High-throughput single-cell transcriptome and gene mutation integration analysis method
An integrated analysis and single-cell technology, applied in the direction of biochemical equipment and methods, microbial measurement/testing, etc., can solve problems such as not suitable for comprehensive promotion, cumbersome operation, and inability to realize direct identification of tumor cell populations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
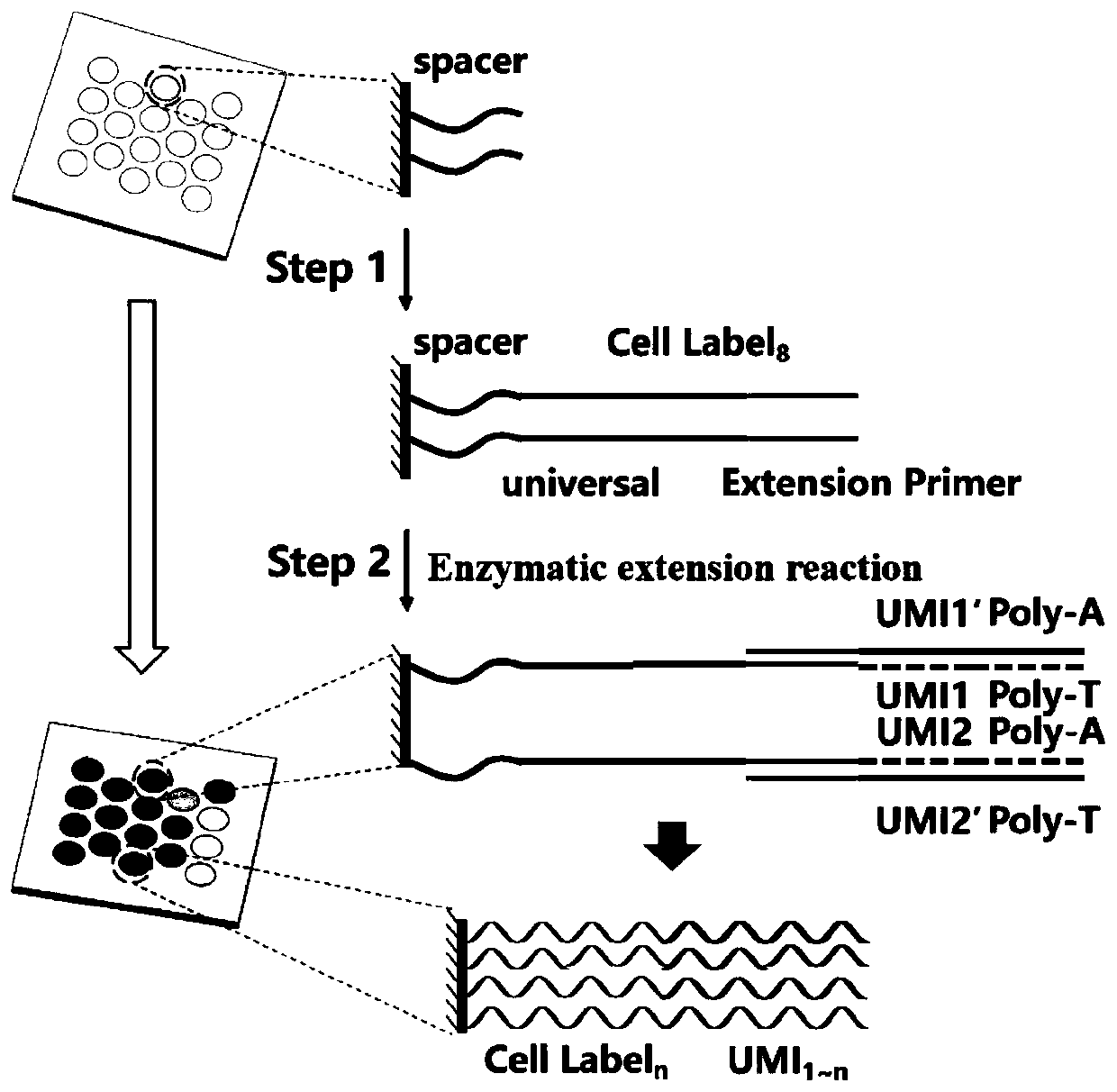
[0039] Embodiment 1 provides a high-throughput single-cell coding chip
[0040] The chip is provided with a plurality of microwells on its substrate, each of which has a unique spatial coordinate code, and several known nucleic acid sequences for capturing target RNA are modified in the microwells, so The nucleic acid sequence includes a cell label for marking the cell from which the RNA originates and a molecular label for marking the bound RNA, and the cell label of each microwell is in one-to-one correspondence with the spatial coordinate code.
[0041] Wherein, the nucleic acid sequence also includes a Spacer sequence, a universal primer sequence as a primer binding region during PCR amplification, and Ploy T. In a further preferred embodiment, the modified nucleic acid sequence in each microwell is not less than 10 6 strip. A molecular tag is a known random nucleic acid sequence.
[0042] Here, the microwells have a size and shape that can accommodate only a single cel...
Embodiment 2
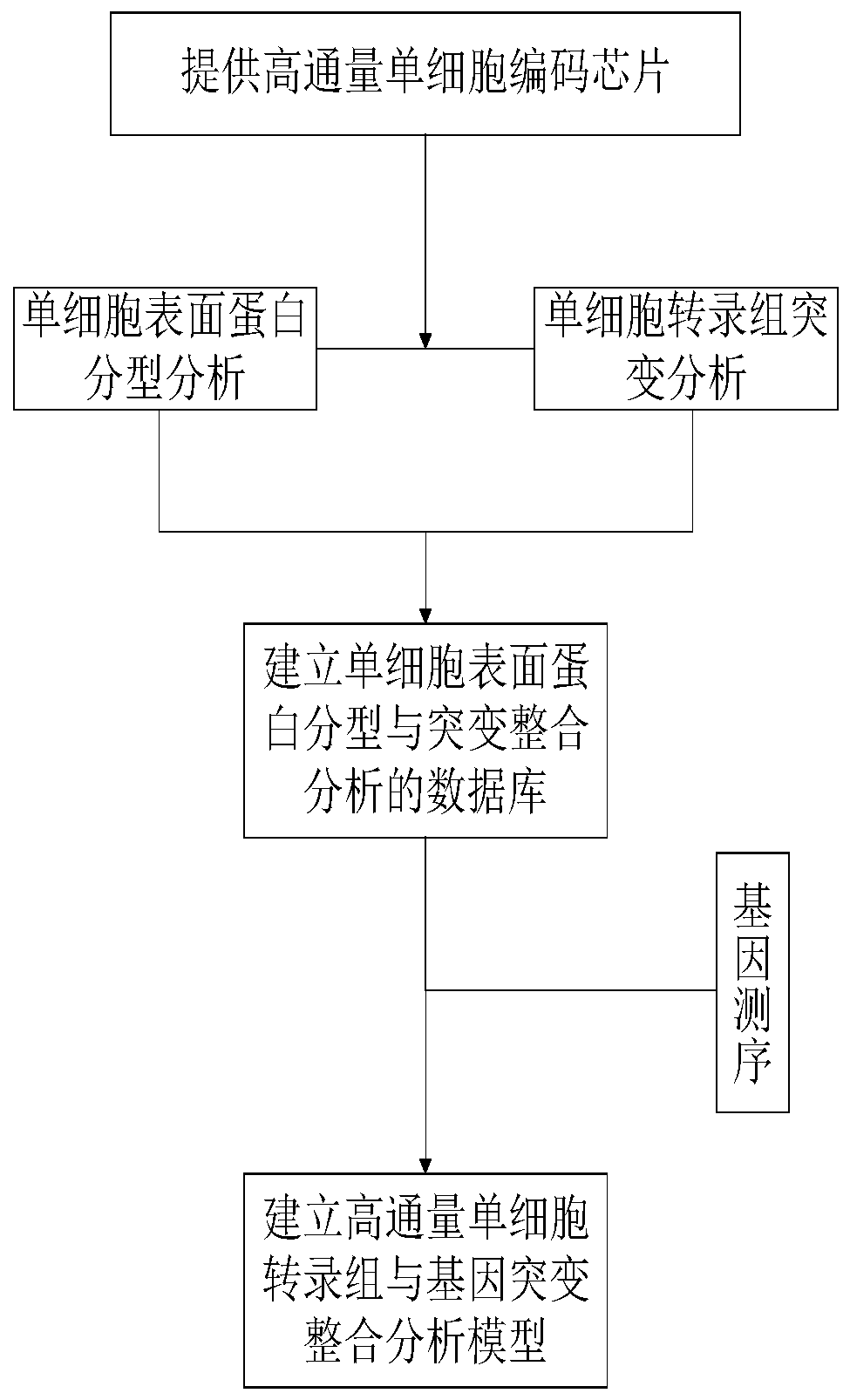
[0052] Example 2 Using the chip of Example 1 to perform integrated analysis of single-cell transcriptome and gene mutation, refer to figure 1 , including the following steps:
[0053] 1. Single cell surface protein typing analysis:
[0054] The target gene of the cell is fluorescently labeled in advance, and then the cell is added to the chip, and the single cell is captured by the micropore of the chip, incubated, and then the fluorescence image is collected, and the position of the micropore on the fluorescence image is positioned. Fluorescent image analysis is used to identify the location of specific cells containing the target gene, and the cell protein expression information of each microwell location is obtained. Among them, three-color fluorescence channels are used for fluorescence imaging, and light sources of three different wavelengths are used to achieve full-band visible light coverage (wavelength 400nm-700nm).
[0055] Wherein, the method for locating the posi...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| depth | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com