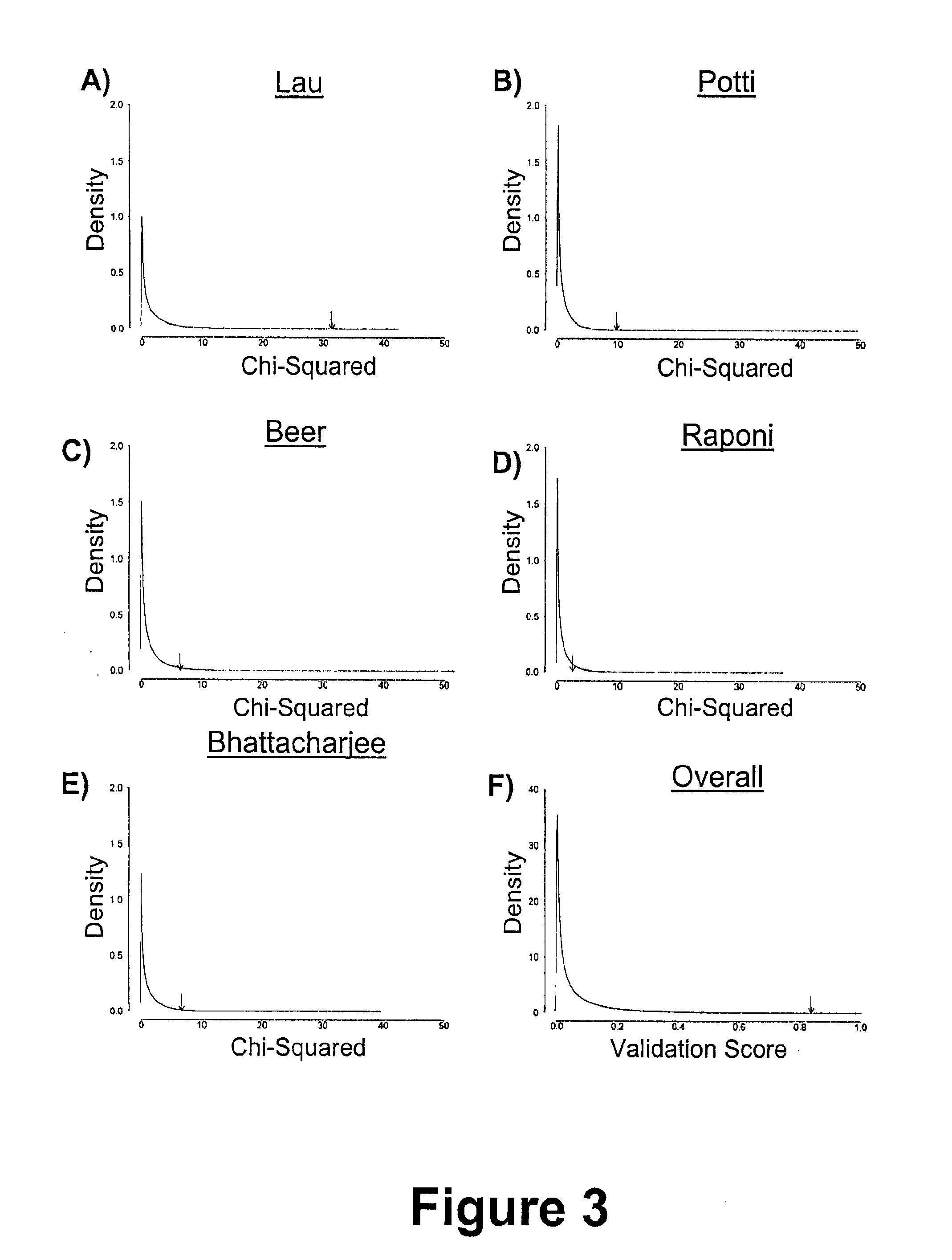
There is provided a method for identifying a biomarker, such as a gene signature, associated with a biological parameter A 6-gene signature for non-small cell lung cancer (NSCLC) is also provided, as well as a method of prognosing or classifying a subject with non-small cell lung cancer into a poor survival group or a good survival group, using said gene signature
Owner:UNIV HEALTH NETWORK
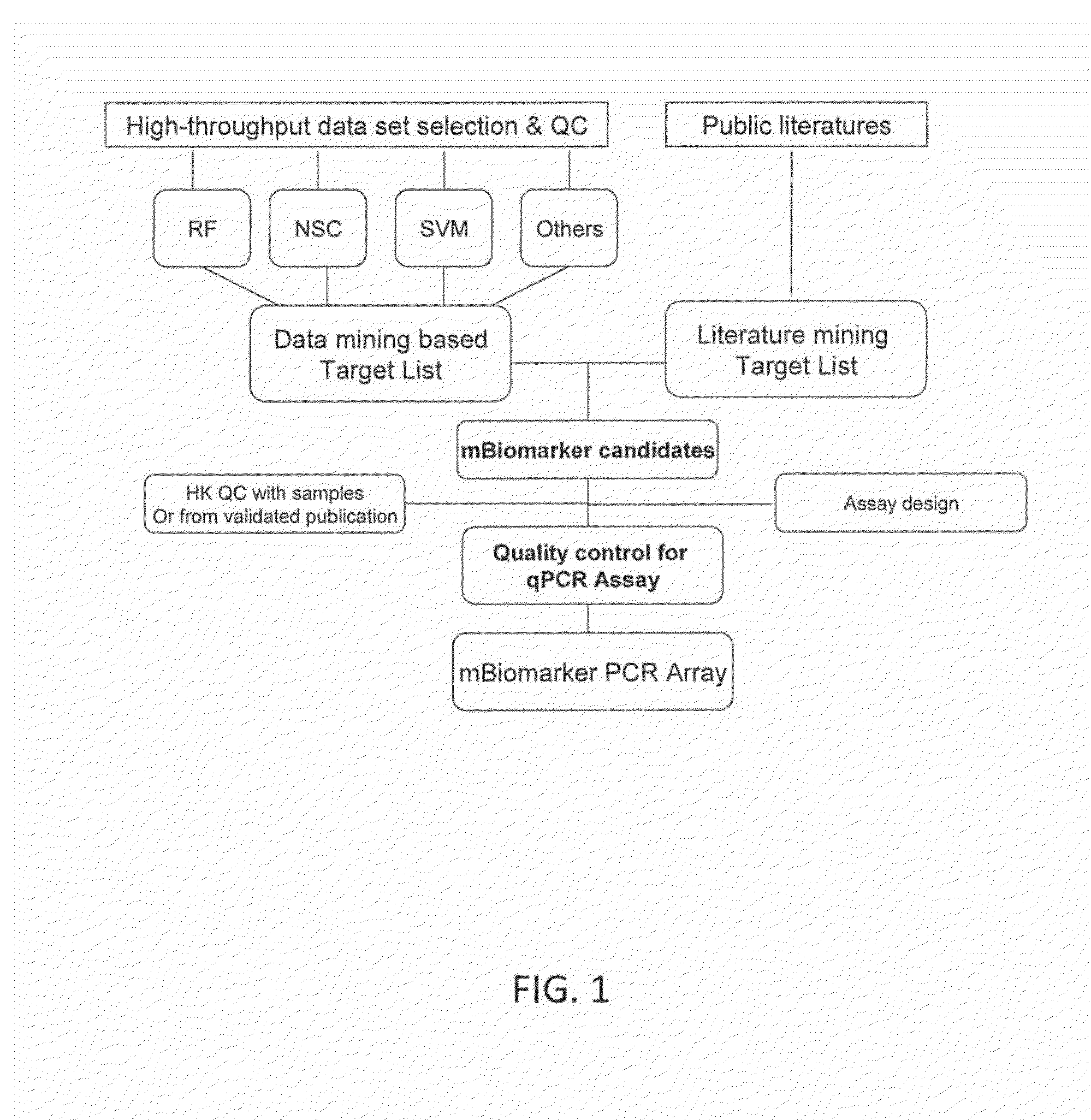
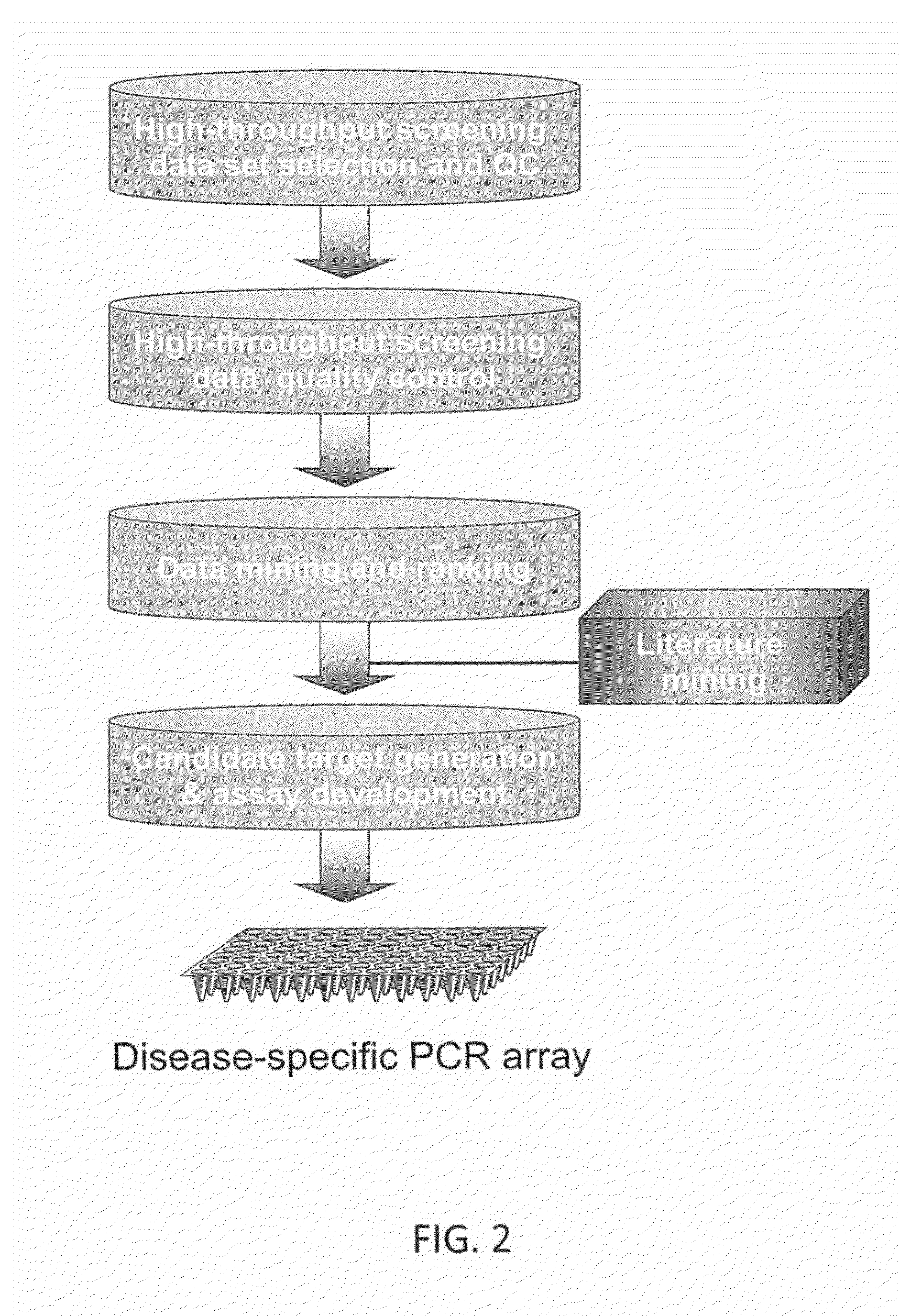
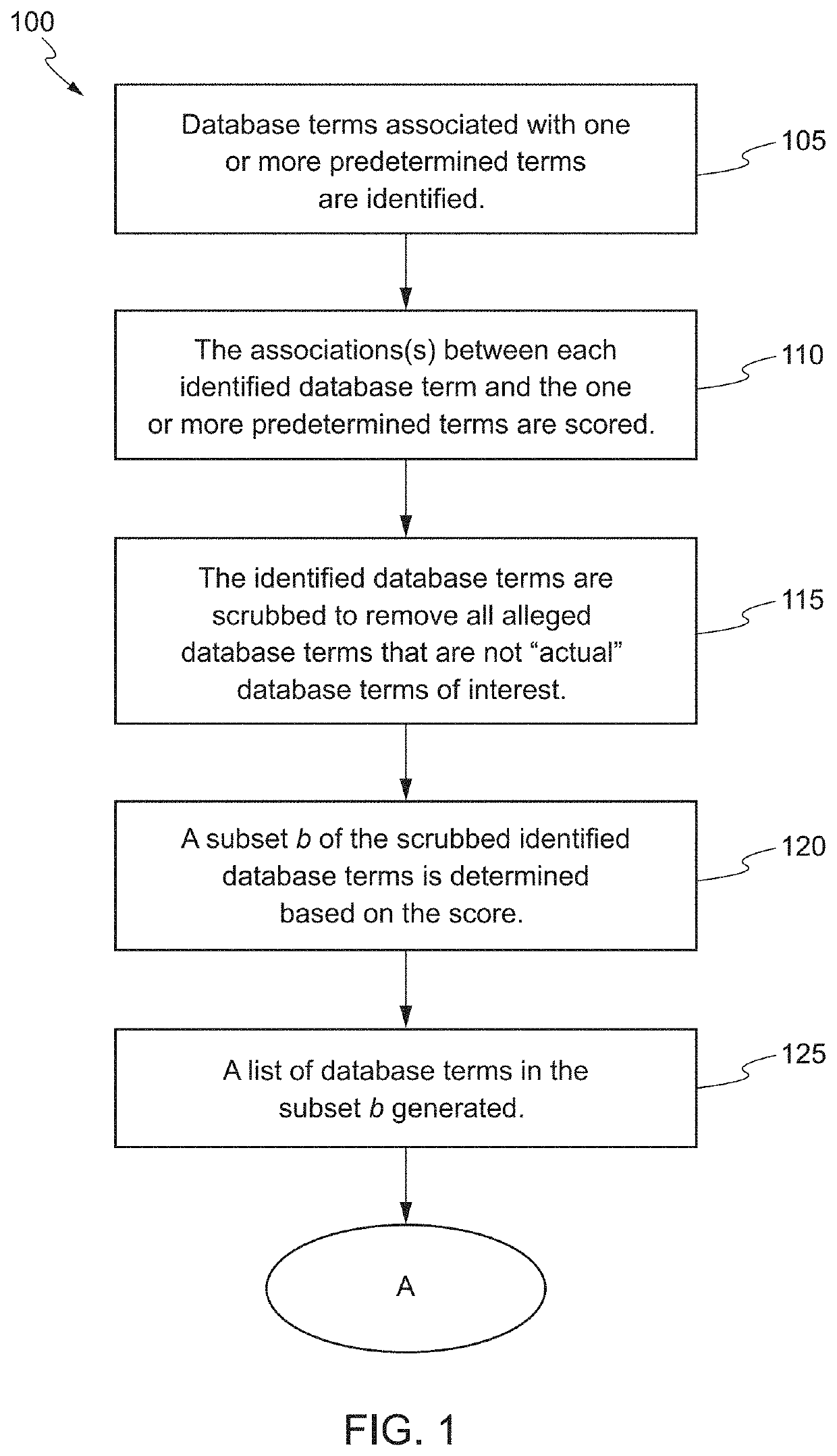
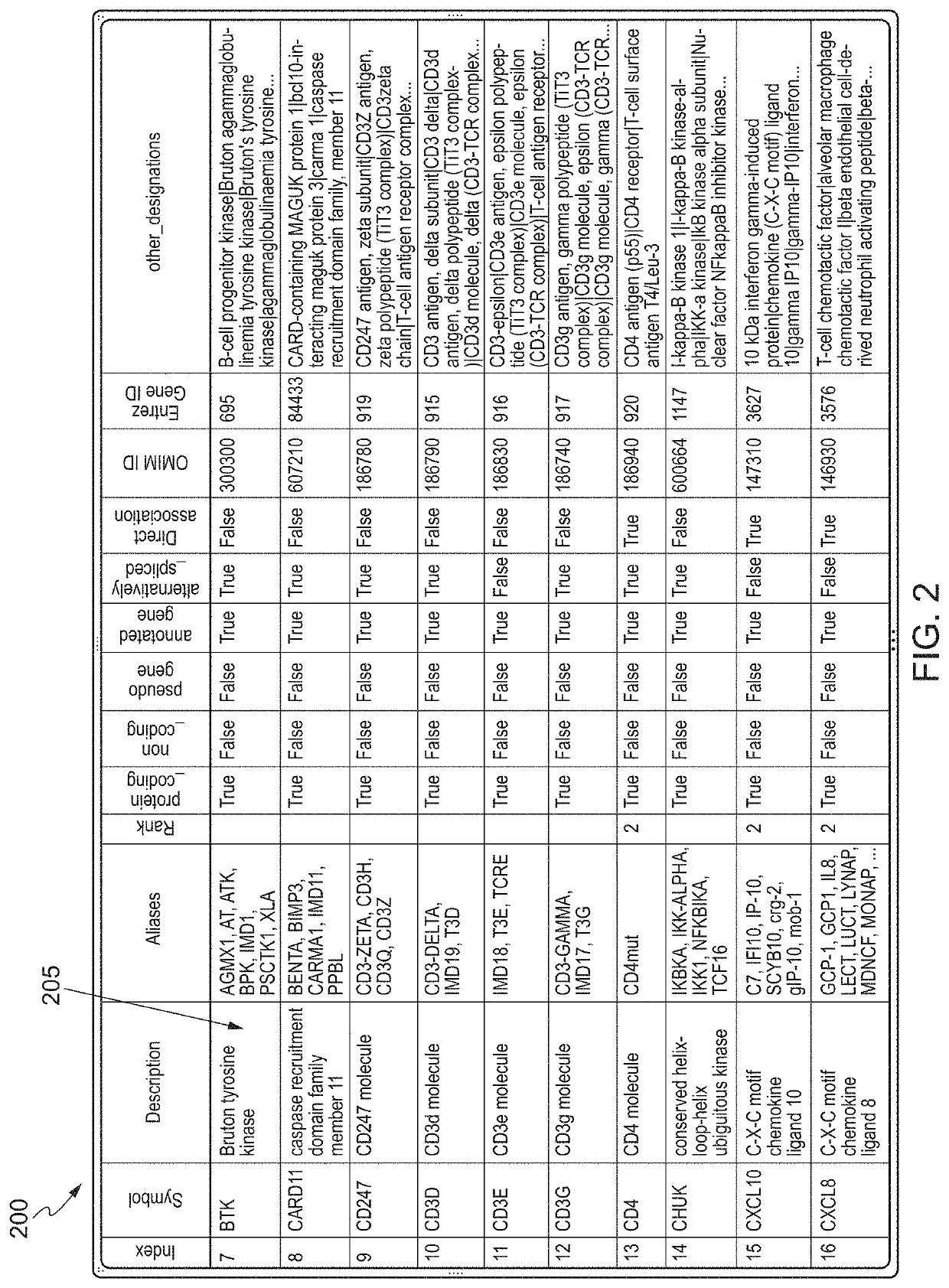
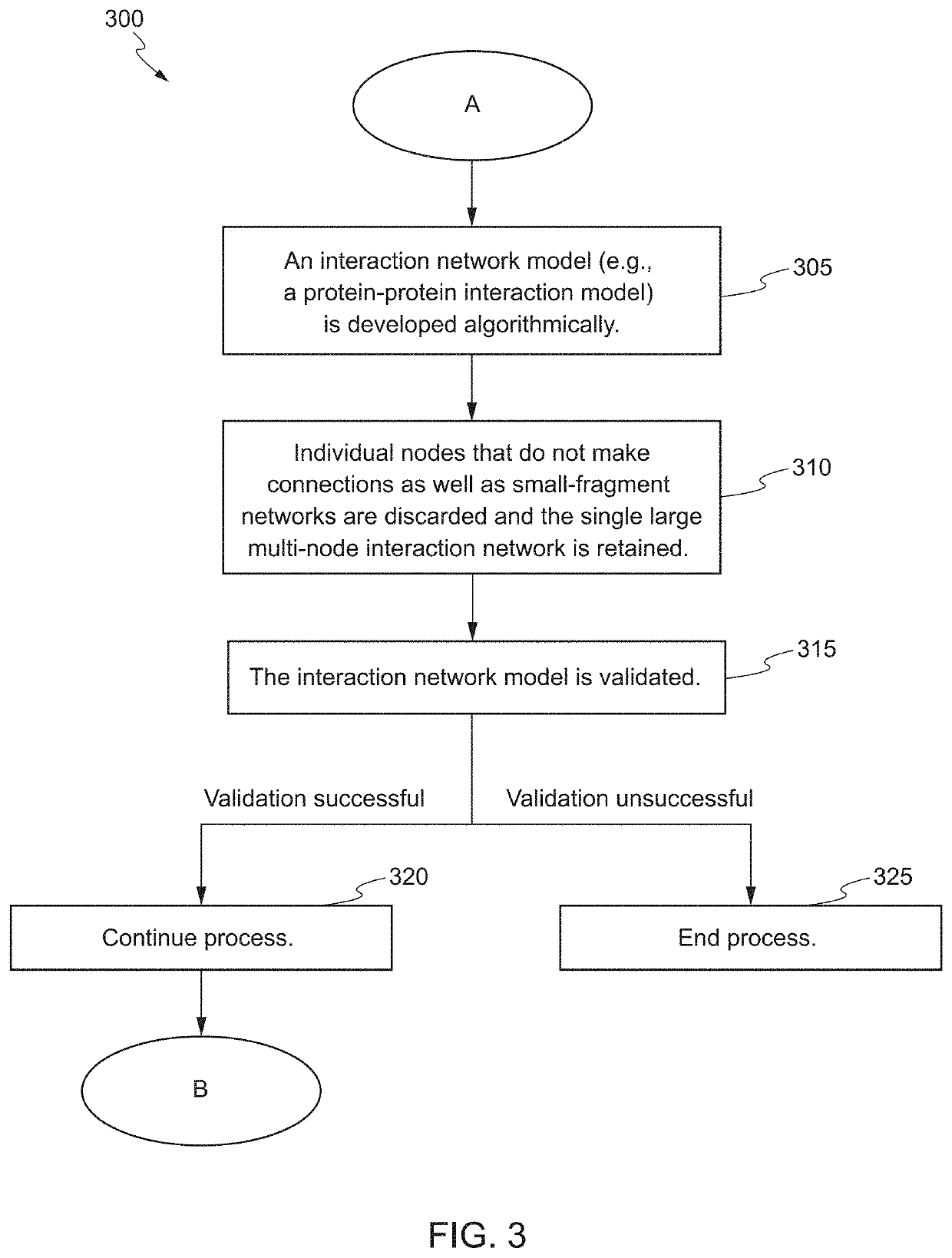
Systems and Methods for Biomarker Identification
ActiveUS20190065665A1Microbiological testing/measurementBiostatisticsKnowledge-based systemsInteraction nets
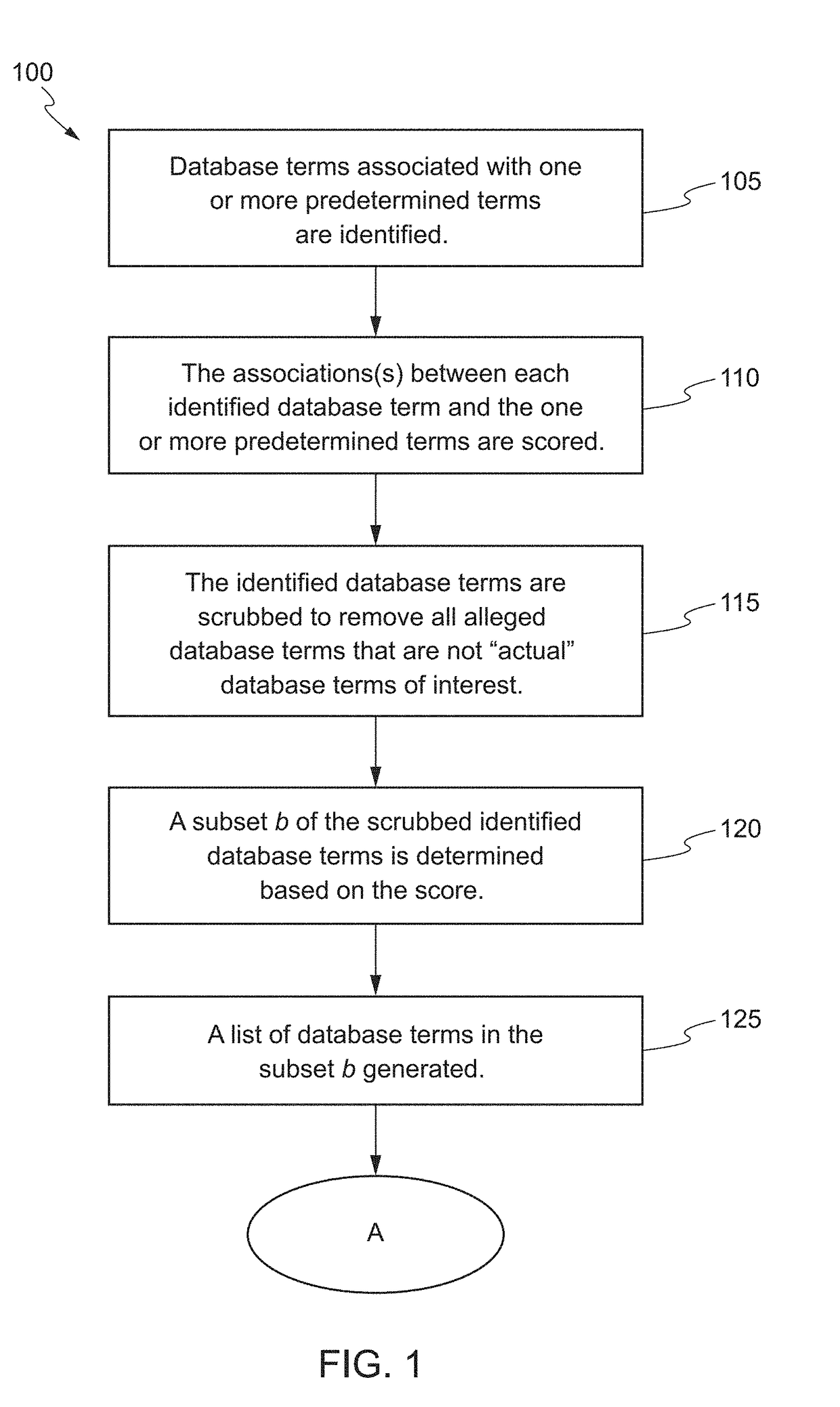
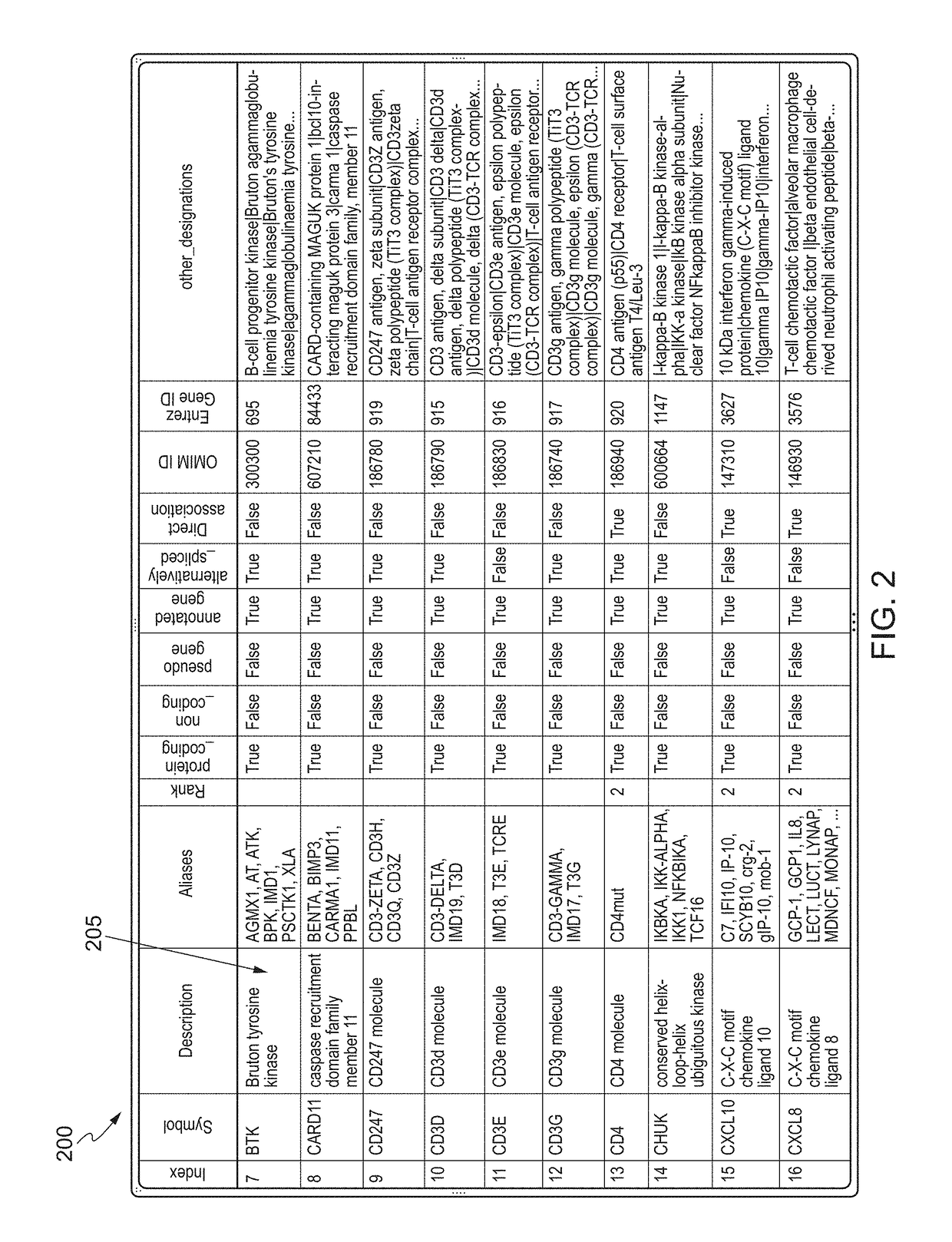
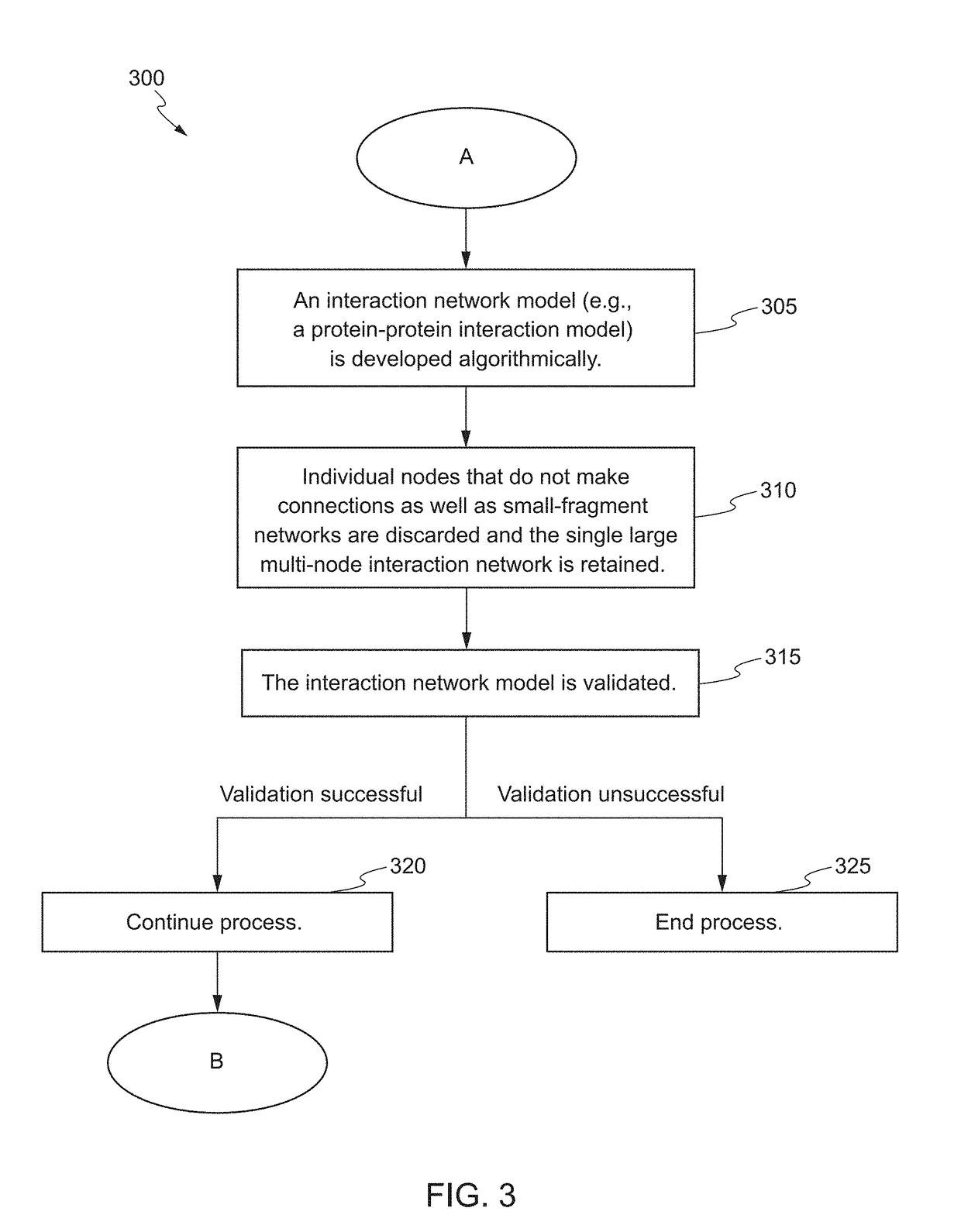
The present invention relates to systems and methods for identifying a biomarker from associative and knowledge based systems and processes. Particularly, aspects of the present invention are directed to a computer implemented method that includes data mining one or more public sources of biomedical text, scientific abstract, or bioinformatic data using queries to identify database terms associated with one or more predetermined terms, scoring association(s) between each of the identified database terms and the one or more predetermined terms, determining a subset b based on the score of the association(s), developing an interaction network model comprising the database terms in subset b, interactions, and additional database terms using a combination of algorithms in a predetermined order, and identifying candidate biomarkers from the interaction network model based on a ranking of the database terms in subset b and the additional database terms in the interaction network model.
Owner:LAB OF AMERICA HLDG
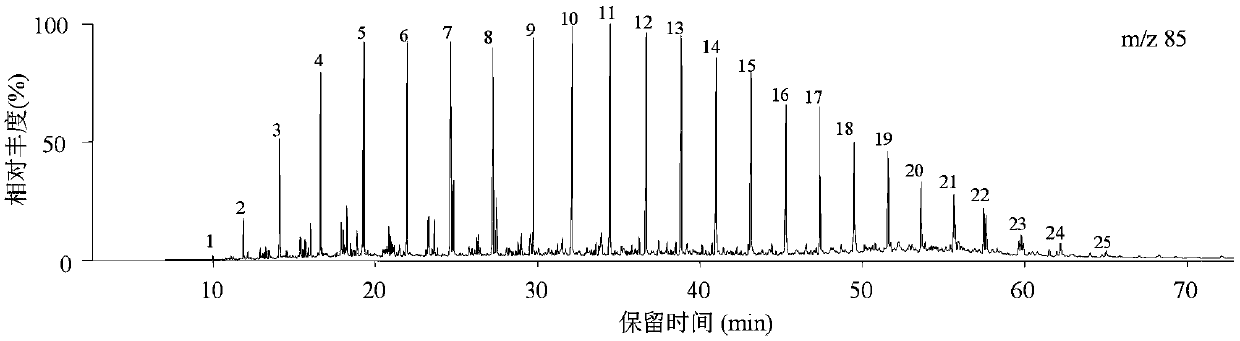
Crude oil density prediction method based on saturated hydrocarbon biomarker parameters
ActiveCN109556990AFew influencing factorsEasy to operateComponent separationComplex mathematical operationsGas chromatography–mass spectrometryPredictive methods
The invention discloses a crude oil density prediction method based on saturated hydrocarbon biomarker parameters, comprising of: collecting crude oil saturated hydrocarbon biomarkers comprising biomarkers of normal paraffin series, terpenoid series, steroid series, and alkyl cyclohexane series for identification and relative quantification; calculating the saturated hydrocarbon parameters of thecrude oil; establishing a fitting formula according to the correlation between the saturated hydrocarbon parameters of the crude oil and the density of ground crude oil; and predicting the density ofthe ground crude oil in a target oil-bearing layer according to the fitting formula. According to the crude oil density prediction method based on the saturated hydrocarbon biomarker parameters, the density of the ground crude oil in the target oil-bearing layer can be predicted through a gas chromatography-mass spectrometry analysis of the saturated hydrocarbon component of the wall core or a small amount of core extract. The crude oil density prediction method based on the saturated hydrocarbon biomarker parameters fits and predicts the segmentations of medium light oil and heavy oil, and has the advantages of being simple to operate, low in cost, high in precision and less in influence factors.
Owner:CHINA NAT OFFSHORE OIL CORP +1
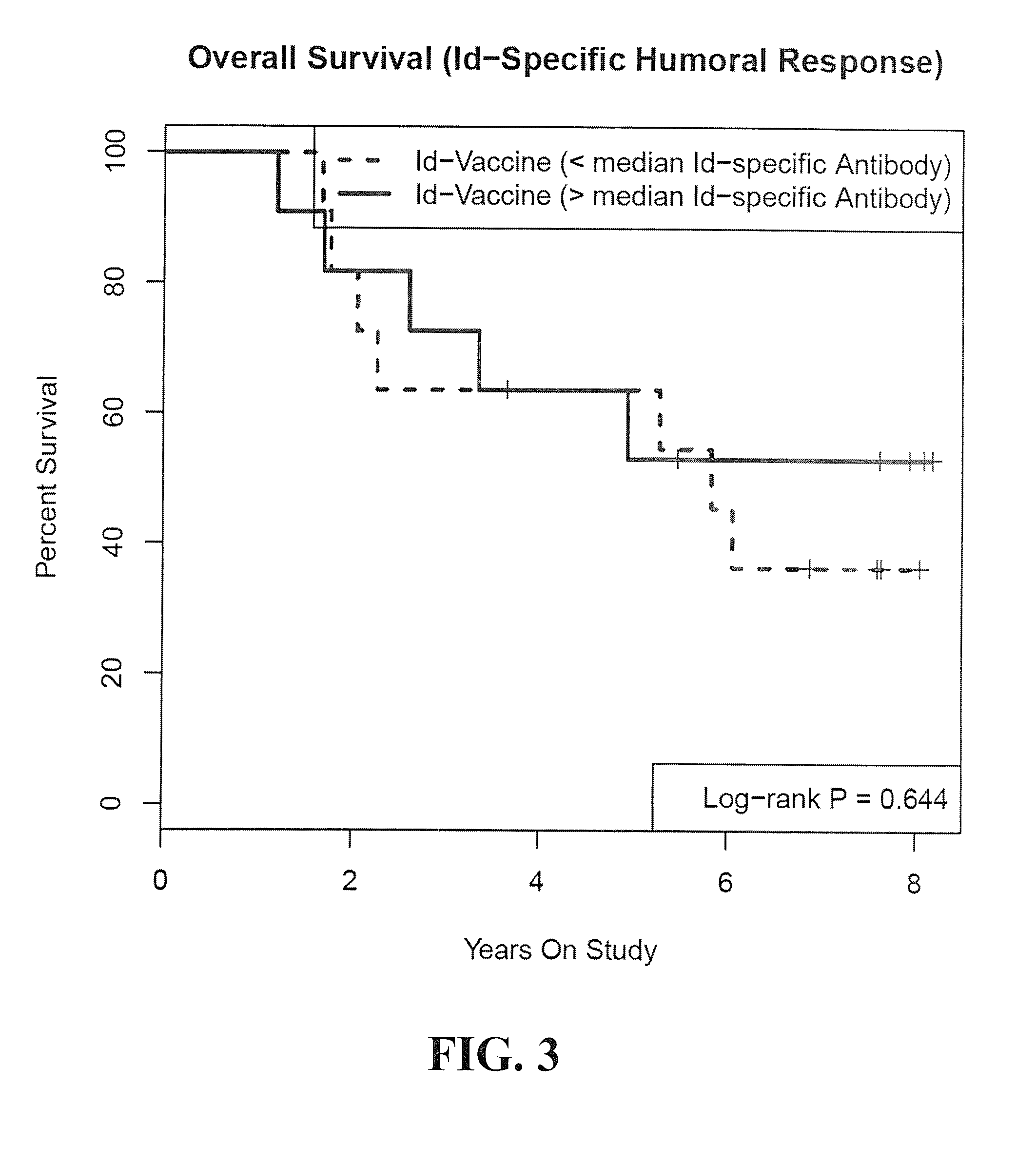
Tumor-specific gm-csf cytokine response as predictor of cancer vaccine effectiveness
InactiveUS20140363456A1Quick comparisonEfficacy of differentSnake antigen ingredientsDisease diagnosisVaccinationBiomarker identification
The present invention relates to methods of treating cancer with a cancer vaccine using granulocyte-macrophage colony-stimulating factor (GM-CSF) as a biomarker; methods of prognosticating an outcome of cancer treatment with a cancer vaccine using GM-CSF as a biomarker; methods for qualifying subjects for cancer vaccination using GM-CSF as a biomarker; methods for comparing the efficacy of two or more cancer vaccine treatments based on GM-CSF response; and kits for the effective treatment of cancer.
Owner:BIOVEST INT +1
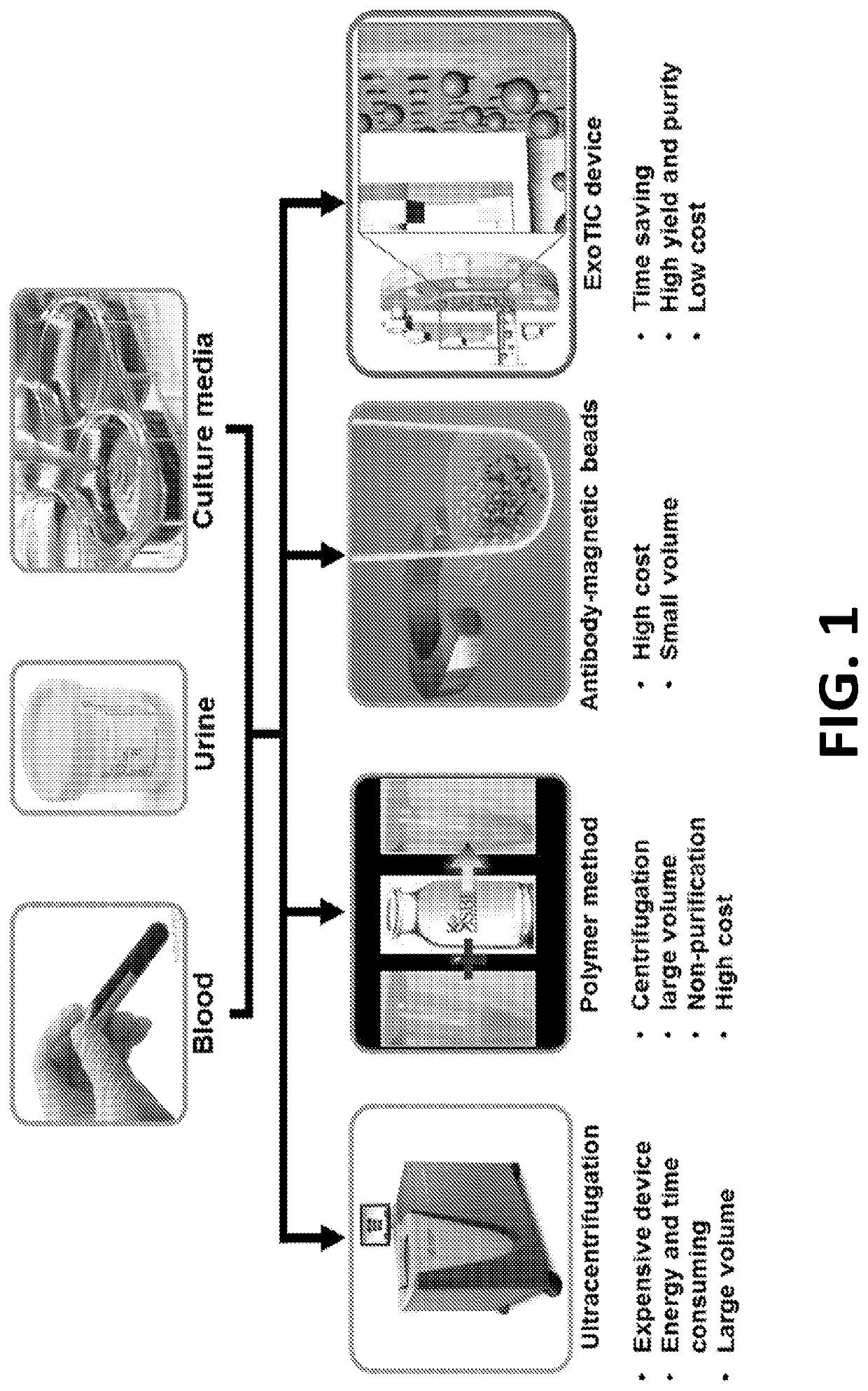
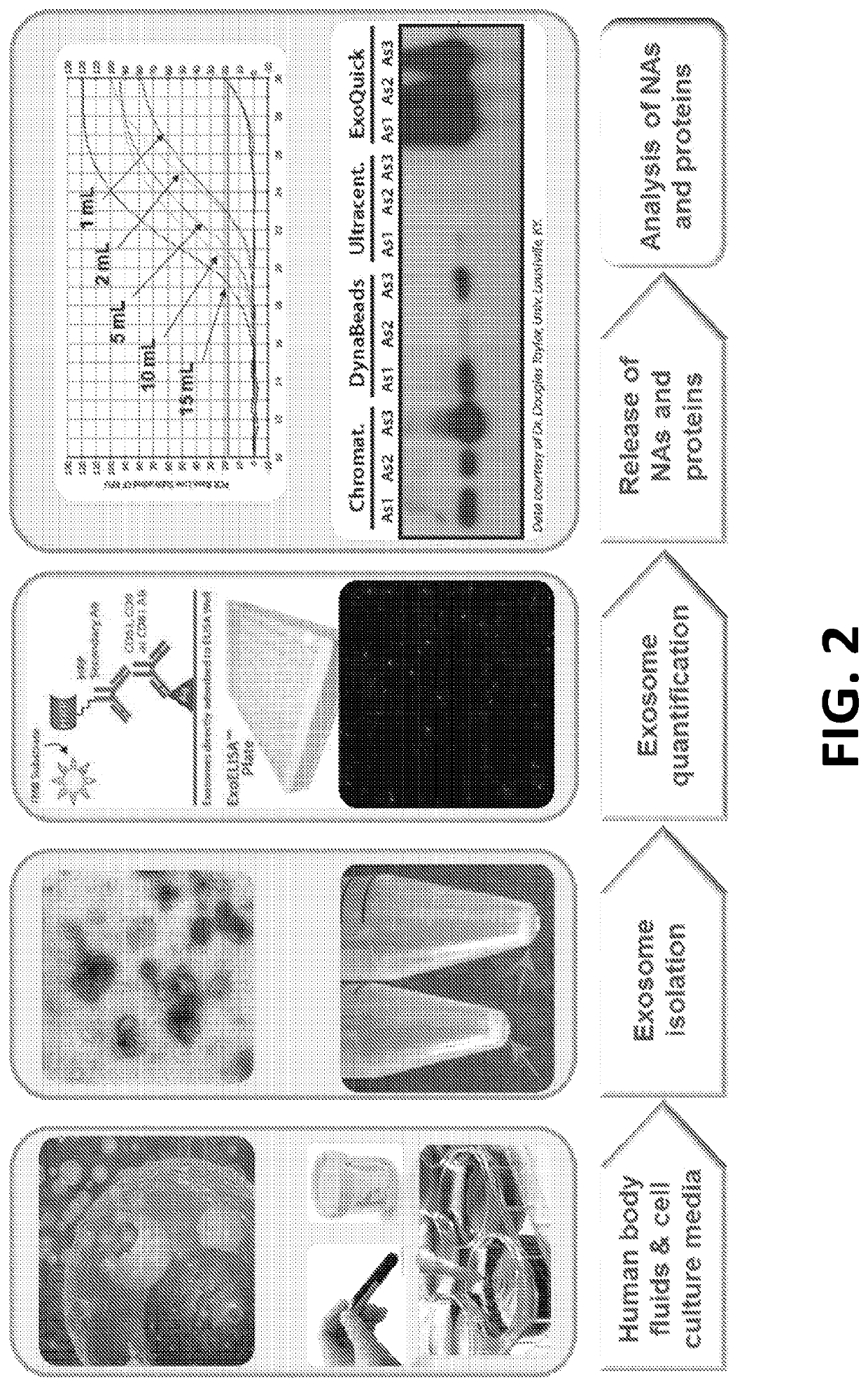
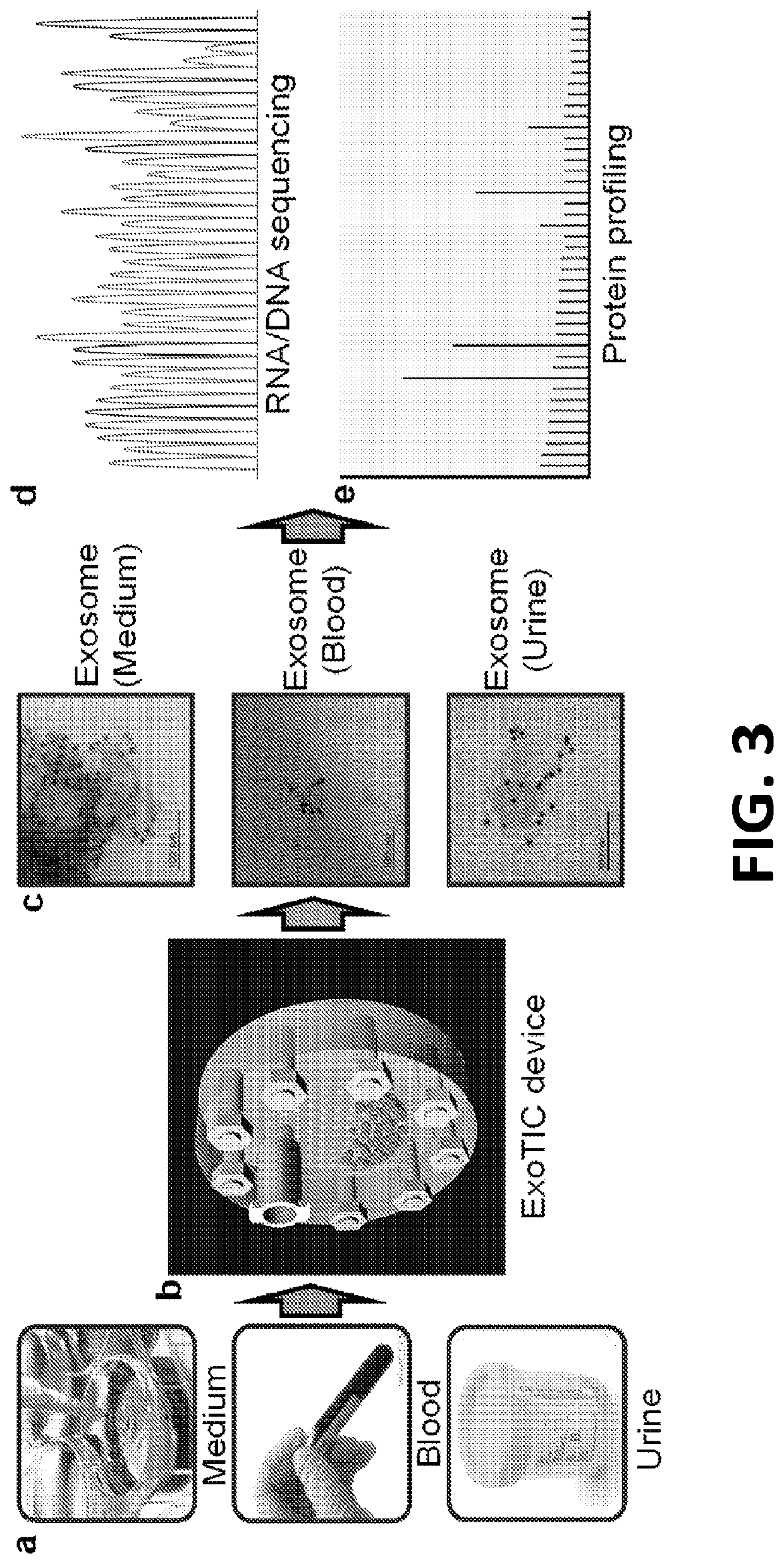
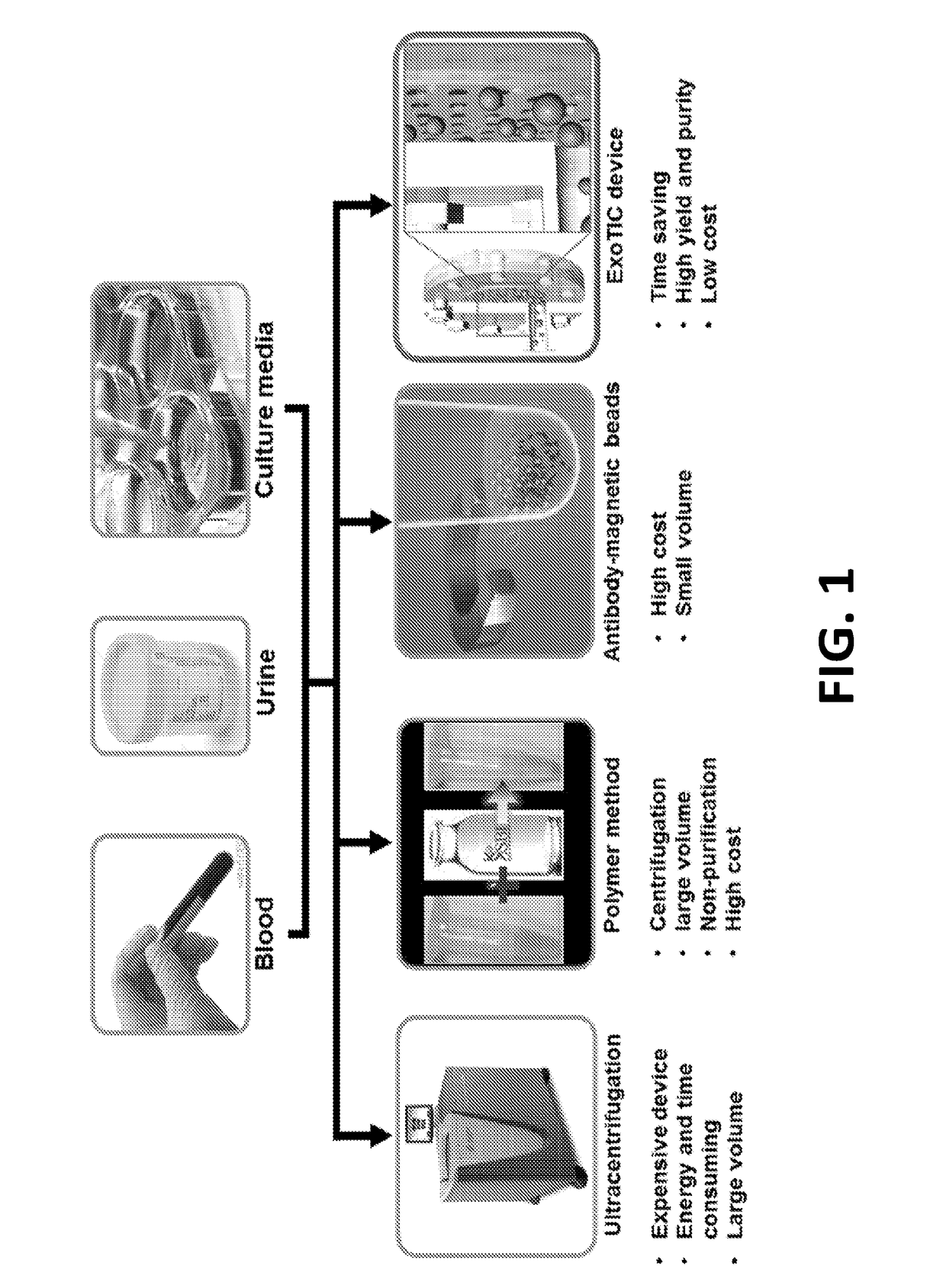
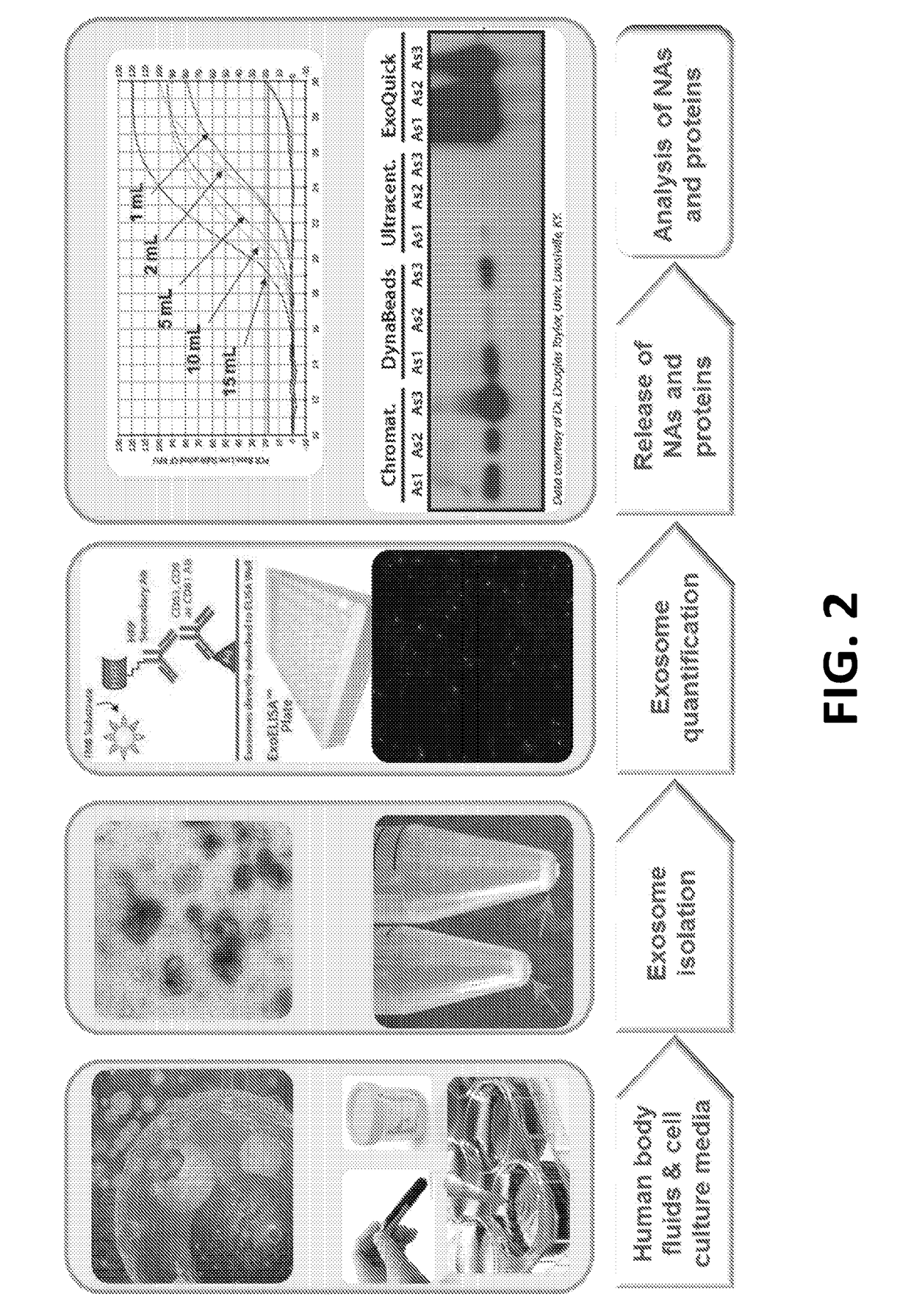
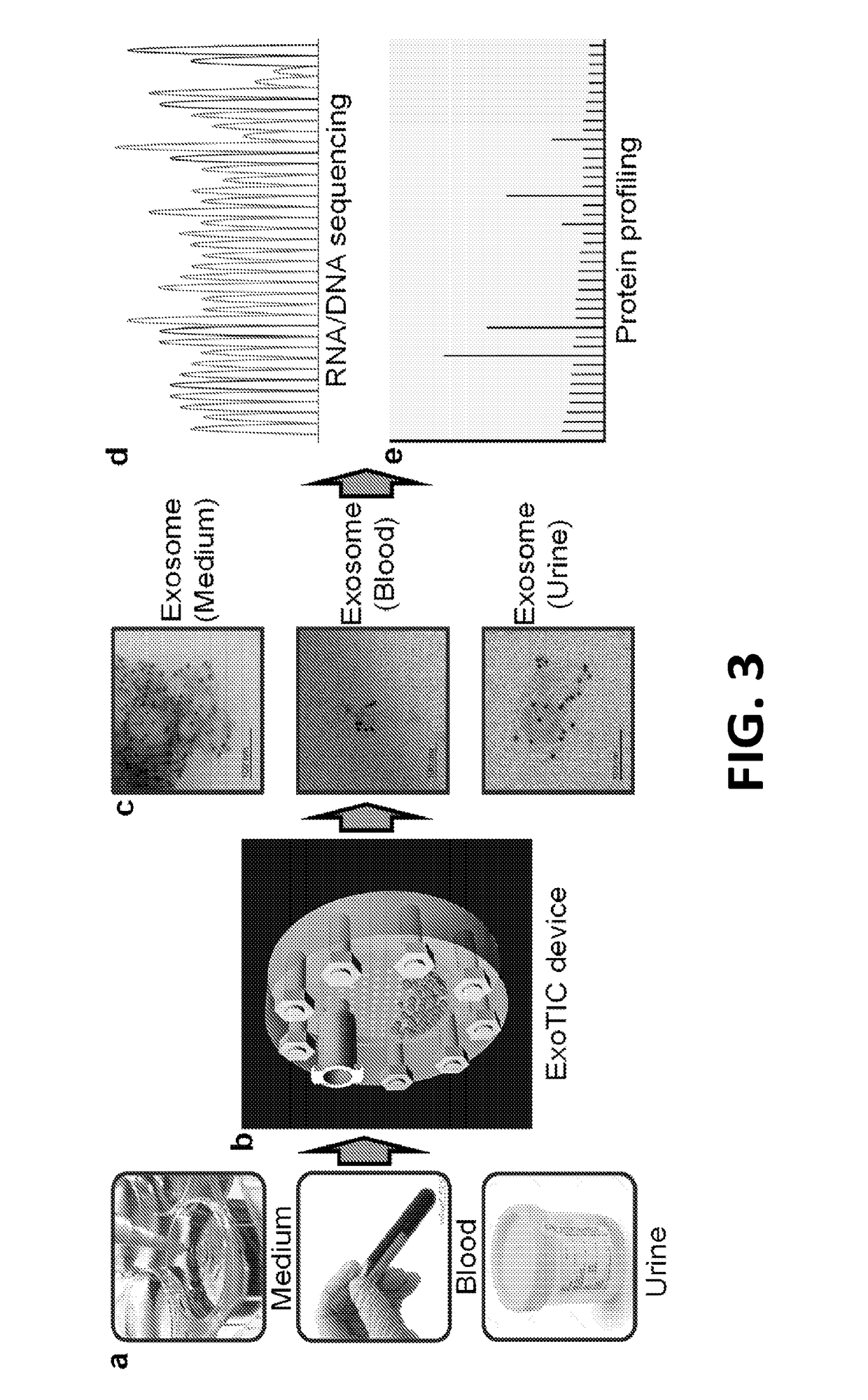
Exosome-Total-Isolation-Chip (ExoTIC) device for isolation of exosome-based biomarkers
ActiveUS11073511B2Facilitate identification of biomarkersRapid and high yield and high purity isolationLaboratory glasswaresDisease diagnosisDiseaseExtracellular vesicle
A device (“the ExoTIC device”) for the isolation of extracellular vesicles from an extracellular vesicle-containing sample in which the sample is flowed through a membrane in a flow chamber to capture and purify the extracellular vesicles on the membrane. The extracellular vesicles may be washed and collected and utilized in any one of a number of ways including, but not limited to, identifying biomarkers of a disease, identifying the presence of a biomarker in a patient to determine whether a patient has a disease, and therapeutically treating existing diseases by re-introducing the extracellular vesicles, potentially modified, back into a body.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
Exosome-Total-Isolation-Chip (ExoTIC) Device for Isolation of Exosome-Based Biomarkers
ActiveUS20190049438A1Low costConvenient amountLaboratory glasswaresDisease diagnosisExtracellular vesicleBiomarker identification
A device (“the ExoTIC device”) for the isolation of extracellular vesicles from an extracellular vesicle-containing sample in which the sample is flowed through a membrane in a flow chamber to capture and purify the extracellular vesicles on the membrane. The extracellular vesicles may be washed and collected and utilized in any one of a number of ways including, but not limited to, identifying biomarkers of a disease, identifying the presence of a biomarker in a patient to determine whether a patient has a disease, and therapeutically treating existing diseases by re-introducing the extracellular vesicles, potentially modified, back into a body.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
Systems and methods for biomarker identification
ActiveUS10861583B2Microbiological testing/measurementBiostatisticsKnowledge-based systemsInteraction nets
The present invention relates to systems and methods for identifying a biomarker from associative and knowledge based systems and processes. Particularly, aspects of the present invention are directed to a computer implemented method that includes data mining one or more public sources of biomedical text, scientific abstract, or bioinformatic data using queries to identify database terms associated with one or more predetermined terms, scoring association(s) between each of the identified database terms and the one or more predetermined terms, determining a subset b based on the score of the association(s), developing an interaction network model comprising the database terms in subset b, interactions, and additional database terms using a combination of algorithms in a predetermined order, and identifying candidate biomarkers from the interaction network model based on a ranking of the database terms in subset b and the additional database terms in the interaction network model.
Owner:LAB OF AMERICA HLDG
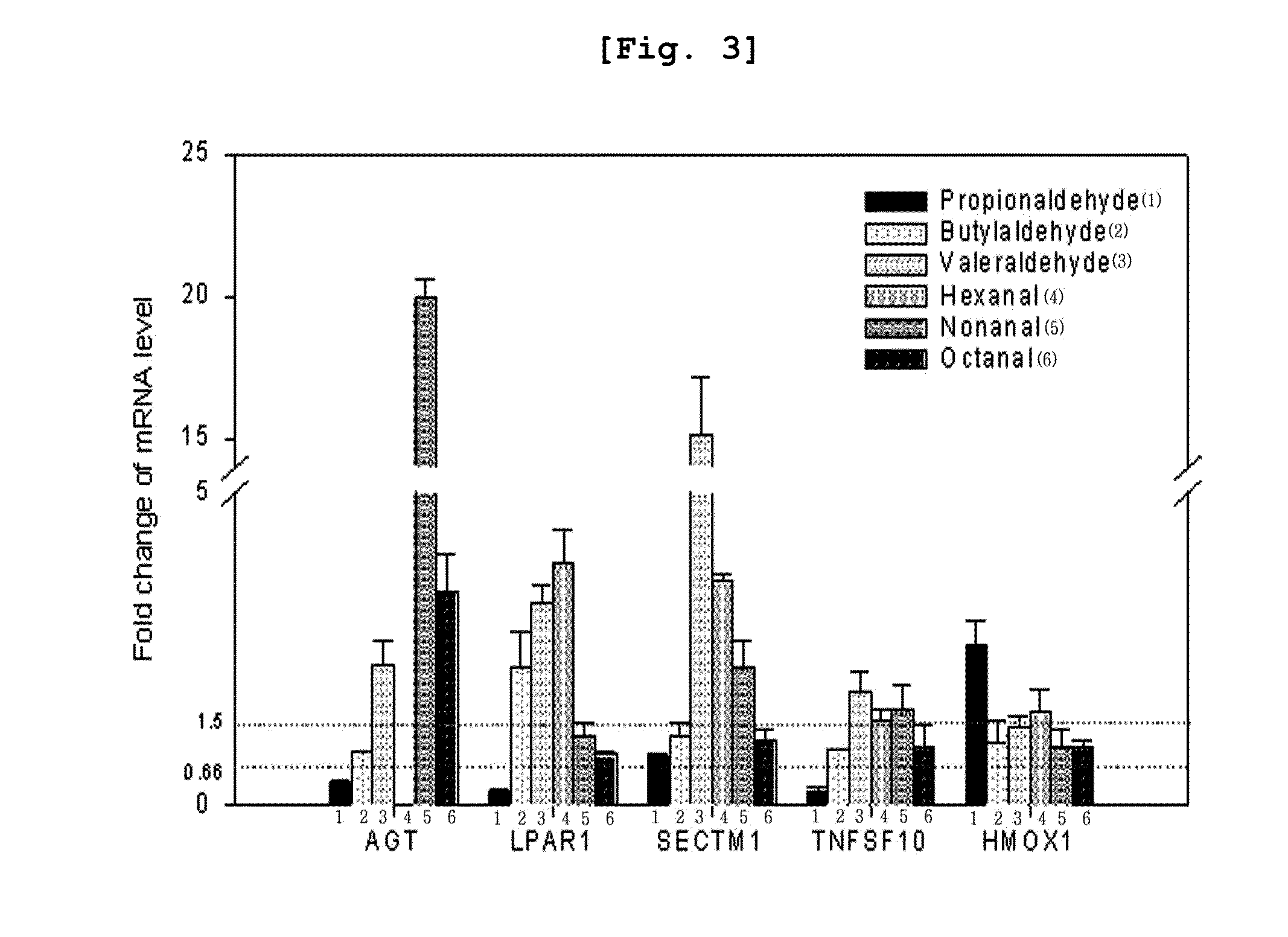
Specific biomarker for identificaton of exposure to propionaldehyde and the method of identification using the same
InactiveUS20130281311A1Microbiological testing/measurementLibrary screeningDNA Microarray ChipBiomarker identification
The present invention relates to a biomarker for the identification of specific exposure to propionaldehyde which is one of volatile organic compounds exposed in the environment, and a method for the identification of specific exposure to propionaldehyde using the same, precisely a biomarker which is up-regulated or down-regulated specifically by propionaldehyde and a method for the identification of specific exposure to propionaldehyde using the biomarker. The biomarker of the present invention is the reacted genes selected by using DNA microarray chip, which can be effectively used for the monitoring and evaluation of propionaldehyde contamination in the environment samples and at the same time as a tool for the investigation of the toxic mechanism induced specifically by propionaldehyde.
Owner:KOREA INST OF SCI & TECH
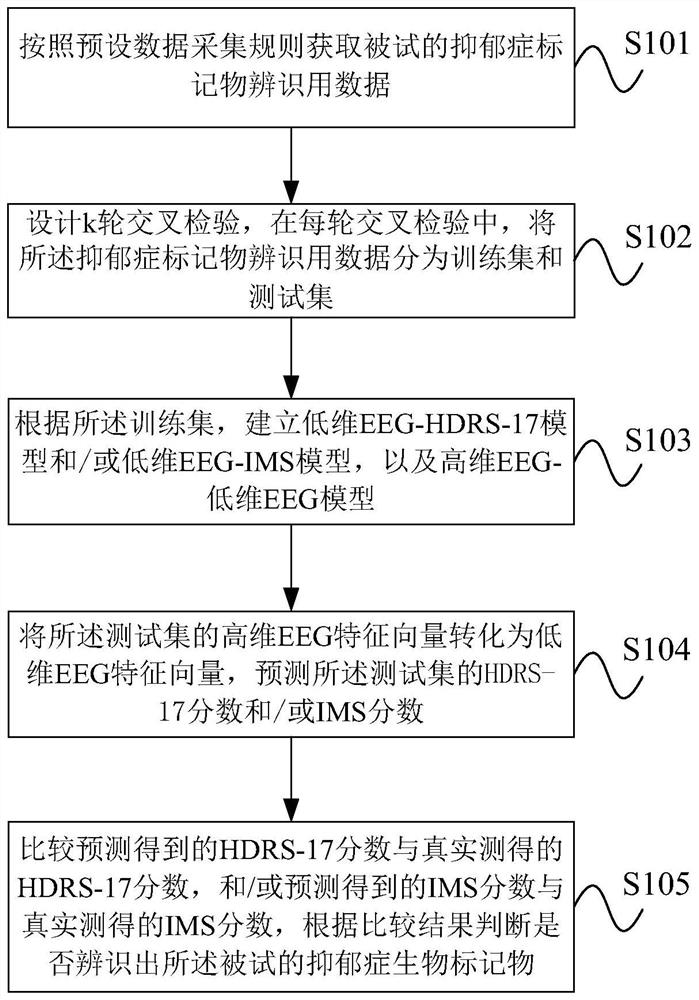
Depression biomarker identification method based on non-invasive electroencephalogram signal
PendingCN112568912ACancel noiseAids in identifying researchDiagnostic signal processingSensorsFeature vectorMedicine
The invention provides a depression biomarker identification method based on a non-invasive electroencephalogram signal. The method comprises the following steps: acquiring tested depression marker identification data according to a preset data acquisition rule, wherein the data includes an HDRS-17 score and / or an IMS score, and EEG signals corresponding to the HDRS-17 score and / or the IMS score;designing k rounds of cross inspection, and in each round of cross inspection, dividing depression marker identification data into a training set and a test set; establishing a low-dimensional EEG-HDRS-17 model and / or a low-dimensional EEG-IMS model and a high-dimensional EEG-low-dimensional EEG model according to the training sets; converting the high-dimensional EEG feature vector of the test sets into a low-dimensional EEG feature vector by using the high-dimensional EEG-low-dimensional EEG model, and predicting an HDRS-17 score and / or an IMS score of the test set by using the low-dimensional EEG-HDRS-17 model and / or low-dimensional EEG-IMS model; and comparing the predicted HDRS-17 score with an actually measured HDRS-17 score, and / or comparing the predicted IMS score with an actuallymeasured IMS score, and judging whether the depression biomarker to be tested is identified or not according to a comparison result.
Owner:陈盛博
Crude Oil Density Prediction Method Based on Saturated Hydrocarbon Biomarker Parameters
ActiveCN109556990BFew influencing factorsEasy to operateComponent separationComplex mathematical operationsAlkaneGas liquid chromatographic
A crude oil density prediction method based on saturated hydrocarbon biomarker parameters, comprising: collecting crude oil saturated hydrocarbon biomarkers, including n-alkanes series, terpanes series, sterane series and alkylcyclohexane series biomarkers identification and Relative quantification; calculate crude oil saturated hydrocarbon parameters; establish a fitting formula based on the correlation between crude oil saturated hydrocarbon parameters and surface crude oil density; predict the surface crude oil density of the target oil-bearing formation according to the fitting formula. The purpose of the present invention is to predict the surface crude oil density of the target oil-bearing layer system by gas chromatography-mass spectrometry analysis of the saturated hydrocarbon components of the wall core or a small amount of rock core extract. The invention performs fitting and forecasting of medium and light oil and heavy oil in sections, and has the advantages of simple operation, low cost, high precision and few affected factors.
Owner:CHINA NAT OFFSHORE OIL CORP +1
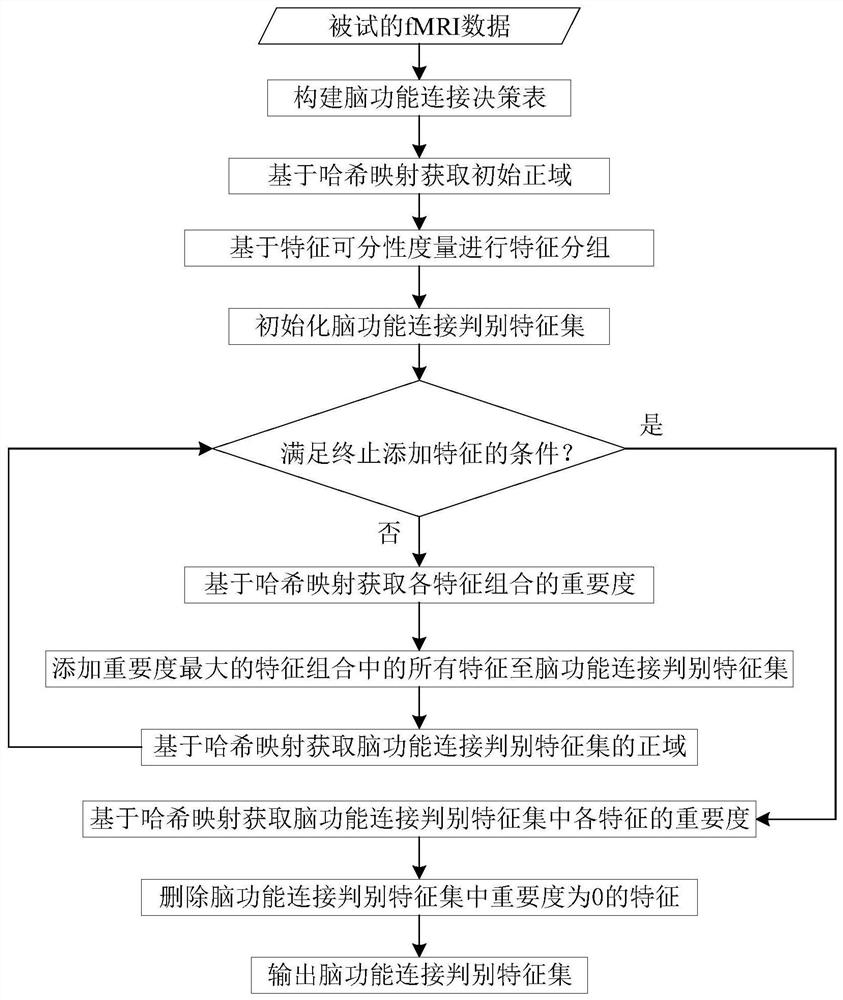
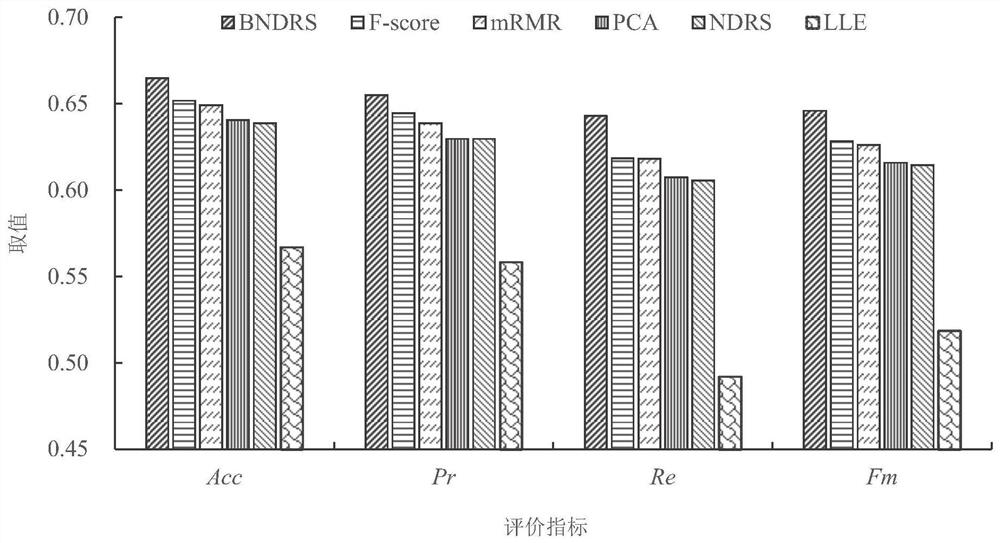
Brain function connection biomarker identification method based on neighborhood decision rough set
PendingCN113919429ADigital data information retrievalCharacter and pattern recognitionPattern recognitionBiomarker identification
The invention provides a brain function connection biomarker recognition method based on a neighborhood decision rough set, and relates to the two fields of brain science and the neighborhood decision rough set. the neighborhood decision rough set capable of effectively processing continuous and high-noise data is introduced for the first time aiming at the characteristics of continuity and high noise of the brain function connection data to improve the accuracy of brain function connection biomarker recognition; the efficiency of the neighborhood decision rough set for identifying the brain function connection biomarker is ensured by reducing a feature search space and quickly generating equivalence classes, namely, the coarse-grained feature search space of the neighborhood decision rough set is generated by grouping brain function connection features by using feature separability measurement; and the equivalence class of each sample is quickly obtained by using the symmetry of the Hash mapping and the neighborhood relationship. The method can accurately and rapidly obtain brain function connection discrimination characteristics with strong classification capability, and is expected to provide more accurate biomarkers for diagnosis of neurological and psychiatric diseases.
Owner:BEIJING UNIV OF TECH
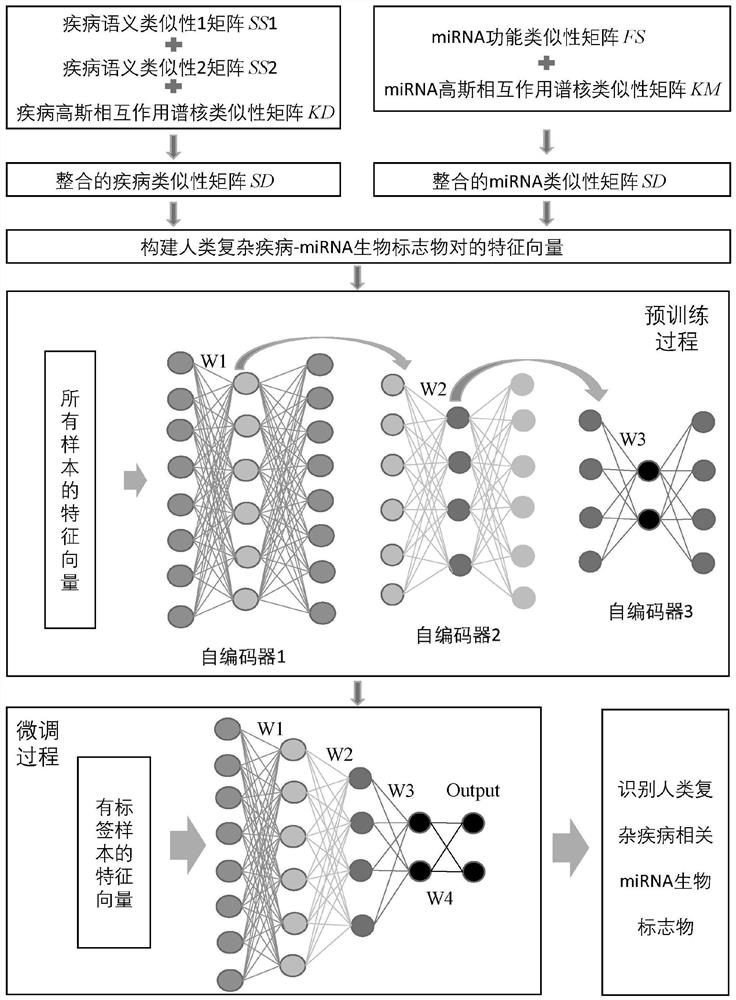
miRNA biomarker identification method and system based on stack auto-encoder
PendingCN113743589AOvercome the limitation of not being able to fully utilize the information of unlabeled samplesOvercome limitationsSemantic analysisMedical automated diagnosisBiomarker identificationBiologic marker
The invention discloses a miRNA biomarker identification method and system based on a stack auto-encoder. The method comprises the following steps: collecting known human complex diseases and associated information of miRNA from a database, and storing the associated information in a matrix form; calculating the similarity between the diseases; calculating the similarity between the miRNAs; constructing a feature vector of the human complex disease-miRNA biomarker pair; building a stack auto-encoder neural network and pre-training neural network parameters; performing fine tuning on the stack auto-encoder neural network by using a labeled complex disease-miRNA sample; and identifying the miRNA biomarker related to the human complex disease based on the trained stack auto-encoder neural network. According to the method, the information of the labeled samples and the information of the unlabeled samples are fully utilized, and the limitation that the information of the unlabeled samples cannot be fully utilized in a traditional machine learning method is overcome.
Owner:CHINA UNIV OF MINING & TECH
Biomarker identification method based on multiple networks
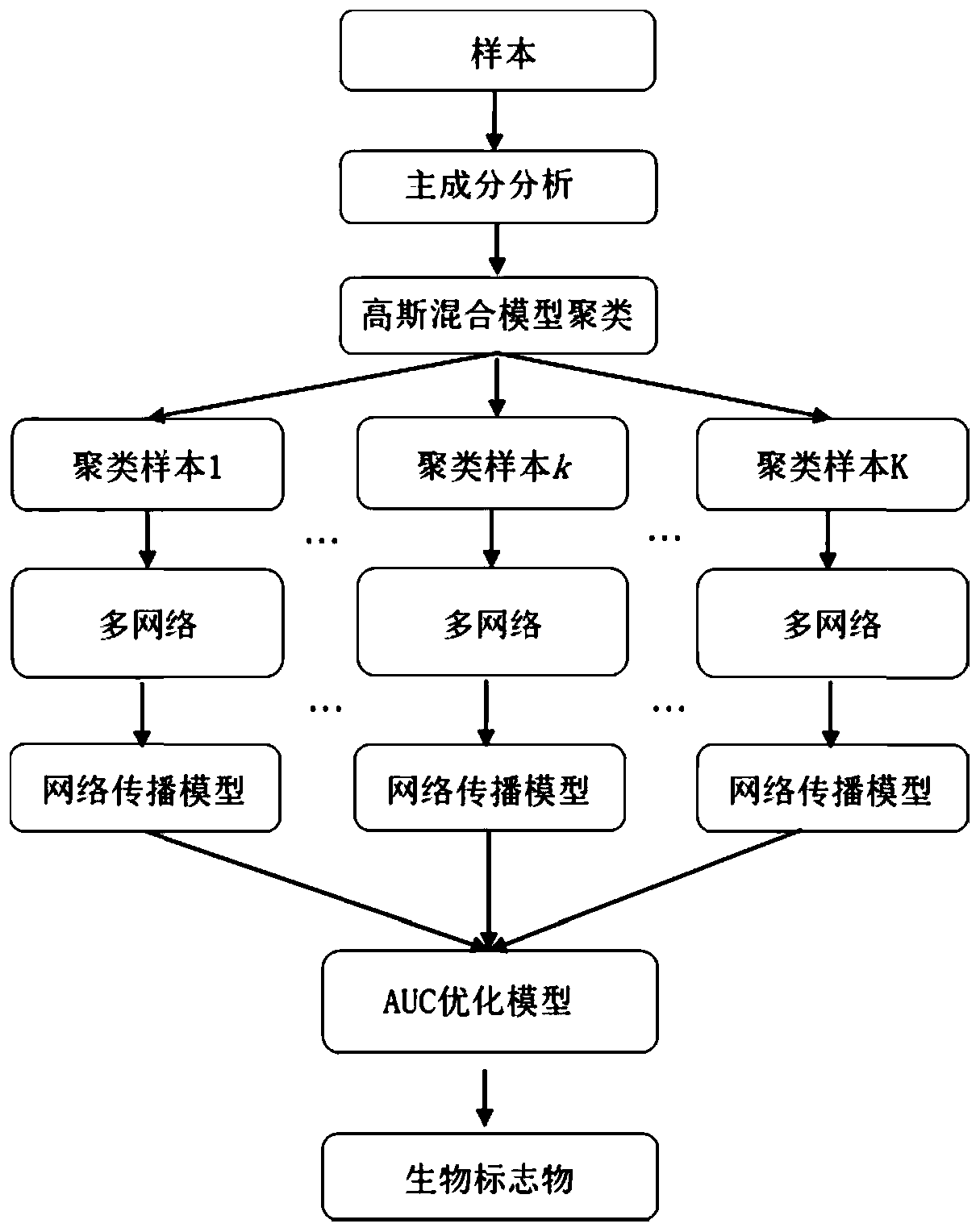
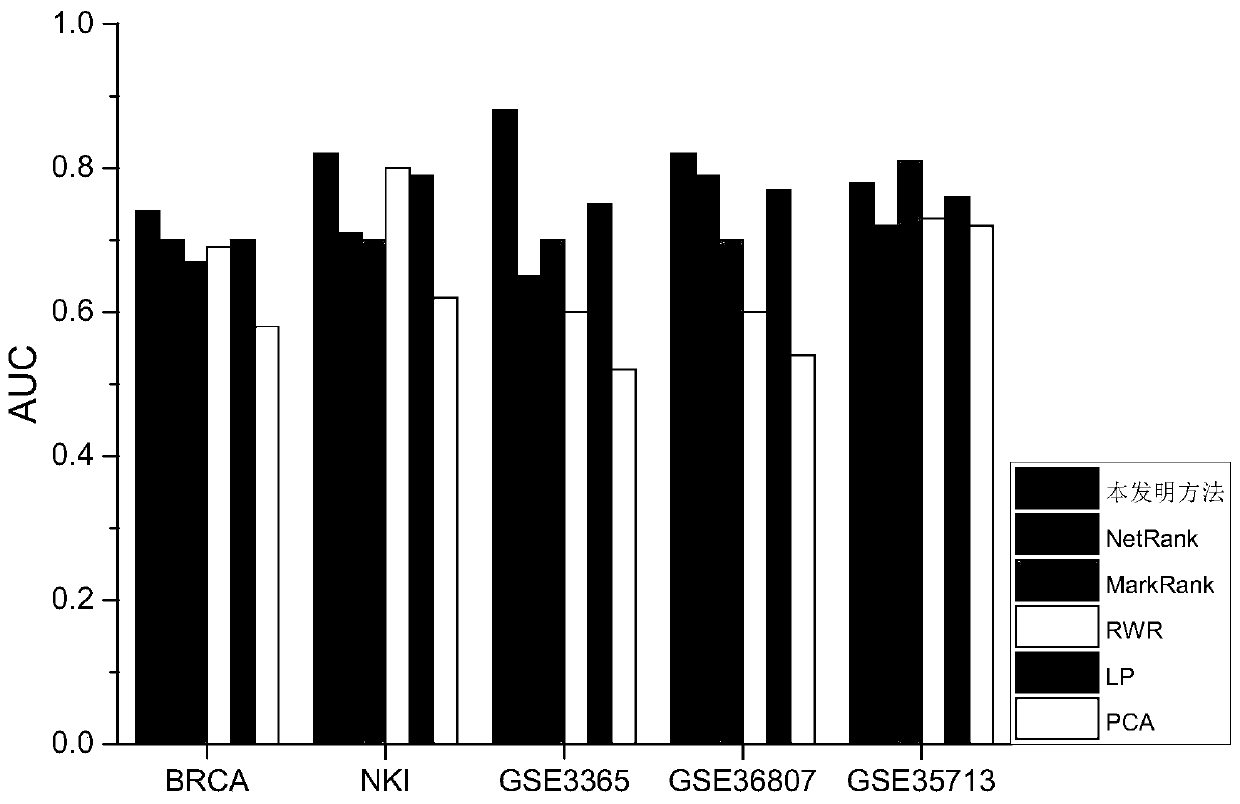
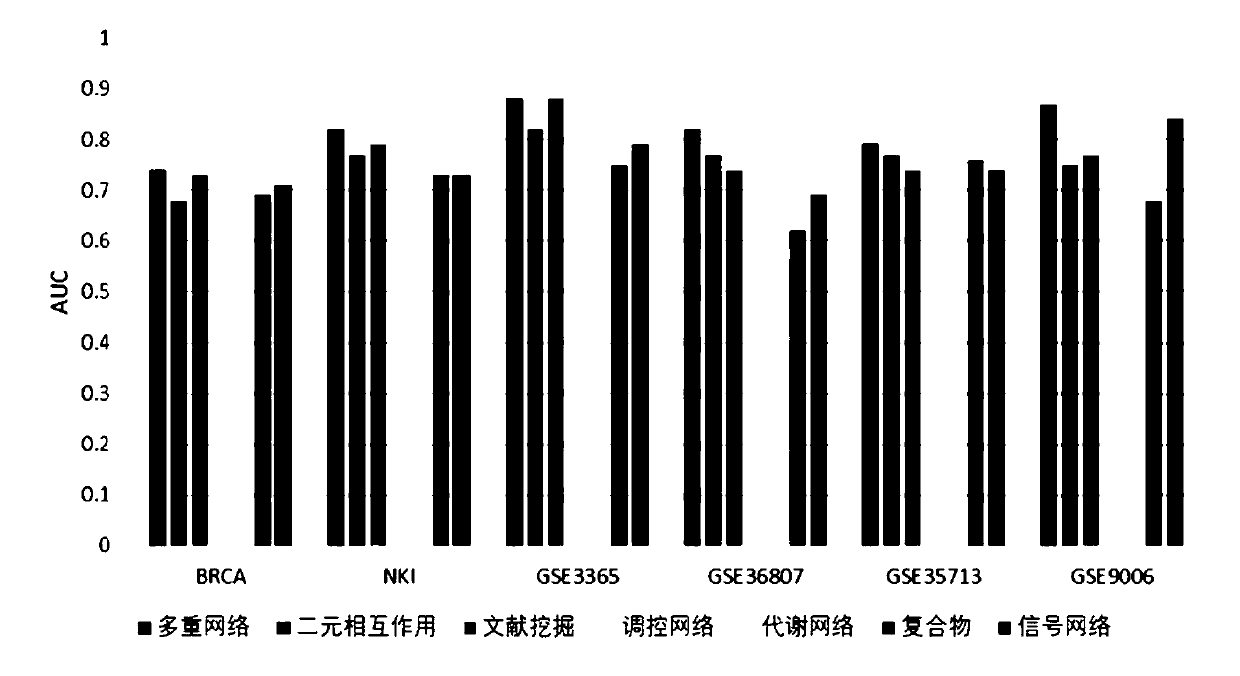
ActiveCN110797083AImprove classification performanceCharacter and pattern recognitionNeural architecturesPrincipal component analysisBiomarker identification
The invention discloses a biomarker identification method based on multiple networks. Considering the influence of sample heterogeneity, the method disclosed by the invention comprises the following steps of: firstly, standardizing gene expression profile data, carrying out principal component analysis on samples, and clustering the samples through a Gaussian mixture model by utilizing the first two principal components, constructing a network propagation model based on multiple networks to sort all genes in the networks as for each type of samples for preliminarily screening important genes,and in order to obtain the biomarker with the maximum distinguishing capability and the minimum redundancy, further grading and sequencing the genes in the important characteristics obtained in the previous step through an area under curve (AUC) optimization model of a receiver operation characteristic curve to obtain the biomarker. According to the method, the multi-source biological network information is fully utilized, and the biomarker with the maximum distinguishing capacity, the minimum redundancy and the biointerpretability can be effectively recognized and used for heterogeneous complex disease analysis.
Owner:CENT SOUTH UNIV
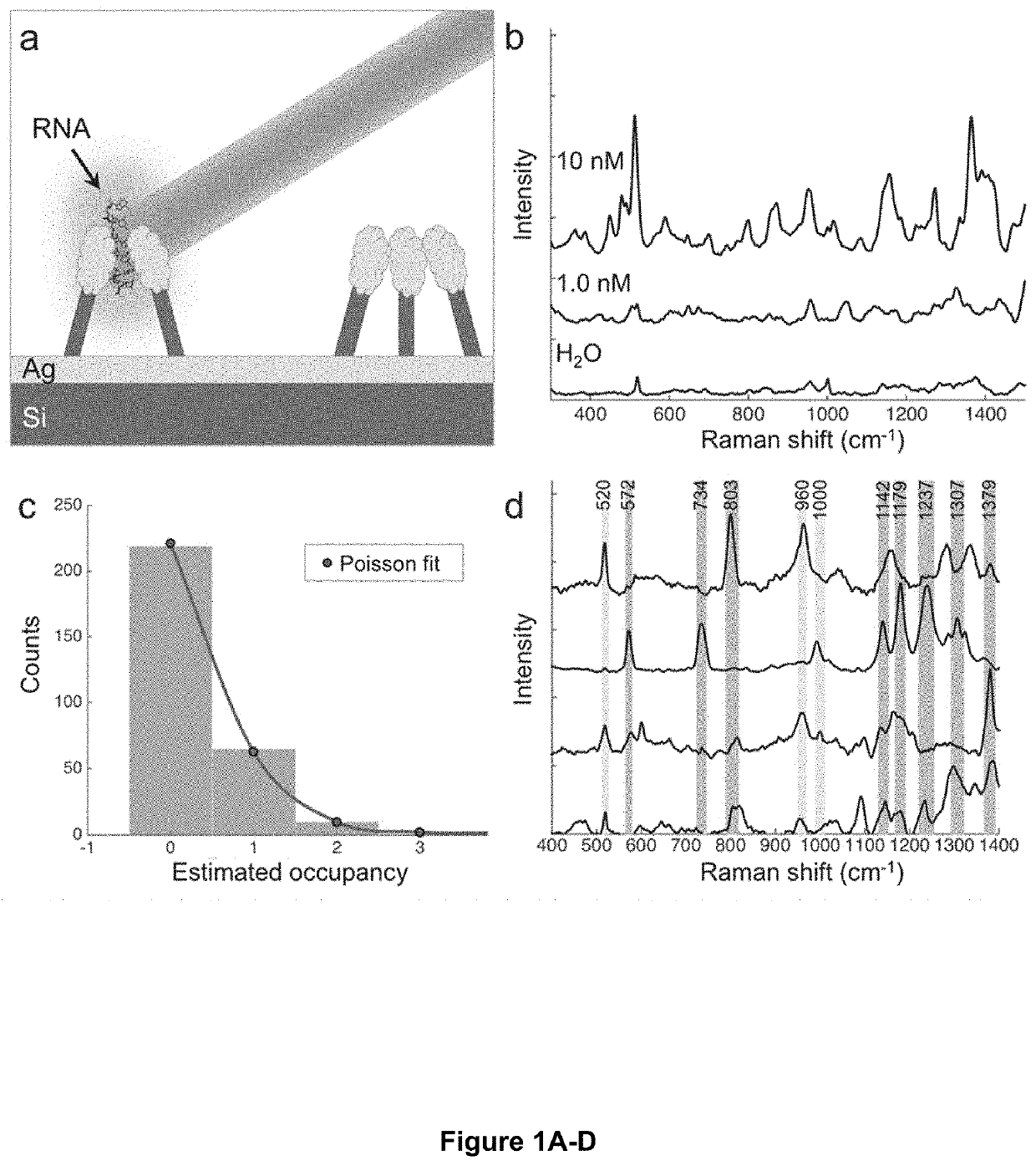
Single-molecule optical sequence identification of nucleic acids and amino acids for combined single-cell omics and block optical content scoring (BOCS): DNA k-mer content and scoring for rapid genetic biomarker identification at low coverage
PendingUS20200240845A1Accurate identificationImprove throughputRadiation pyrometryMaterial analysis by optical meansGenomicsMolecular identification
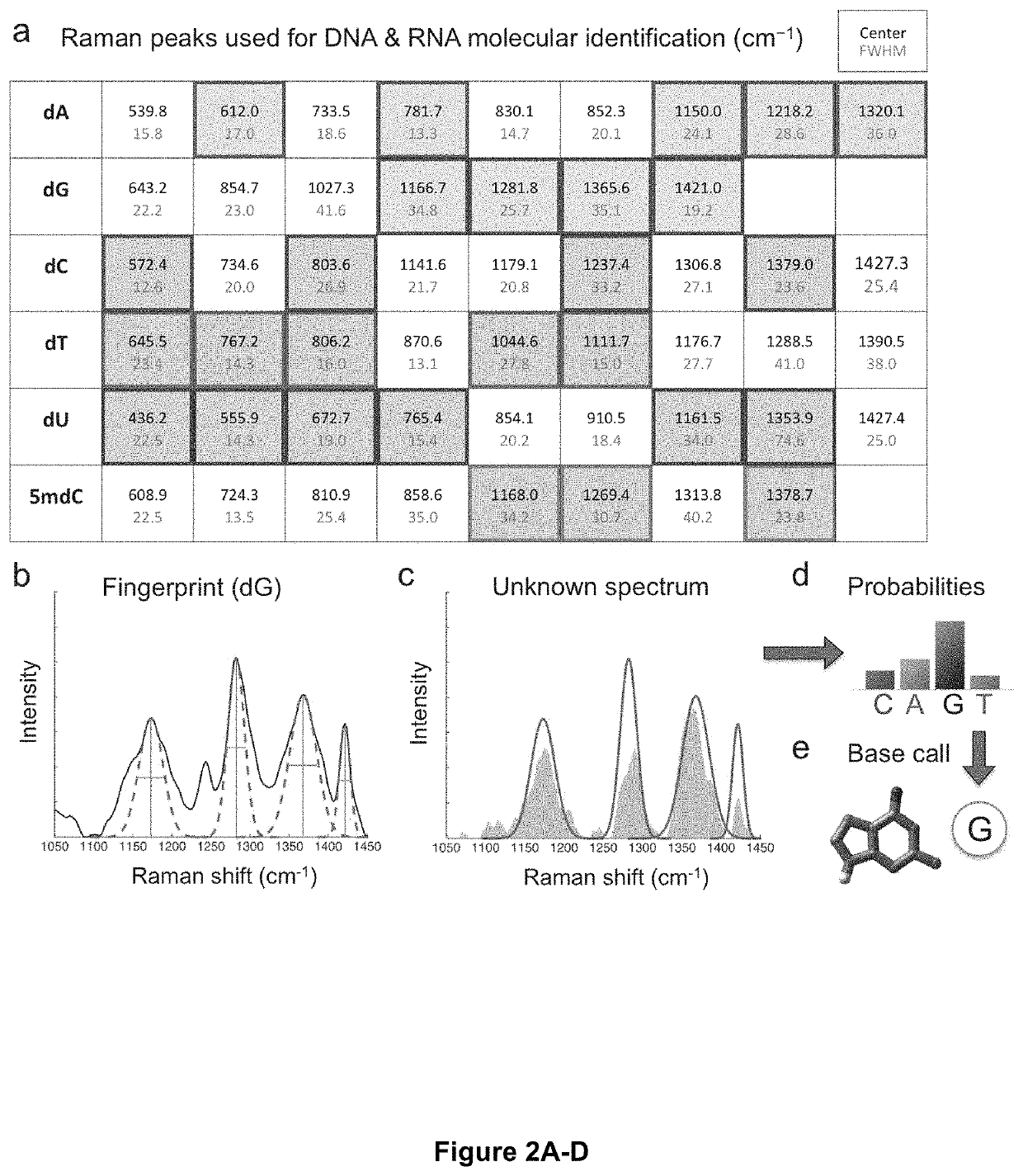
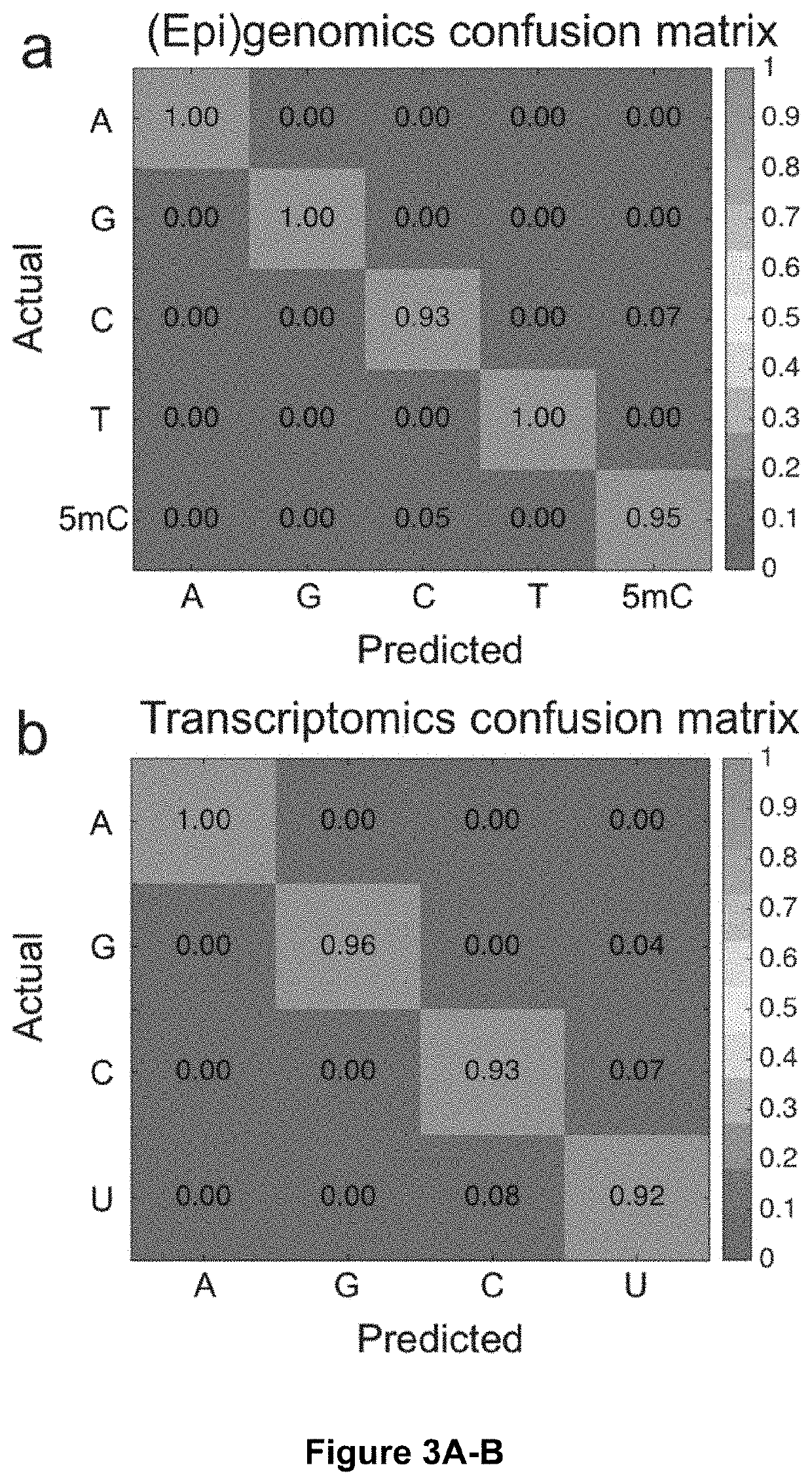
Optical fingerprints for label-free high-throughput (epi)genomics, transcriptomics, and proteomics profiling of single cells. Vibrational spectroscopy signatures combined with a molecular identification algorithm rooted in machine learning enables identification of nucleic acids and amino acids, and their molecular variations, thereby identifying genetic variation by mapping heterogeneity and identifying low copy-number variants. Additional embodiments include the BOCS algorithm which takes measurements of DNA k-mer content from high-throughput single-molecule Raman spectroscopy measurements and maps them to gene databases for probabilistic determination of genetic biomarkers at low coverages. Starting with a log of measured k-mer content blocks (B1 . . . Bn as shown) and a genetic biomarker database (excerpts from the MEGARes antibiotic resistance database are shown), the blocks are individually aligned to each gene in the database based on content. This alignment consists of finding all match locations for the k-mer block content within a gene via translating through the gene one nucleotide at a time and looking at fragments of length k. For each block, a raw probability can be calculated for each gene based on the number of matches for the k-mer block content within the gene, length of the k-mer block, and length of the gene (calculation shown in the schematic). As more blocks are analyzed, probabilities are compounded and genes in the database are ranked. The gene(s) from which the Raman-analyzed k-mer blocks originate quickly generate the top probabilities and can often be determined in coverages <<1.0, meaning that only a small fraction of the gene blocks need to be analyzed for identification of a specific genetic biomarker.
Owner:UNIV OF COLORADO THE REGENTS OF
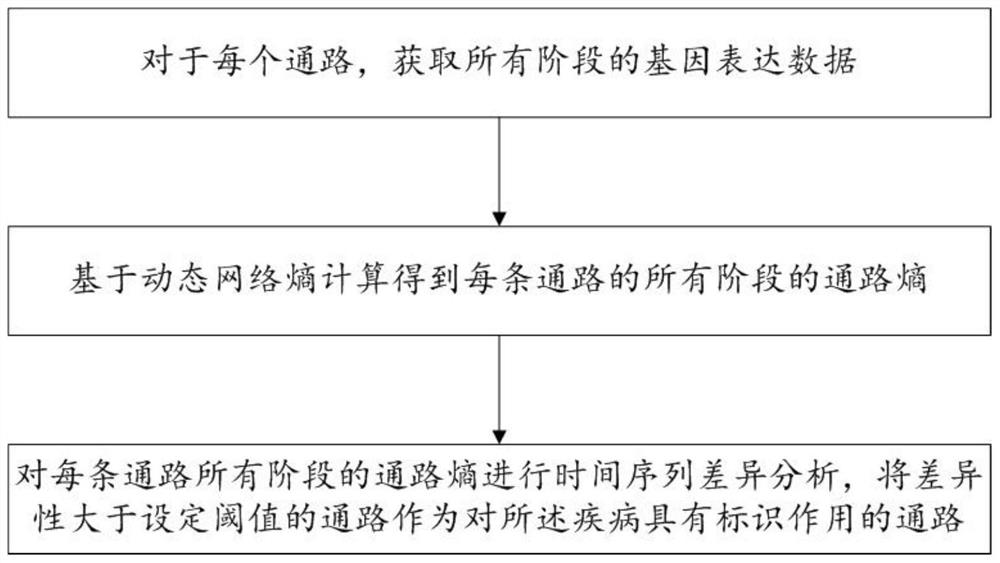
Biomarker identification method and system based on dynamic network entropy
PendingCN113889180AInteraction data sources are accurate and reliableEasy accessBiostatisticsCharacter and pattern recognitionDiseaseStatistical analysis
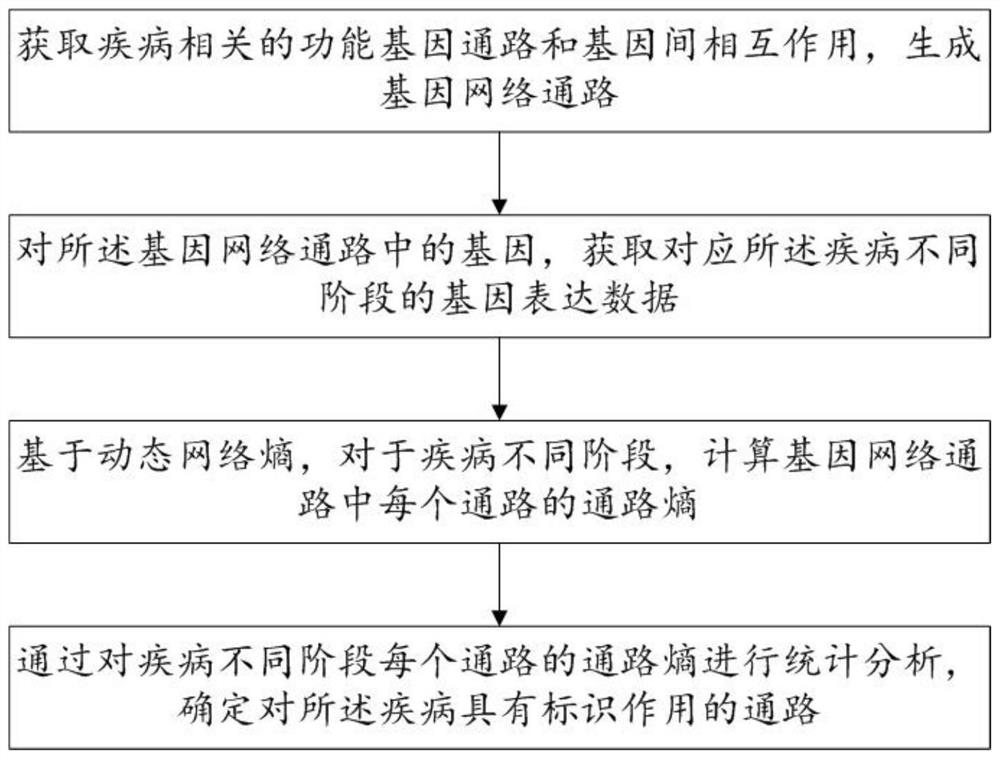
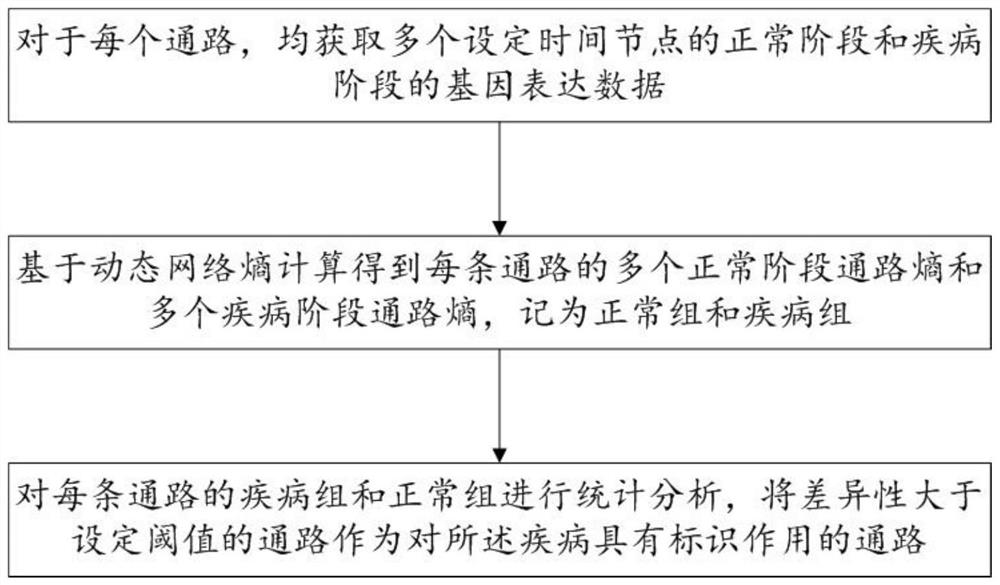
The invention discloses a biomarker identification method and system based on dynamic network entropy, and the method comprises the following steps: obtaining a functional gene pathway related to a disease and interaction between genes, and generating a gene network pathway; acquiring gene expression data corresponding to different stages of the disease for the genes in the gene network pathway; based on the dynamic network entropy, for different stages of the disease, calculating the pathway entropy of each pathway in the gene network pathways; and performing statistical analysis on the channel entropy of each channel in different stages of the disease to determine a channel with an identification effect on the disease. According to the method, the gene network pathway is constructed, information measurement is carried out on pathways in different stages of the disease by adopting entropy, and the gene pathway biomarkers related to the disease progress can be accurately identified by analyzing the change of the entropy.
Owner:SHANDONG UNIV
Human tumor xenotransplantation model construction method and application thereof
PendingCN114375901AHigh tumor formation rateTumor formation time shortenedAnimal husbandryHuman tumorBiomarker identification
The invention relates to a human tumor xenotransplantation model construction method and application thereof. According to the construction method of the humanized tumor xenotransplantation model of the narrative hamster based on the IL2RG gene knockout immunodeficiency, provided by the invention, the personalized humanized narrative hamster tumor xenotransplantation model is successfully established by transplanting tumor cells or tissues of a patient. The method is high in tumor formation rate and short in tumor formation time, and more human-derived tumor samples can be obtained. The constructed human-derived tumor xenotransplantation model retains matrix heterogeneity, histological characteristics, molecular diversity and microenvironment of primary tumors, can be widely applied to the aspects of preclinical drug effect personalized screening evaluation, biomarker identification and the like, and lays a foundation for tumor fundamental research and drug evaluation.
Owner:HENAN UNIV OF CHINESE MEDICINE
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com