Method for analyzing double cells in single-cell transcriptome data
A single-cell, double-cell technology, applied in the field of double-cell analysis, can solve the problems of cumbersome steps, high knowledge background, false positives and false negatives, and achieve the effect of improving efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
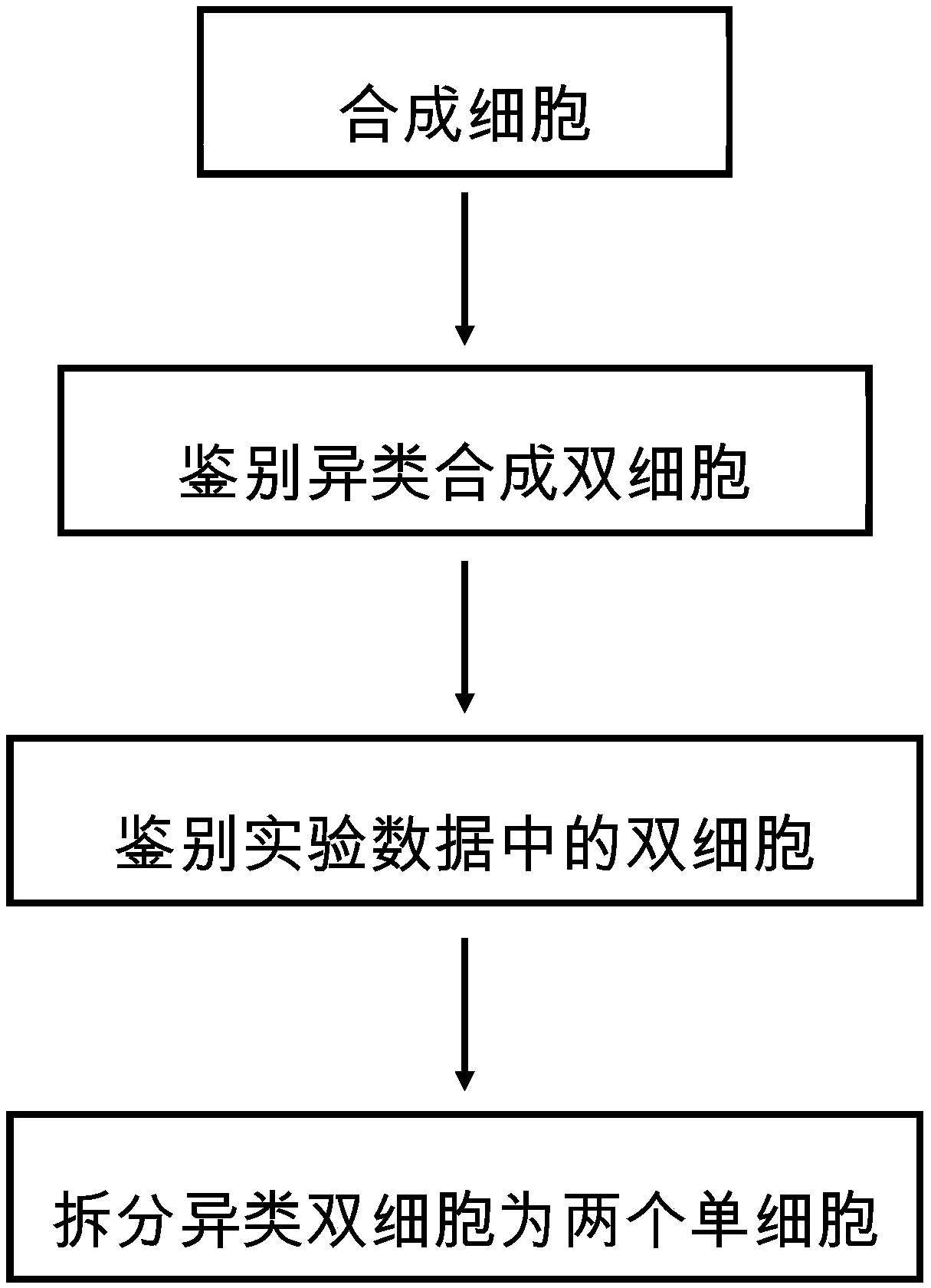
[0027] This embodiment discloses a method for automatically identifying double cells and splitting the cell expression profiles in the double cells. The principle is to carry out identification and analysis of double cells based on computer simulation, which mainly includes the following four processes:
[0028] Synthetic Cells: Synthesize cell expression profiles based on experimental data through computer simulations. The experimental data is the detected cell data. Since the proportion of double cells in the experimental data is very low, most of them are single cells, so most of the synthetic cells will be obtained by superimposing two random source single cells in the detected single cell data. into, that is, the formation of synthetic twin cells.
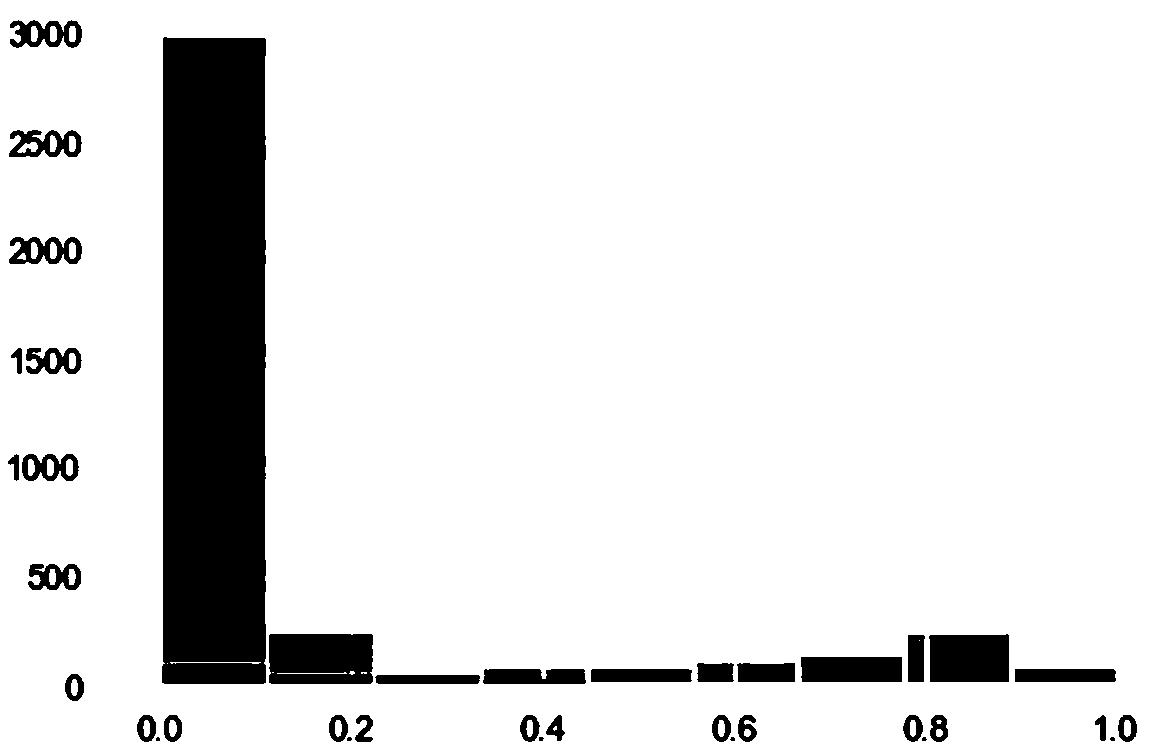
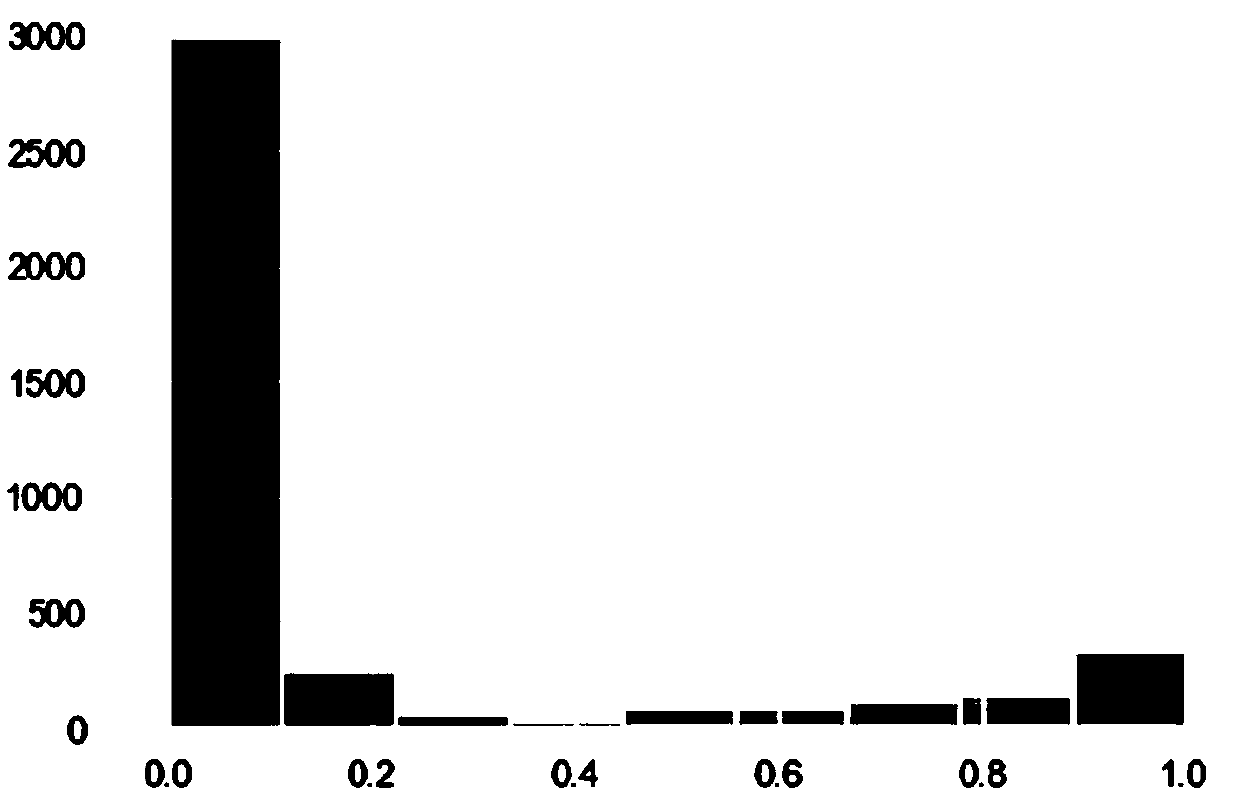
[0029] There are two types of twin cells, homogeneous twin cells are formed by mixing 2 cells belonging to the same cell type, and heterogeneous twin cells are formed by mixing 2 cell types of different kinds. Therefore, the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com