GENE EXPRESSION ANALYSIS METHOD USING TWO DIMENSIONAL cDNA LIBRARY
a gene expression and two-dimensional technology, applied in the field of gene expression analysis methods, can solve the problems that the expression levels of a number of genes cannot be observed, and the satisfactory method has not yet been developed, and achieve the effect of simple and efficient detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
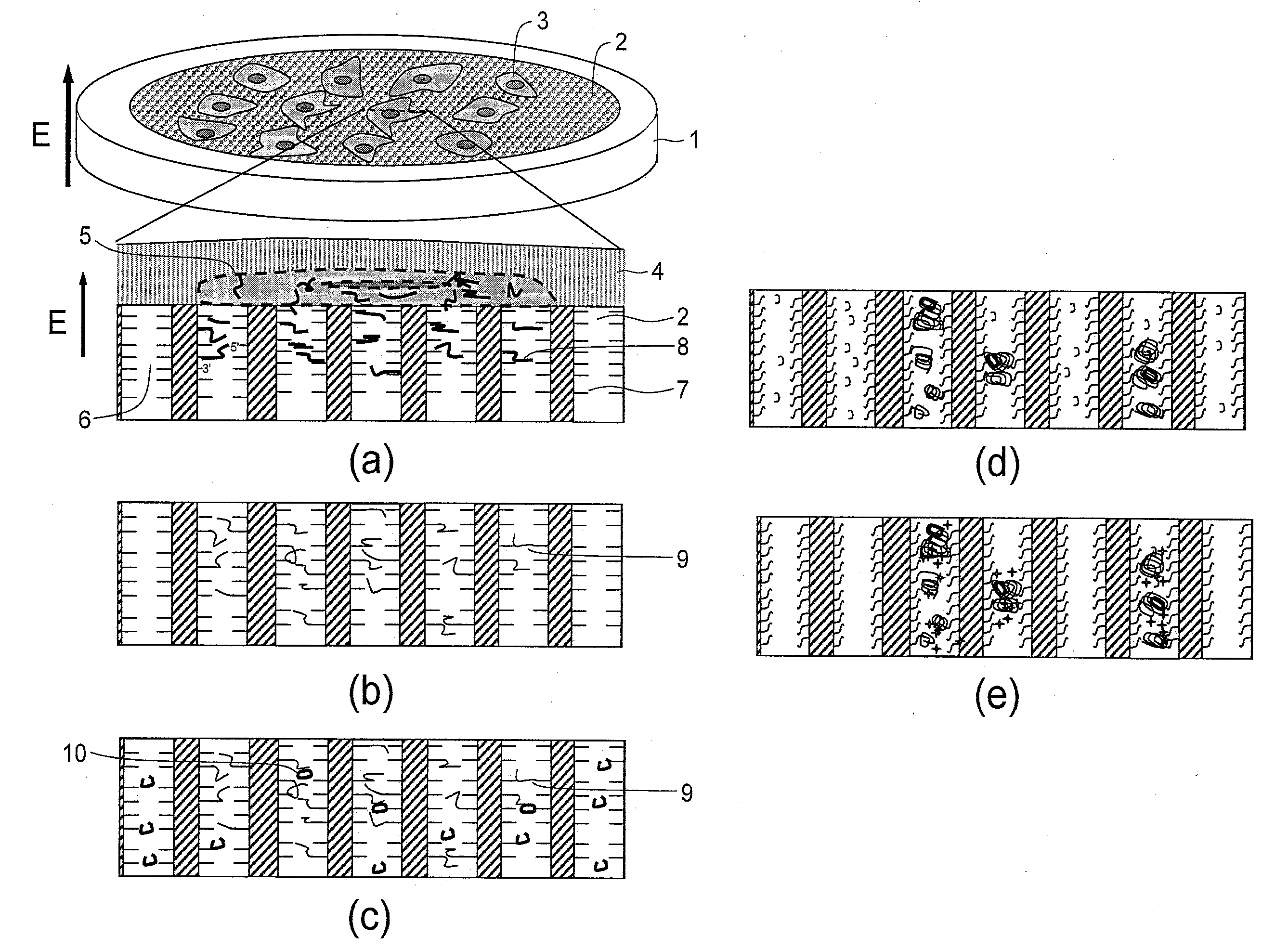
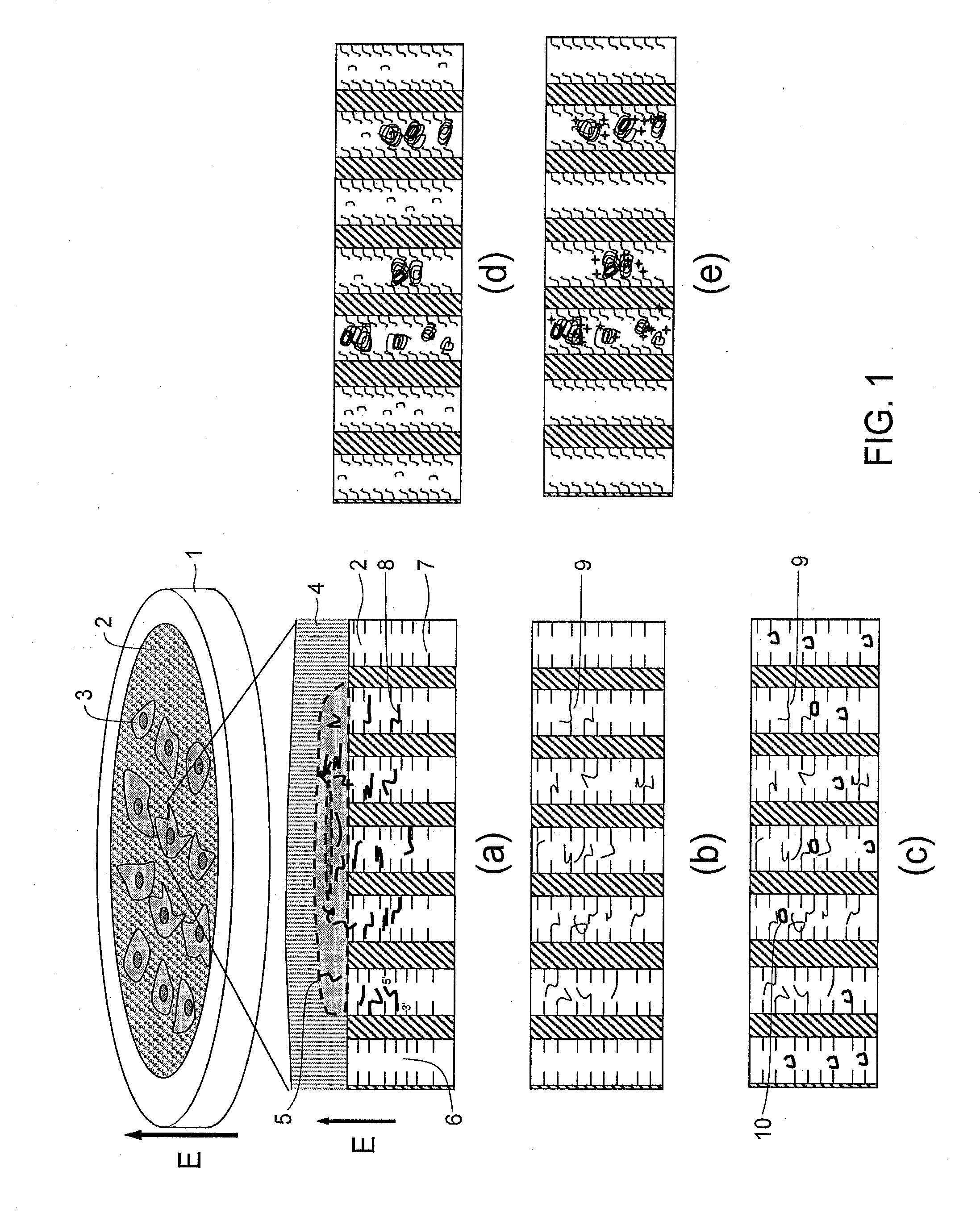
[0113]This Example is an example of using a two-dimensional cDNA library sheet, which was constructed in a pore array sheet, from sheet-type cells while keeping positional information of mRNA contained therein. Hereinafter, a sheet in which a number of pores are two-dimensionally formed will be referred to as a pore array sheet and a pore array sheet in which a cDNA library is formed will be referred to as a cDNA library sheet. A conceptual figure of an example of the method is shown in FIG. 1.
[0114]This method contains: a step of extracting and trapping mRNA, while keeping cell positional information, by a DNA probe within a sheet (FIG. 1 (a)); a step of preparing a cDNA library by reverse transcription within the sheet (FIG. 1 (b)); a step of determining presence of a target cDNA by hybridizing a DNA probe (padlock probe), which is capable of hybridizing with the target cDNA at both ends, with the target cDNA and preparing a ring-form DNA probe (referred to as a ring probe) throug...
example 2
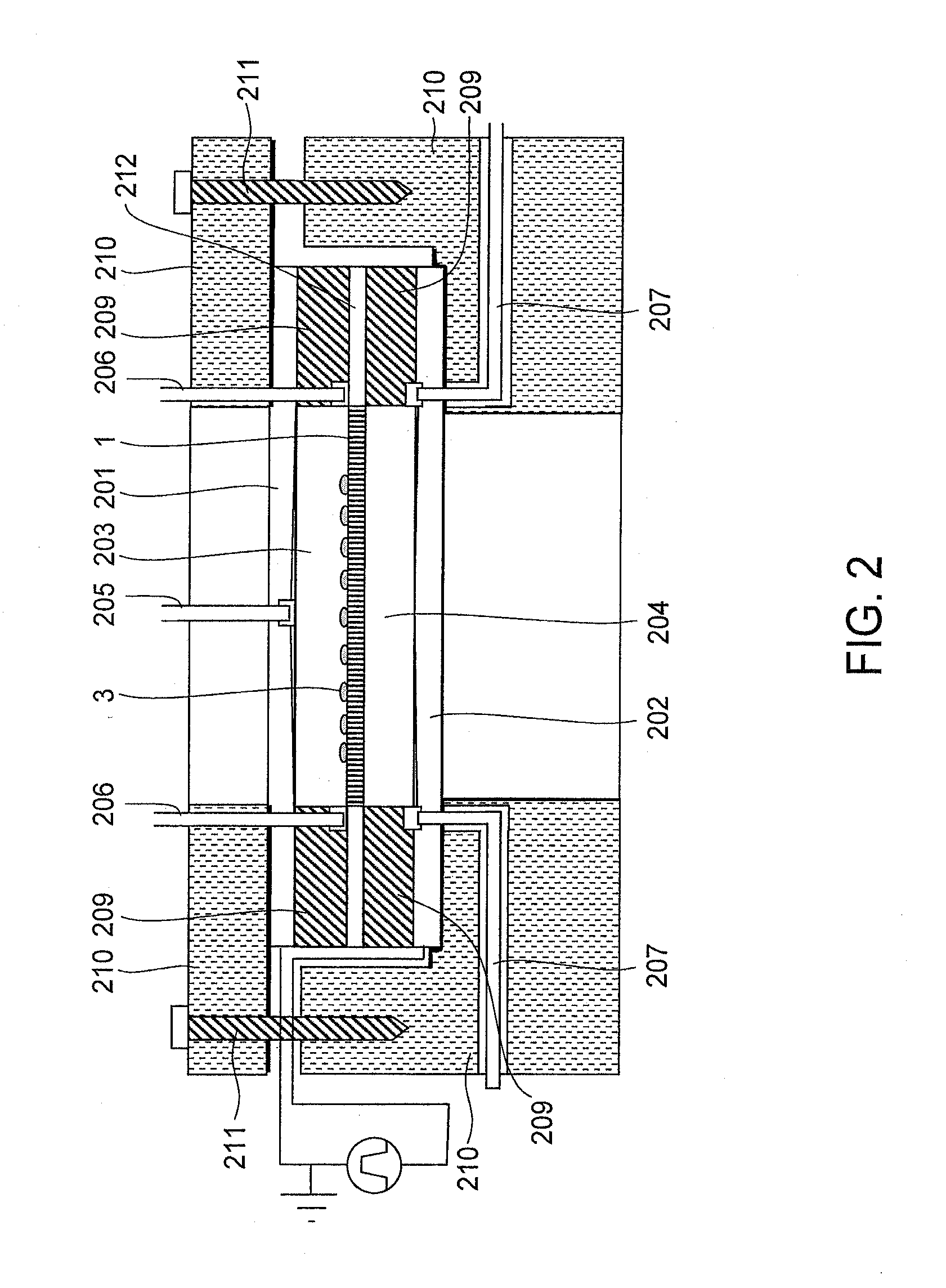
[0147]This Example is outlined in FIG. 11. This is an example of using a two-dimensional cDNA library sheet, which was constructed from a tissue slice sample 1101 in a pore array sheet. A frozen tissue slice was cut by a microtome into a film-form sample having a thickness of about 5 to 20 μm. As shown in FIG. 11, the tissue slice sample 1101 is mounted on a pore array sheet 1 made of alumina and immediately covered with a low melting-point agarose containing a cell lysis reagent. The agarose solution remains in a state of solution at 35° C. The sheet 1 is immediately gelatinized by cooling it to 4° C. Thereafter, the sheet is set in the reaction cell shown in FIG. 2, and mRNA is extracted in the same manner as Example 1.
[0148]A solution, which is prepared by mixing a low melting-point agarose gel (SeaPrep Agarose; gelatinization temperature is 19° C., melting point is 45° C. at an agarose concentration of 2%) and a cell lysis reagent, is used to lyse cell membrane and cell tissue w...
example 3
[0171]In this Example, unlike Example 1 or 2 in which a sheet having pores was used as a cDNA library sheet, another support such as a membrane was used to prepare a cDNA library.
[0172]As the membrane, a membrane rarely adsorbing a protein such as a cellulose acetate membrane, a nitrocellulose membrane or a membrane formed of a mixture of these and a nylon membrane can be available. In the Example, a case where a cellulose acetate membrane (Whatman) having a thickness of 115 μm, a pore size of 0.2 μm and a diameter of 25 mm will be described.
[0173]As shown in FIG. 18, to the surface of a membrane fiber 1802 within a membrane 1801, poly-T probes 1803 are immobilized by the silane coupling treatment in the same manner as in Example 1. At the same time, the MPC treatment is performed in the same manner. Next, the membrane is set in the same reaction cell as in Example 1 or 2. A tissue slice sample 1101 is mounted and the same treatments are performed. As a result, mRNA can be trapped b...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
| thickness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com