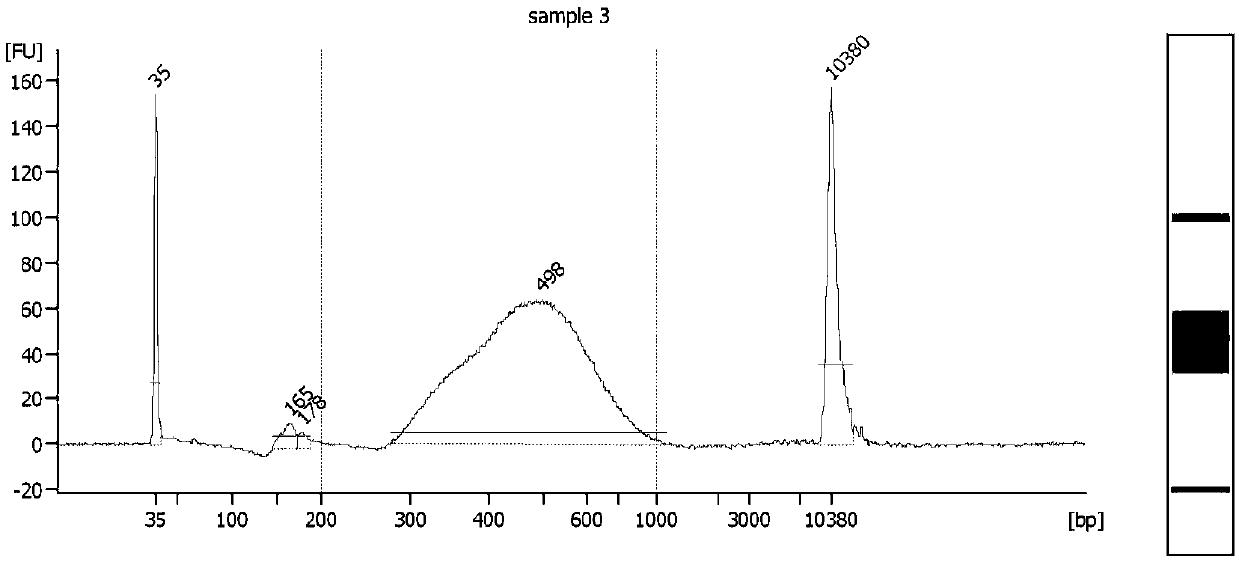
High-throughput single-cell transcriptome sequencing method and kit
A transcriptome sequencing and single-cell technology, applied in the field of single-cell sequencing, can solve the problems of inability to carry out large-scale research work, difficult detailed analysis of different cell subpopulations, constrained throughput, cost, etc., to reduce mutual contamination of microbeads , Convenient single-cell sequencing, and low-cost effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0058] 1. Prepare reagents
[0059] (1) Cell resuspension
[0060] Prepare 5 mL of PBS-BSA with a final concentration of 0.01% BSA; wash the H1975 cell line with PBS-BSA, and resuspend the cells in PBS-BSA for later use.
[0061] (2) Cleavage and reverse transcription reaction reagent preparation
[0062] 1mL of lysis and reverse transcription reaction reagents include: 10% Triton-X 10μL, 0.1M DTT 125μL, RnaseOUT 75μL, 10Mm each dNTPs 50μL, 5×RT buffer400μL, Super Script II ReverseTranscriptase 75μL, Template Switch Oligo 20μL, Rnase-free H 2 O 265 μL.
[0063] Wherein, Template Switch Oligo is the sequence shown in SEQ ID NO.5,
[0064] SEQ ID NO.5: 5'-AAGCAGTGGTATCAACGCAGAGTGAATGGG-3'
[0065] In Template Switch Oligo, the penultimate 2nd and 3rd bases are modified with riboguanosine, and the last base is modified with LNA.
[0066] (3) Microbead resuspension
[0067] In this example, the microbeads have primer sequences with poly(T) random sequences, which can capture...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com