Preparation method of random sgRNA library of enzymic method target sequence
A target sequence and library technology, which is applied in the field of enzymatic target sequence random sgRNA library preparation to achieve the effects of small preference, uniform coverage and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
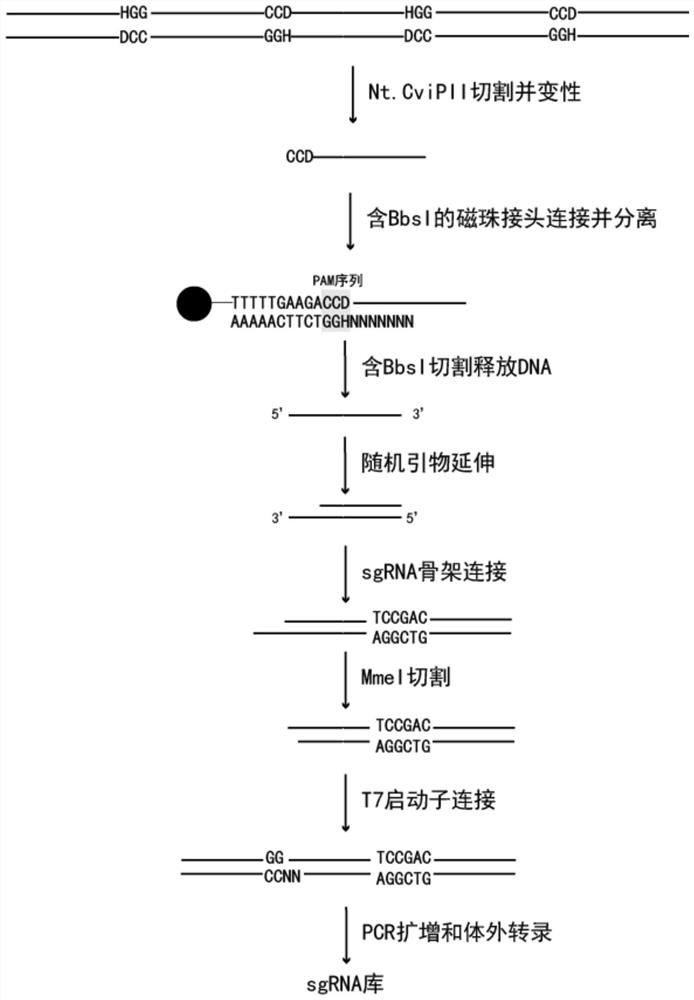
[0038] Example 1: ERSP process
[0039] (1) Preparation of magnetic beads containing Bbs linker
[0040] BbsI-F and BbsI-R were dissolved in 100mM NaCl to 100μM, mixed with equimolar concentration, reacted at 95℃ for 5min, and decreased by 1℃ per minute. After the reaction, the annealed linker was diluted to 10 μM with water. Take 20 μL of the diluted adapter, add 5 μL of streptavidin magnetic beads C1 (Thermo), and rotate and mix at room temperature for 30 min. Wash the magnetic beads 3 times with 100mM NaCl.
[0041] (2) Recognition of restriction enzyme cutting of PAM sequence:
[0042] Table 2 restriction enzyme digestion system
[0043] components Dosage human genomic DNA 1μg 10×rCutSmart buffer 1μL Nt.CviPII (NEB) 0.5-2U Hydrate to 10μL
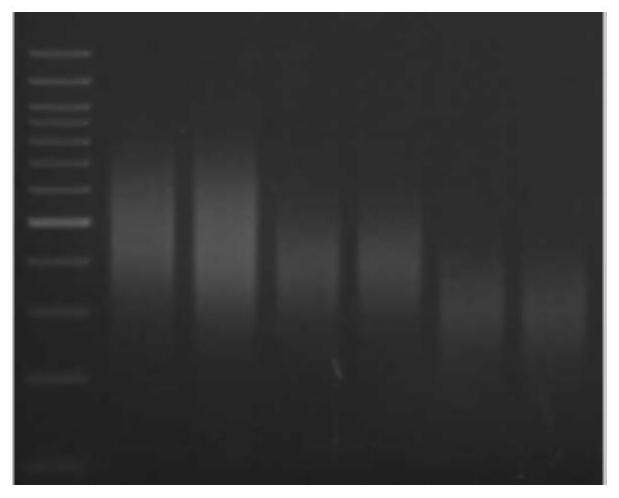
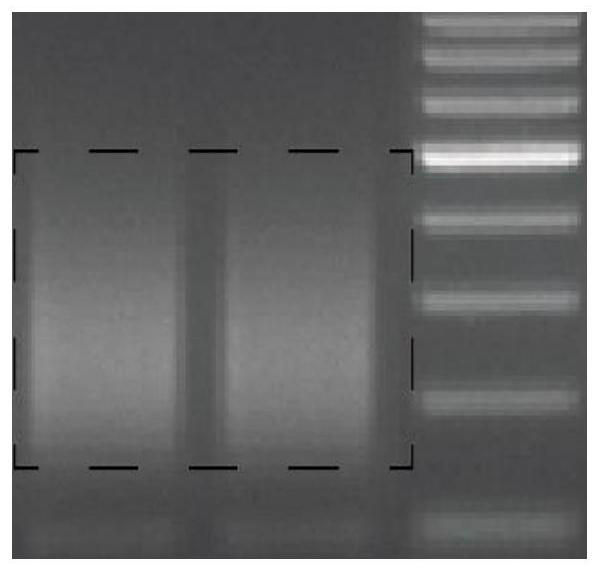
[0044]React at 37°C for 30 minutes, react at 98°C for 10 minutes, and place on ice immediately. The effects of different input amounts of Nt.CviPII on DNA fragmentation are as follows: ...
Embodiment 2
[0076] Example 2: Application of ERSP on removal of rRNA and host genome.
specific Embodiment approach
[0077] In this example, we first prepared a random sgRNA library covering 18SrRNA and 28S rRNA using human RNA as a template according to the ERSP method in Example 1; using human genomic DNA as a template, prepared a random sgRNA library covering human genomic DNA Library, referred to as ERSP library, and applied to rRNA removal and host genome removal. The specific implementation is as follows:
[0078] Table 10 sgRNA library and Cas9 protein pre-assembly:
[0079] components Dosage ERSP library 2-10μg Cas9 (NEB) 0.2-0.5μg 500mM NaCl 1μL total capacity 5μL
[0080] 37°C for 30min.
[0081] Table 11 CRISPR cleavage
[0082] components Dosage Cas9-sgRNA library 5μL RNA or DNA library 1-100ng 10×NEB buffer 3.1 1μL total capacity 10μL
[0083] 37°C for 30-90min, 90°C for 10min.
[0084] (5) Library amplification
[0085] Table 12
[0086] components Dosage The above re...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com