Linker sequence and method for detecting ultra-low frequency mutation of target sequence
A linker sequence and target sequence technology, which is applied in the field of linker sequences with ultra-low frequency mutations, to achieve the effect of reducing the dosage and solving the heterogeneity of tumor cells
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
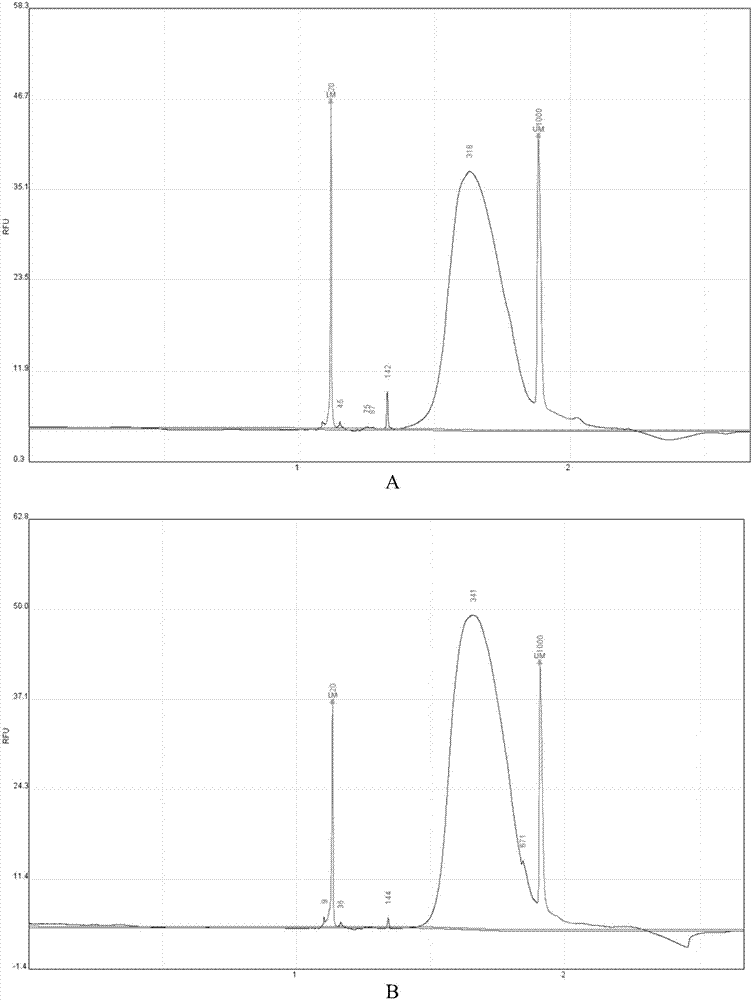
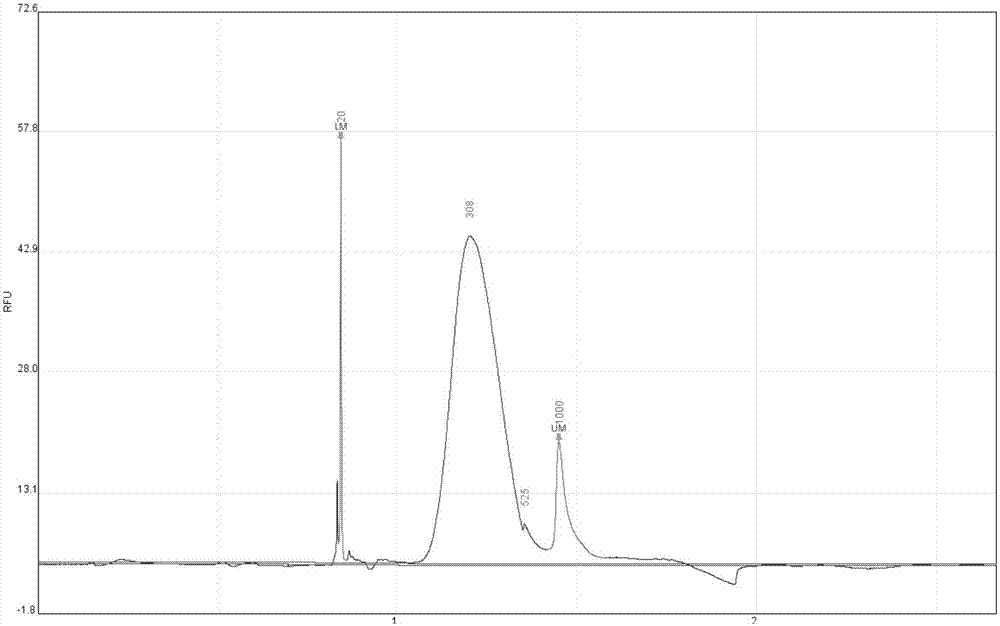
[0053] In the embodiment of the present invention, a certain amount of tagged molecules are obtained by performing end-repair, A addition, adapter ligation, fragment enrichment and capture on the cfDNA standard HD780, which is then sequenced on the computer, combined with The later bioanalytical method corrects the positive and negative chains and analyzes the real mutation information.
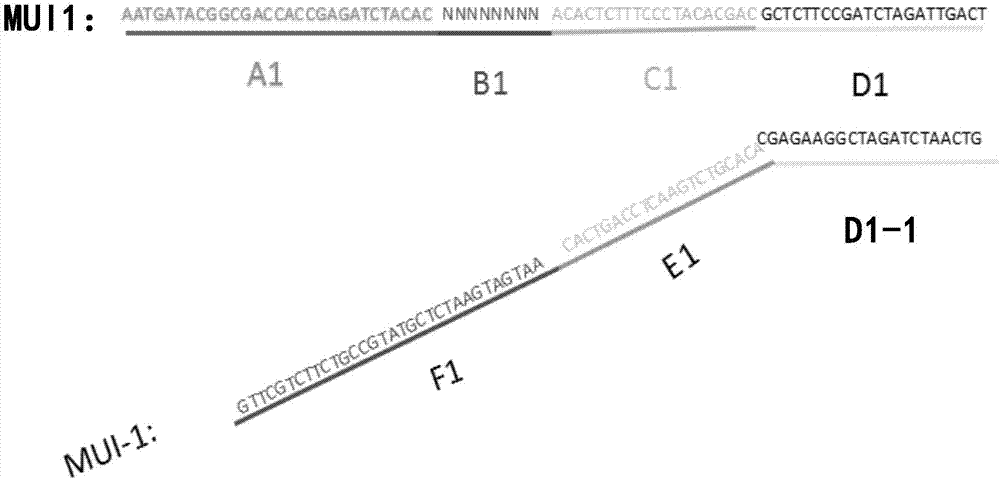
[0054] Design and synthesis of linker sequences with molecular tags:
[0055] MID1:
[0056] AATGATACGGCGACCACCGAGATCTACACNNNNNNNNACACTCTTTCCCTACACGACGCTCTTCCGATCTAGATTGACT (SEQ ID NO. 1)
[0057] MID-1:
[0058] GTCAATCTAGATCGGAAGCAGCACACGTCTGAACTCCAGTCACAATGATGAATCTCGTATGCCGTCTTCTGCTTG (SEQ ID NO. 2)
[0059] MID2:
[0060] AATGATACGGCGACCACCGAGATCTACACNNNNNNNNACACTCTTTCCCTACACGACGCTCTTCCGATCTTACGTGACT (SEQ ID NO. 3)
[0061] MID2-2:
[0062] GTCAGCTAAGATCGGAAGACACACGTCTGAACTCCAGTCACAATGATGAATCTCGTATGCCGTCTTCTGCTTG (SEQ ID NO. 4)
[0063] MID3:
[0064] AATGATACGGCGACCACCGAGATCTACAC...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com