High-throughput single-cell transcriptome and gene mutation integration analysis coding chip
An integrated analysis, single-cell technology, applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc., can solve the problems of high sample demand, cumbersome operation, and not suitable for comprehensive promotion.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0028] The present invention will be further described in detail below in conjunction with the embodiments, so that those skilled in the art can implement it with reference to the description.
[0029] It should be understood that terms such as "having", "comprising" and "including" used herein do not exclude the presence or addition of one or more other elements or combinations thereof.
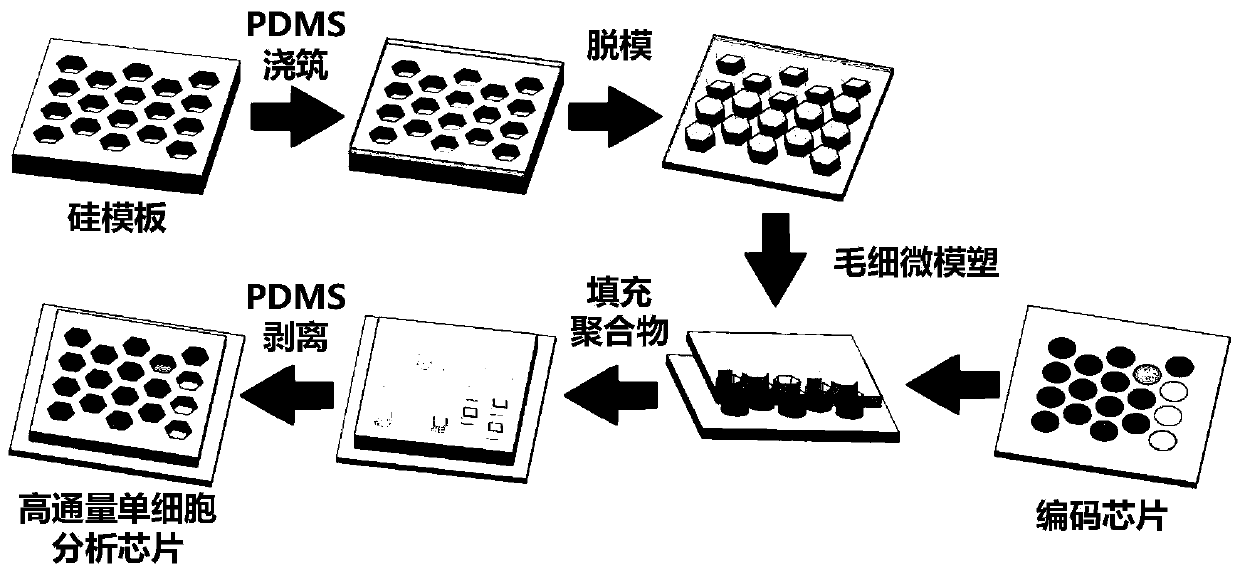
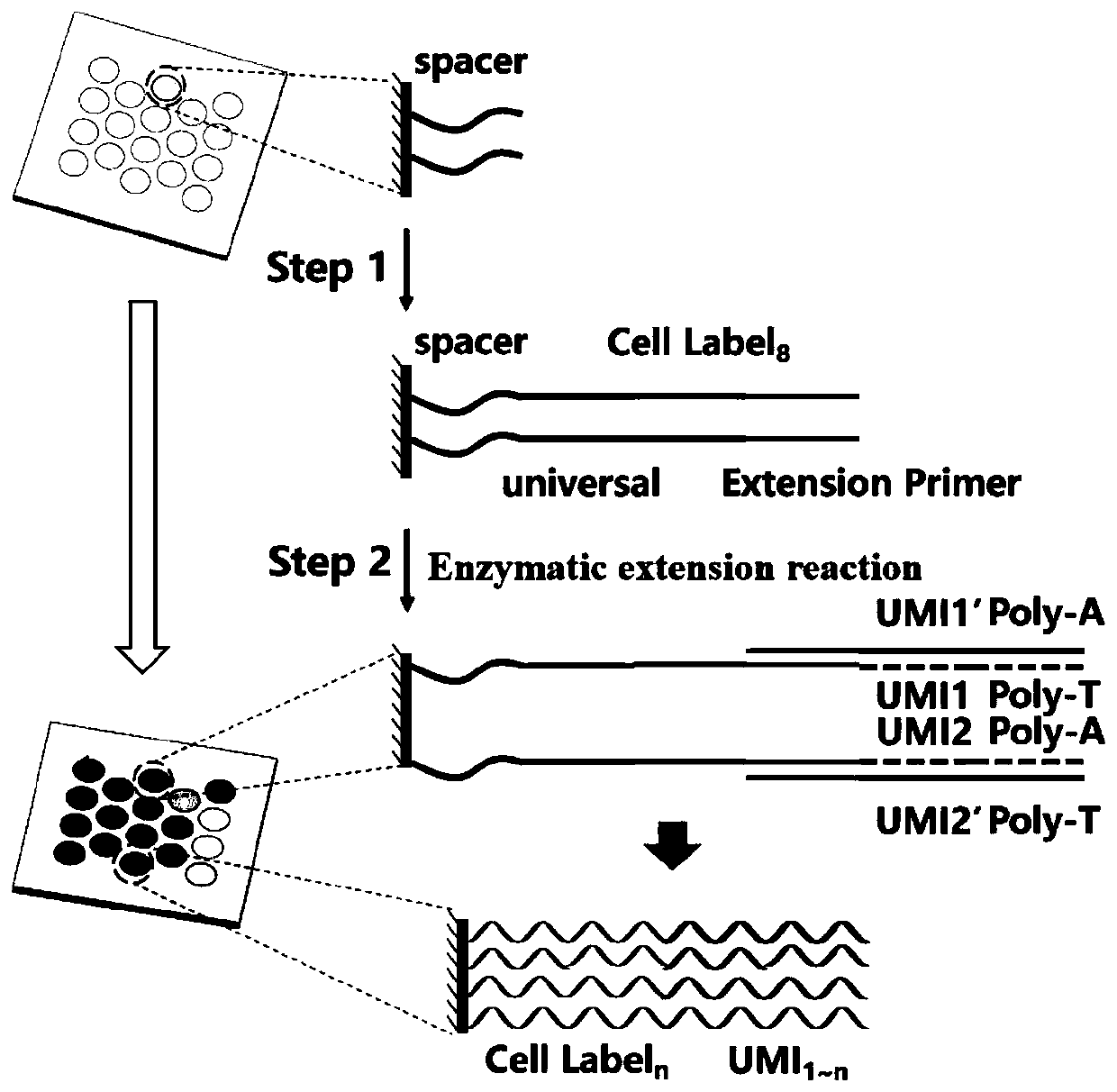
[0030] A high-throughput single-cell transcriptome and gene mutation integrated analysis encoding chip in this embodiment, the chip is provided with multiple microwells on its substrate, and the microwells have a size and shape that can only accommodate a single cell in a microwell , each microwell has a unique spatial coordinate code, and several known nucleic acid sequences are modified in the microwell, and the nucleic acid sequences include:
[0031] Spacer sequence;
[0032] Universal primer sequence, as the primer-binding region during PCR amplification;
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com