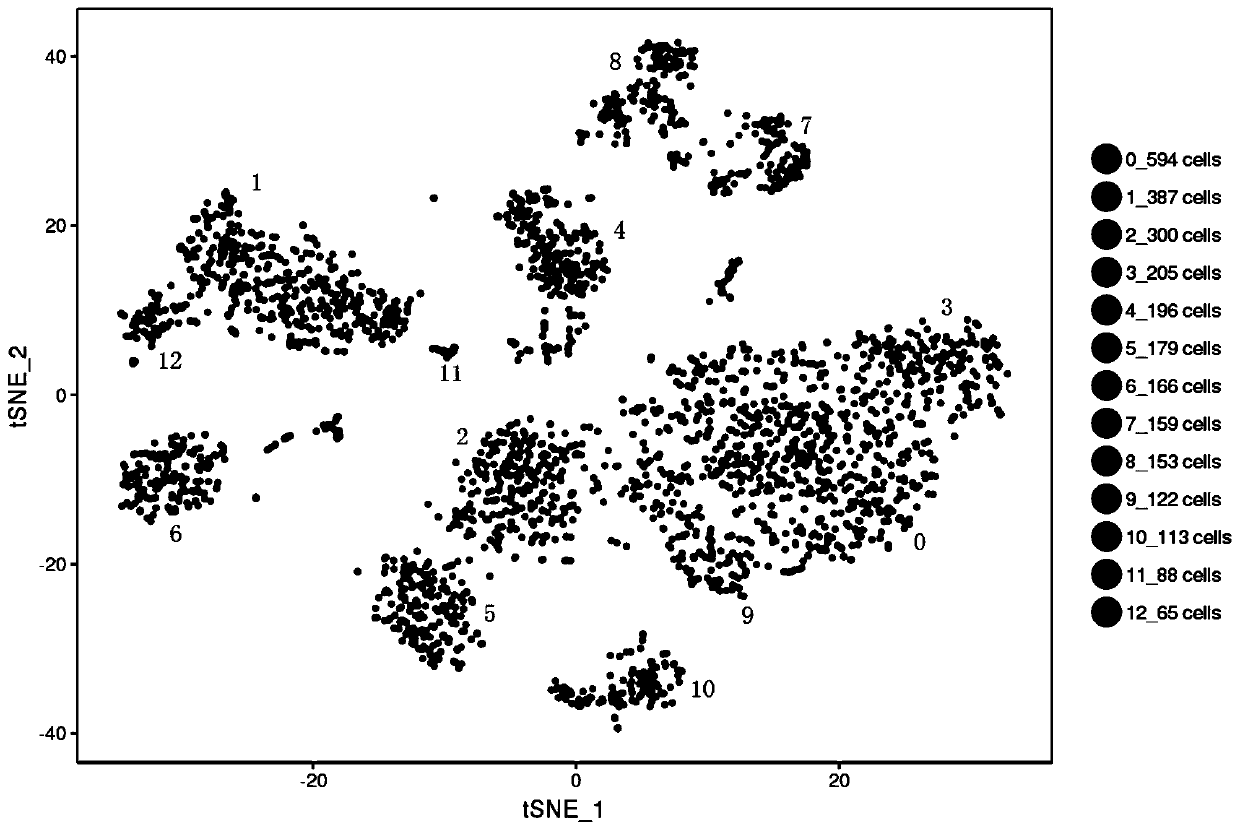
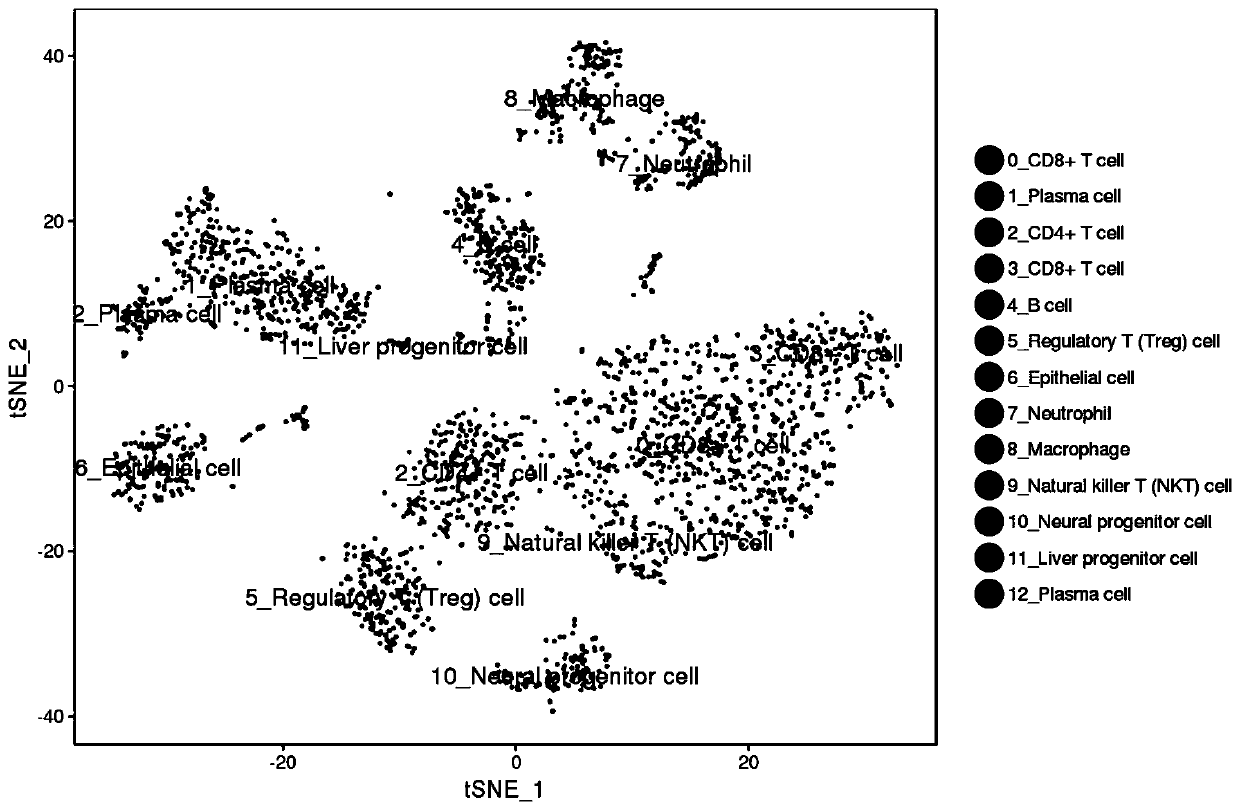
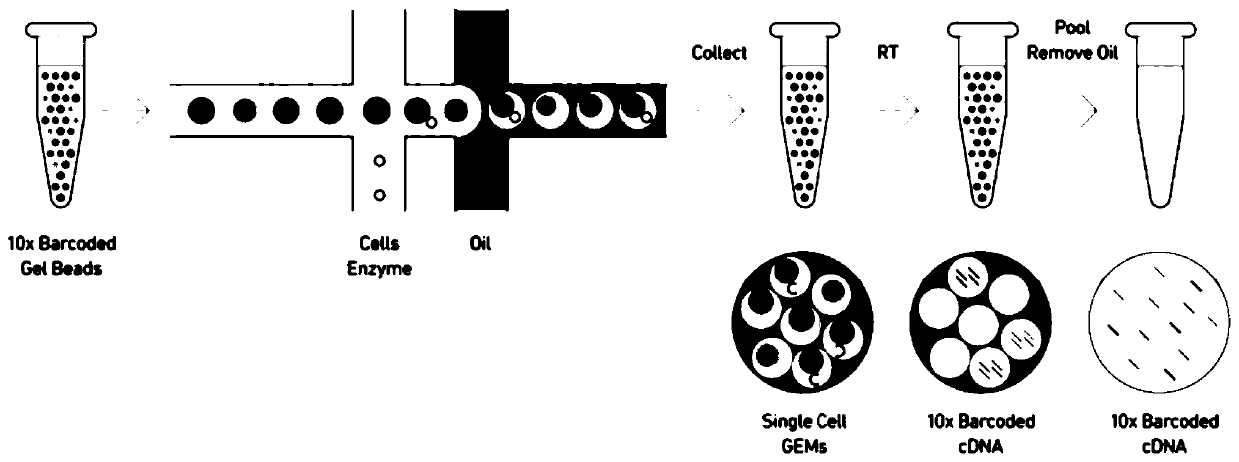
Method for annotating cell identities based on single cell transcriptome clustering results
A transcriptome and single-cell technology, applied in genomics, proteomics, instruments, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] An embodiment of the method for annotating cell identity based on single-cell transcriptome clustering results in the present invention includes the following steps:
[0037] 1) Library construction and sequencing
[0038] A case of fresh nasopharyngeal carcinoma tissue (male, 42 years old, pathologically diagnosed as poorly differentiated squamous cell carcinoma of the nasopharynx) was obtained from the Department of Otorhinolaryngology, Guangzhou Twelfth People's Hospital. Immediately put the cancer tissue into tissue culture medium, keep it on ice, and make cell suspension within two hours. After the cell counter detects that the proportion of viable cells exceeds 90%, it is sent to Guangzhou Sequoma Biotechnology Co., Ltd. for 10X genomics library construction. After the library is built, it will be sent to Beijing Annoroad Gene Technology Co., Ltd. for sequencing. The sequencing platform was Illumina Hiseq X, and the sequencing depth was packet lane sequencing. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com