Patents
Literature
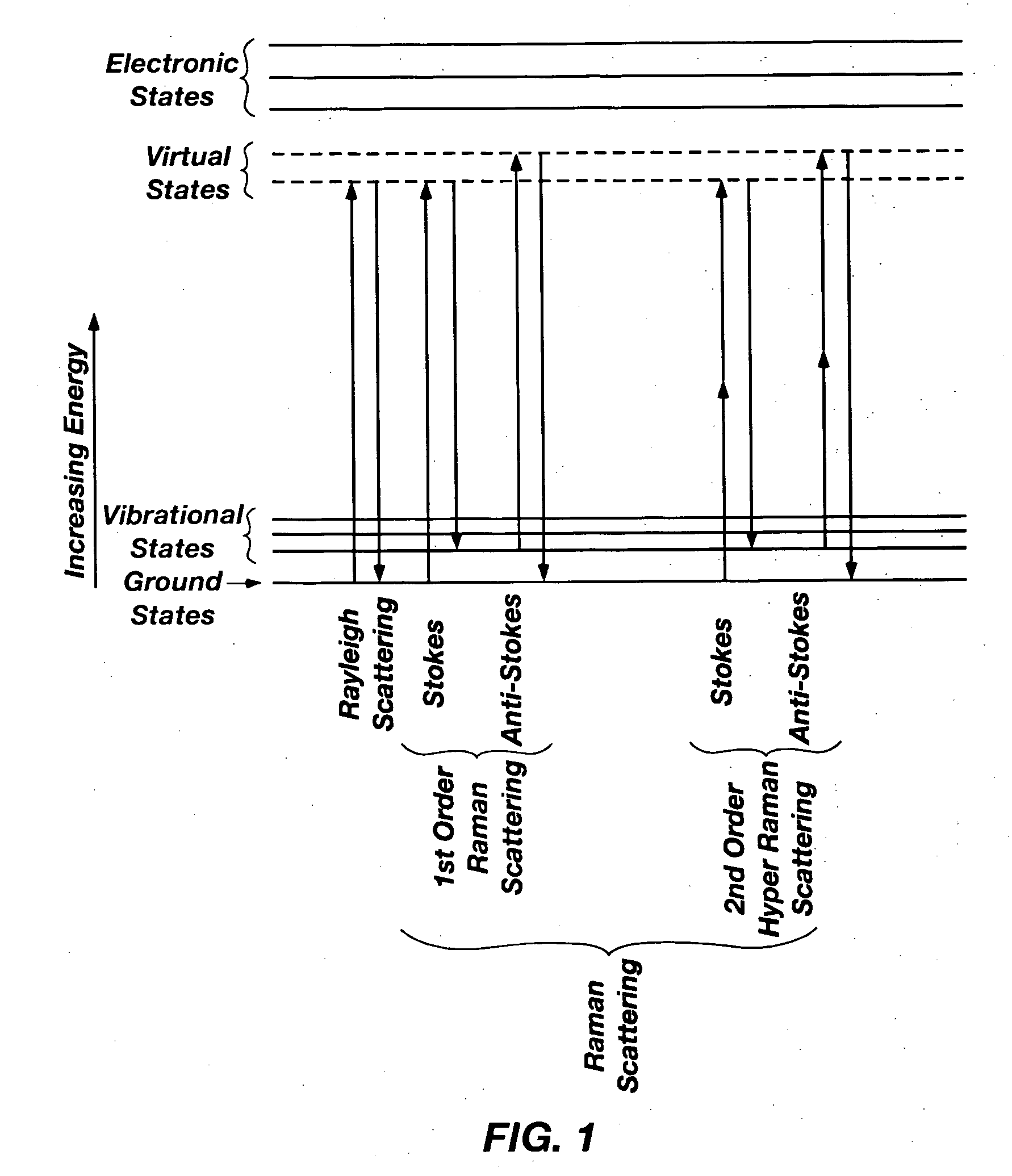
9350results about "Raman scattering" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
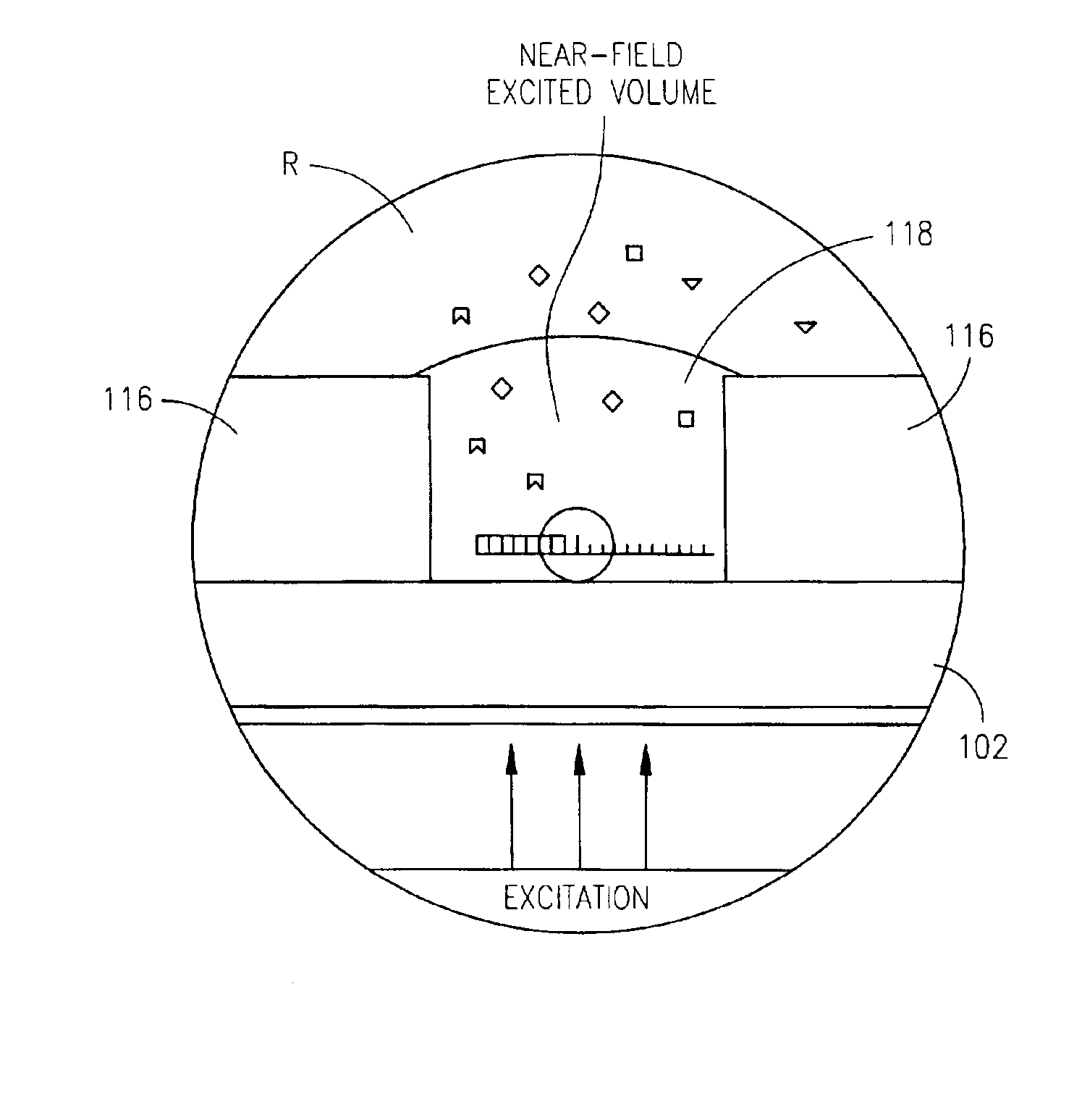
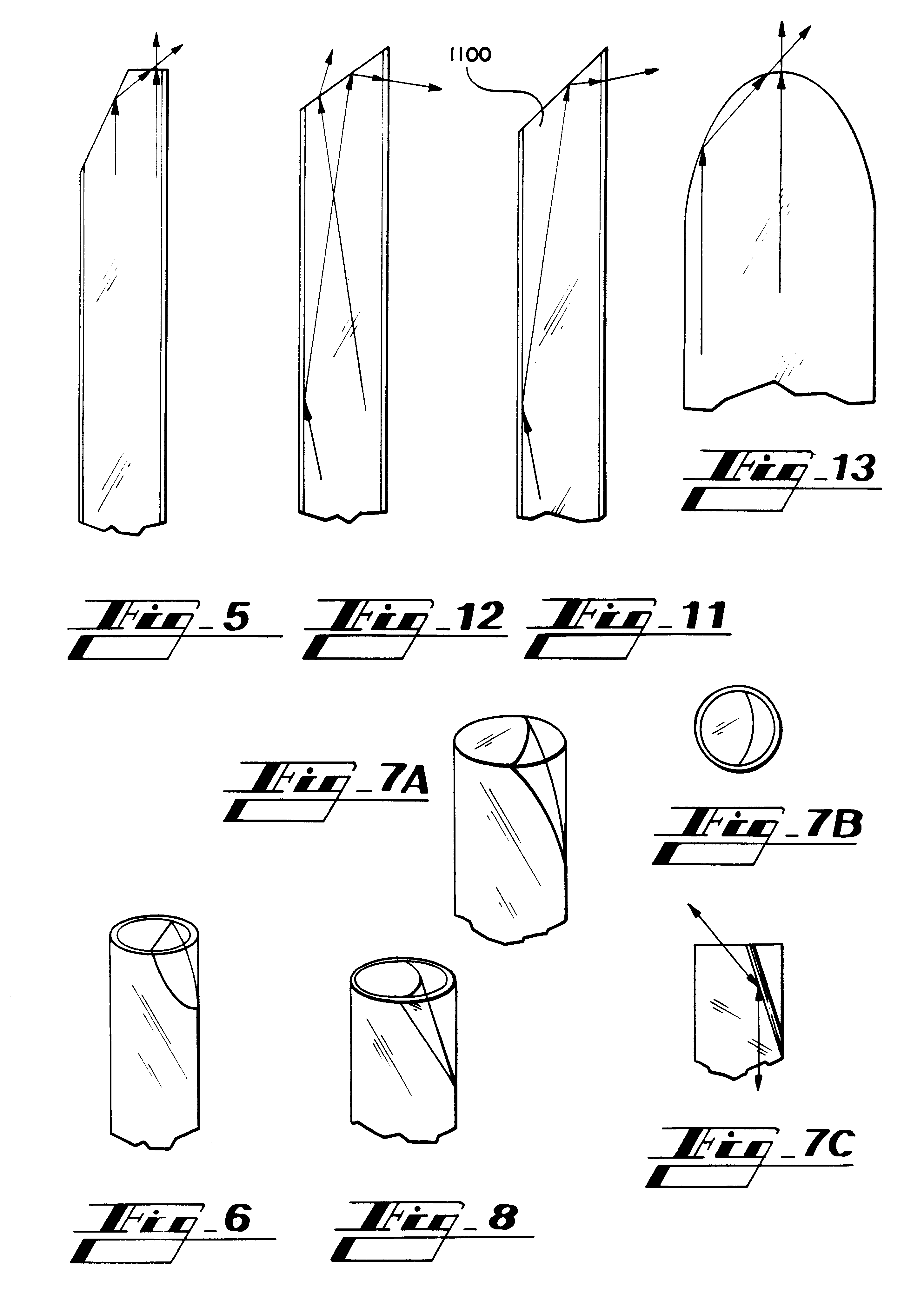
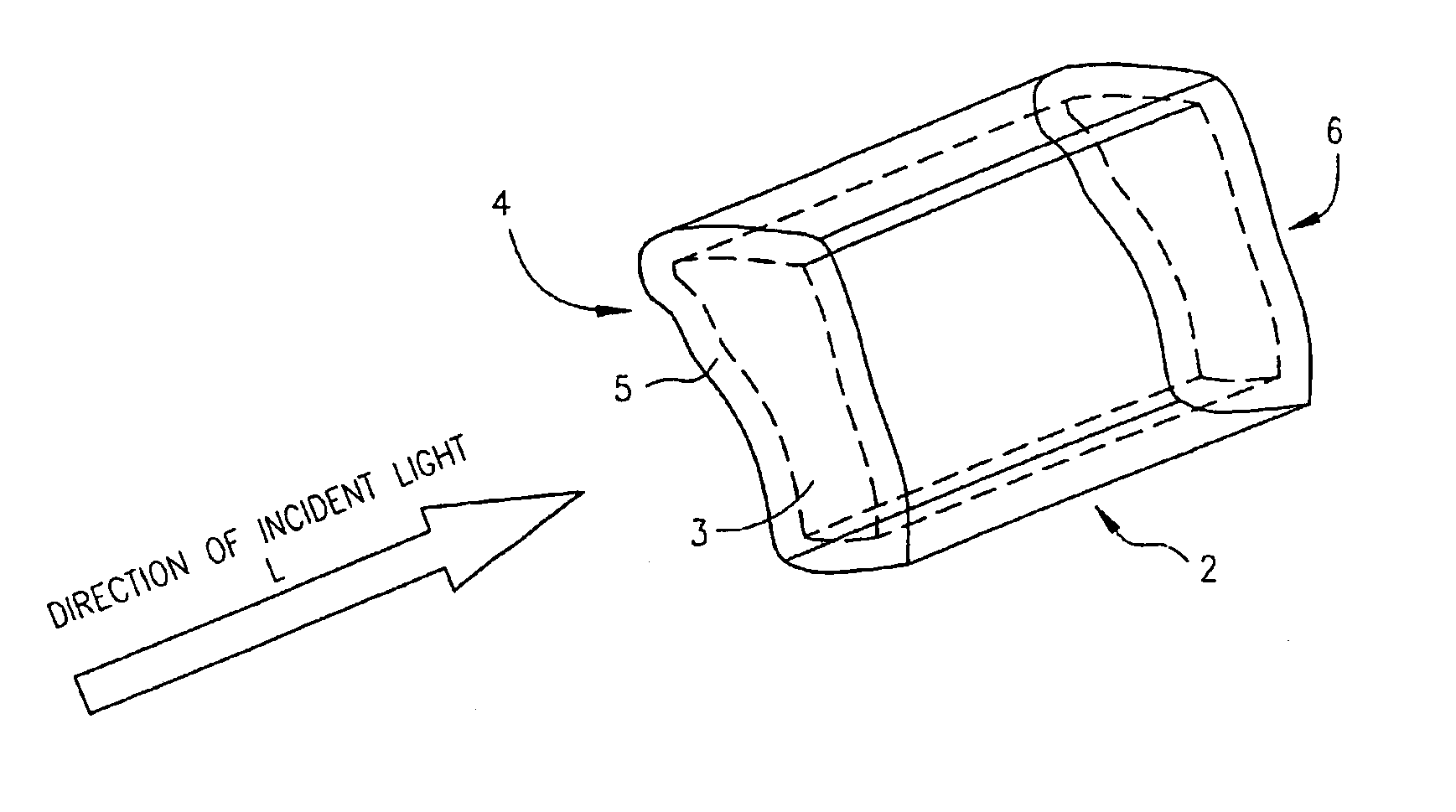
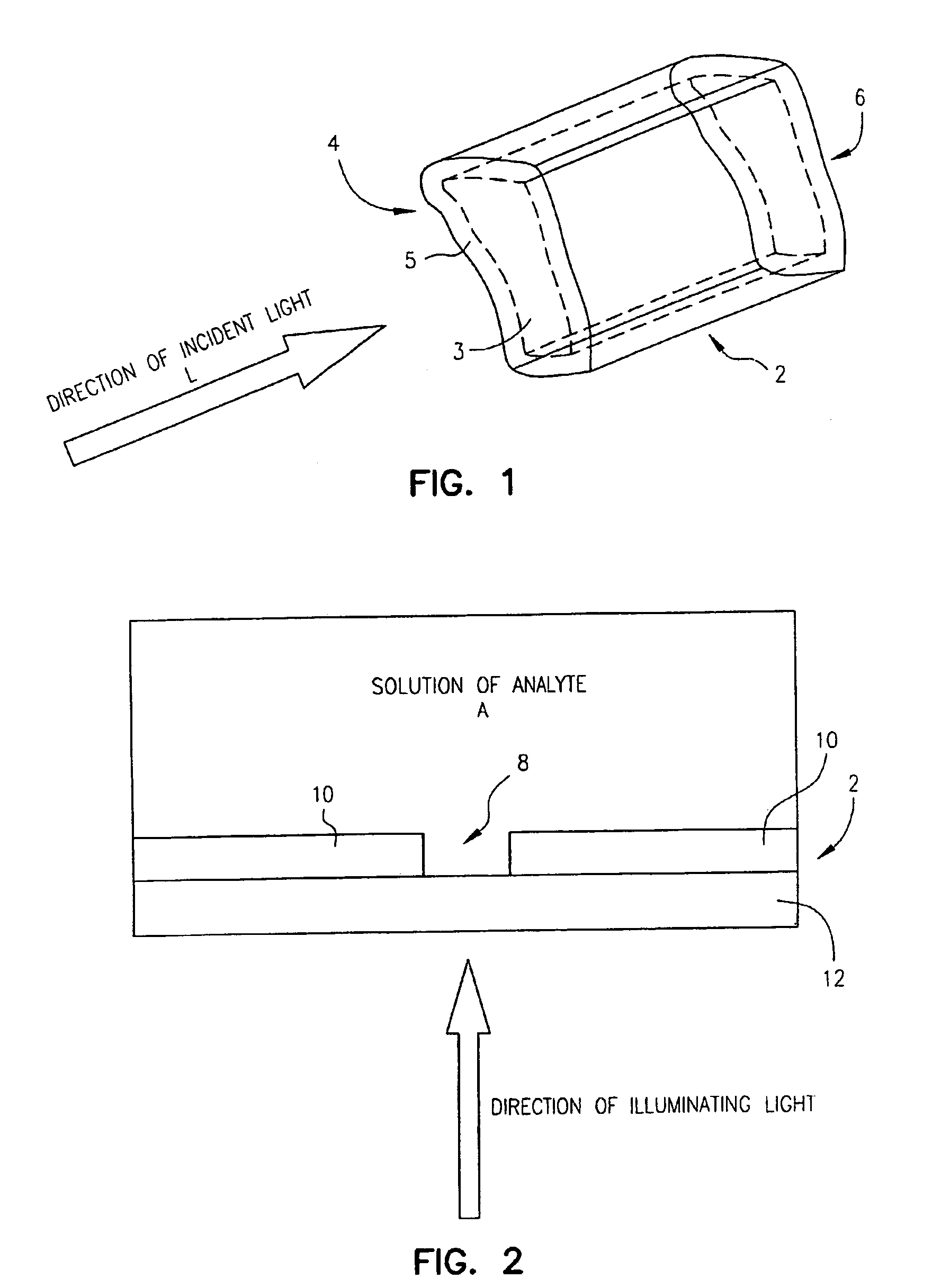
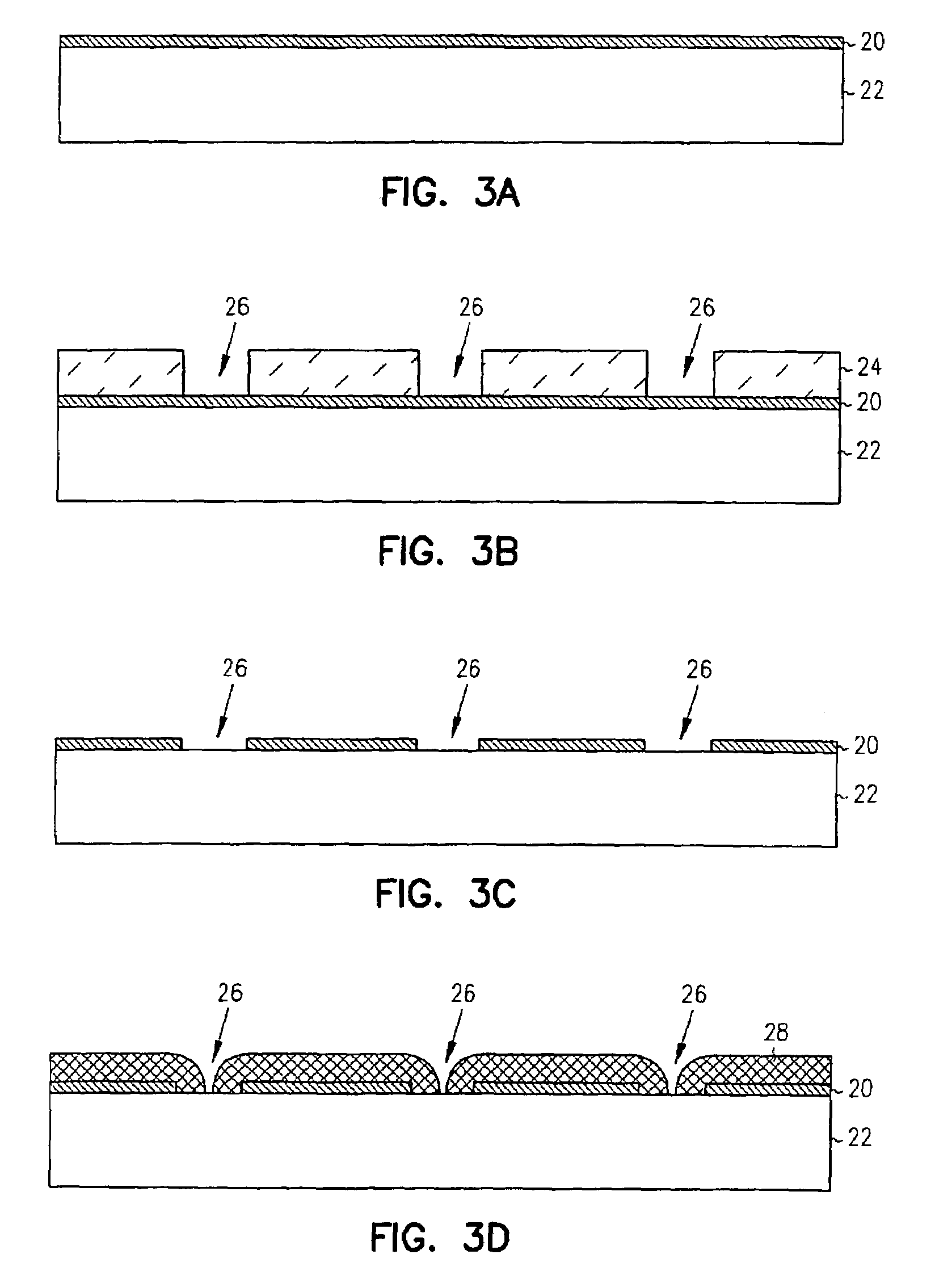
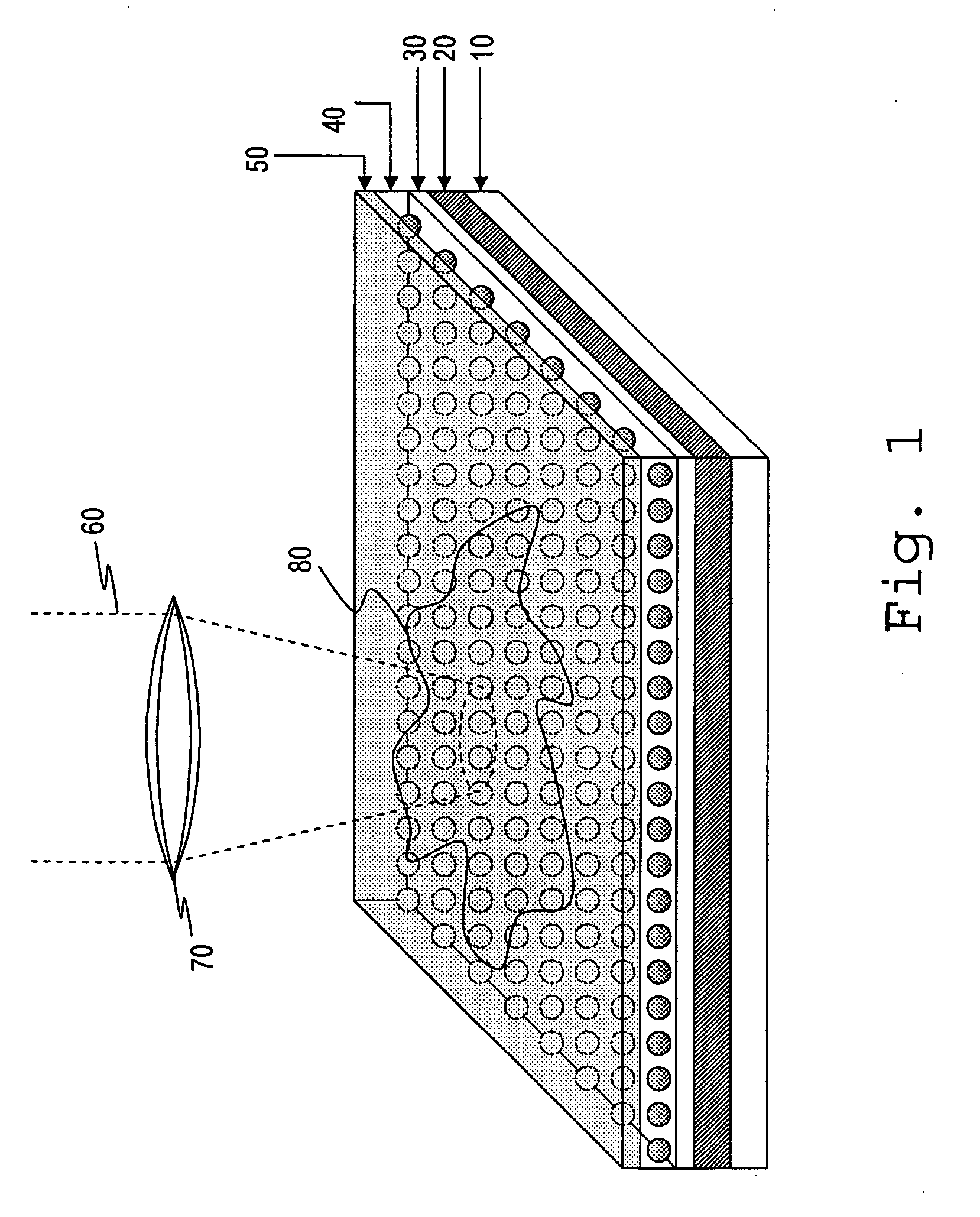
Zero-mode clad waveguides for performing spectroscopy with confined effective observation volumes
InactiveUS6917726B2Effective volumeEasy to useMicrobiological testing/measurementBiological material analysisAnalyteSpectroscopy
The present invention is directed to a method and an apparatus for analysis of an analyte. The method involves providing a zero-mode waveguide which includes a cladding surrounding a core where the cladding is configured to preclude propagation of electromagnetic energy of a frequency less than a cutoff frequency longitudinally through the core of the zero-mode waveguide. The analyte is positioned in the core of the zero-mode waveguide and is then subjected, in the core of the zero-mode waveguide, to activating electromagnetic radiation of a frequency less than the cut-off frequency under conditions effective to permit analysis of the analyte in an effective observation volume which is more compact than if the analysis were carried out in the absence of the zero-mode waveguide.
Owner:CORNELL RES FOUNDATION INC
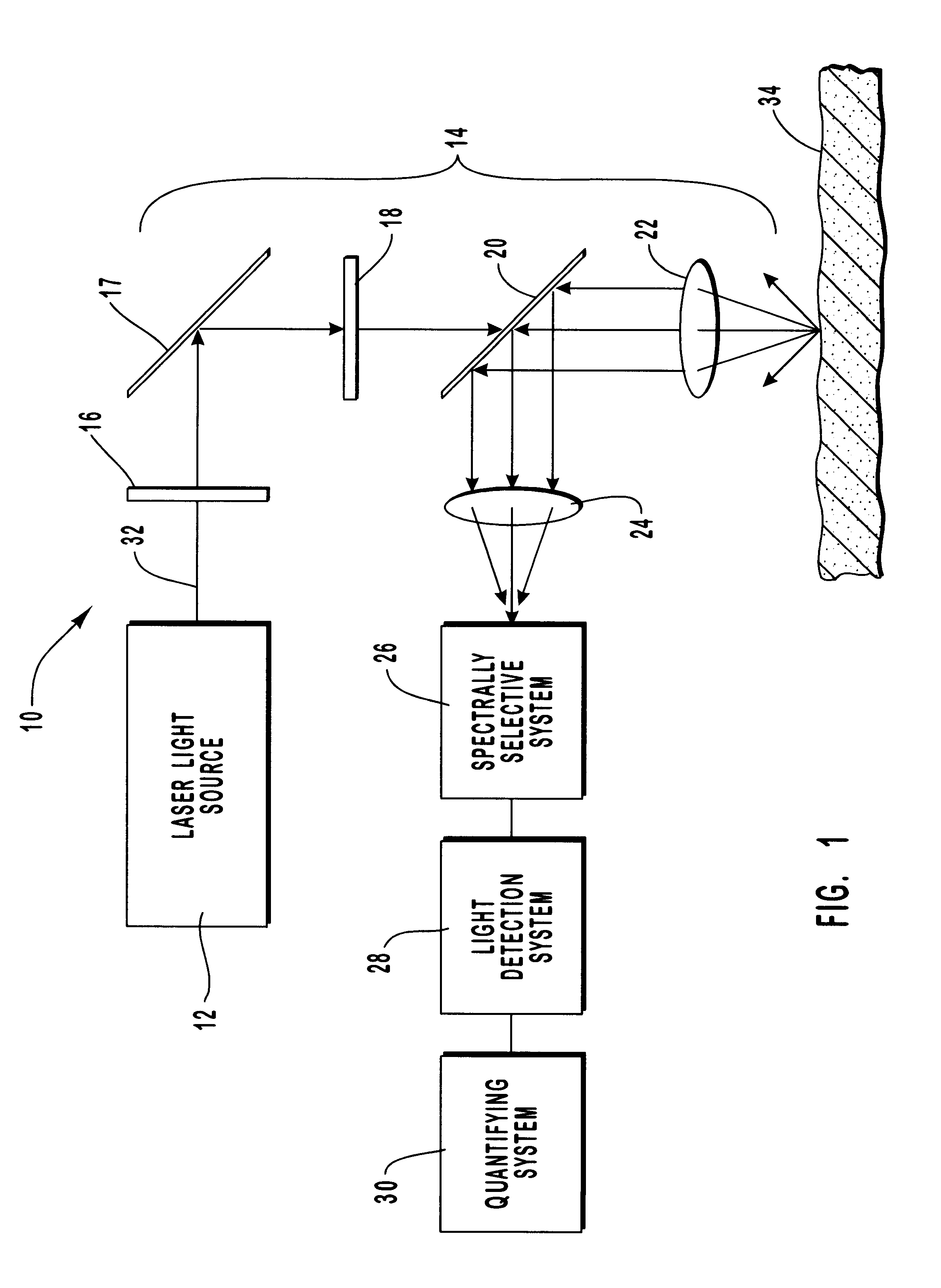
Method and apparatus for noninvasive measurement of carotenoids and related chemical substances in biological tissue
InactiveUS6205354B1Rapid and noninvasive and quantitative measurementRiskRadiation pyrometrySurgeryResonance Raman spectroscopyAntioxidant
A method and apparatus are provided for the determination of levels of carotenoids and similar chemical compounds in biological tissue such as living skin. The method and apparatus provide a noninvasive, rapid, accurate, and safe determination of carotenoid levels which in turn can provide diagnostic information regarding cancer risk, or can be a marker for conditions where carotenoids or other antioxidant compounds may provide diagnostic information. Such early diagnostic information allows for the possibility of preventative intervention. The method and apparatus utilize the technique of resonance Raman spectroscopy to measure the levels of carotenoids and similar substances in tissue. In this technique, laser light is directed upon the area of tissue which is of interest. A small fraction of the scattered light is scattered inelastically, producing the carotenoid Raman signal which is at a different frequency than the incident laser light, and the Raman signal is collected, filtered, and measured. The resulting Raman signal can be analyzed such that the background fluorescence signal is subtracted and the results displayed and compared with known calibration standards.
Owner:UNIV OF UTAH RES FOUND
Methods and apparatus for filtering an optical fiber
InactiveUS6222970B1Improve responseReduce sensitivityCladded optical fibreMaterial analysis by observing effect on chemical indicatorHigh energyPhotonics
Filtering of optical fibers and other related devices. High-energy methods for depositing thin films directly onto the ends of optical fibers can be used to produce high-quality, high-performance filters in quantity at a reasonable cost. These high-quality filters provide the high performance needed for many demanding applications and often eliminate the need for filters applied to wafers or expanded-beam filtering techniques. Having high-quality filters applied directly to optical fiber and faces permits production of high-performance, micro-sized devices that incorporate optical filters. Devices in which these filters may be used include spectroscopic applications including those using fiber optic probes, wavelength division multiplexing, telecommunications, general fiber optic sensor usage, photonic computing, photonic amplifiers, pump blocking and a variety of laser devices.
Owner:CIRREX SYST
Waveguides for performing spectroscopy with confined effective observation volumes
InactiveUS7013054B2Effective volumeEasy to useCladded optical fibreMicrobiological testing/measurementAnalyteSpectroscopy
The present invention is directed to a method and an apparatus for analysis of an analyte. The method involves providing a zero-mode waveguide which includes a cladding surrounding a core where the cladding is configured to preclude propagation of electromagnetic energy of a frequency less than a cutoff frequency longitudinally through the core of the zero-mode waveguide. The analyte is positioned in the core of the zero-mode waveguide and is then subjected, in the core of the zero-mode waveguide, to activating electromagnetic radiation of a frequency less than the cut-off frequency under conditions effective to permit analysis of the analyte in an effective observation volume which is more compact than if the analysis were carried out in the absence of the zero-mode waveguide.
Owner:CORNELL RES FOUNDATION INC
Detection of nucleic acids and nucleic acid units
InactiveUS6127120AReduce riskReducing operator timeSugar derivativesMicrobiological testing/measurementNucleotideBiology
PCT No. PCT / GB96 / 01830 Sec. 371 Date Apr. 21, 1998 Sec. 102(e) Date Apr. 21, 1998 PCT Filed Jul. 25, 1996 PCT Pub. No. WO97 / 05280 PCT Pub. Date Feb. 13, 1997The invention relates to the detection of target nucleic acids or nucleic acid units in a sample, by obtaining a SER(R)S spectrum for a SER(R)S-active complex containing, or derived directly from, the target. The complex includes at least a SER(R)S-active label, and optionally a target binding species containing a nucleic acid or nucleic acid unit. In this detection method, the concentration of the target present in the SER(R)S-active complex, or of the nucleic acid or unit contained in the target binding species in the SER(R)S-active complex, is no higher than 10-10 moles per liter. Additionally or alternatively, one or more of the following features may be used with the method: i) the introduction of a polyamine; ii) modification of the target, and / or of the nucleic acid or nucleic acid unit contained in the target binding species, in a manner that promotes or facilitates its chemi-sorption onto a SER(R)S-active surface; iii) inclusion of a chemi-sorptive functional group in the SER(R)S-active label. The invention also provides SER(R)S-active complexes for use in such a method, a kit for use in carrying out the method or preparing the complexes and a method for sequencing a nucleic acid which comprises the use of the detection method to detect at least one target nucleotide or sequence of nucleotides within the acid.
Owner:RENISHAW DIAGNOSTICS
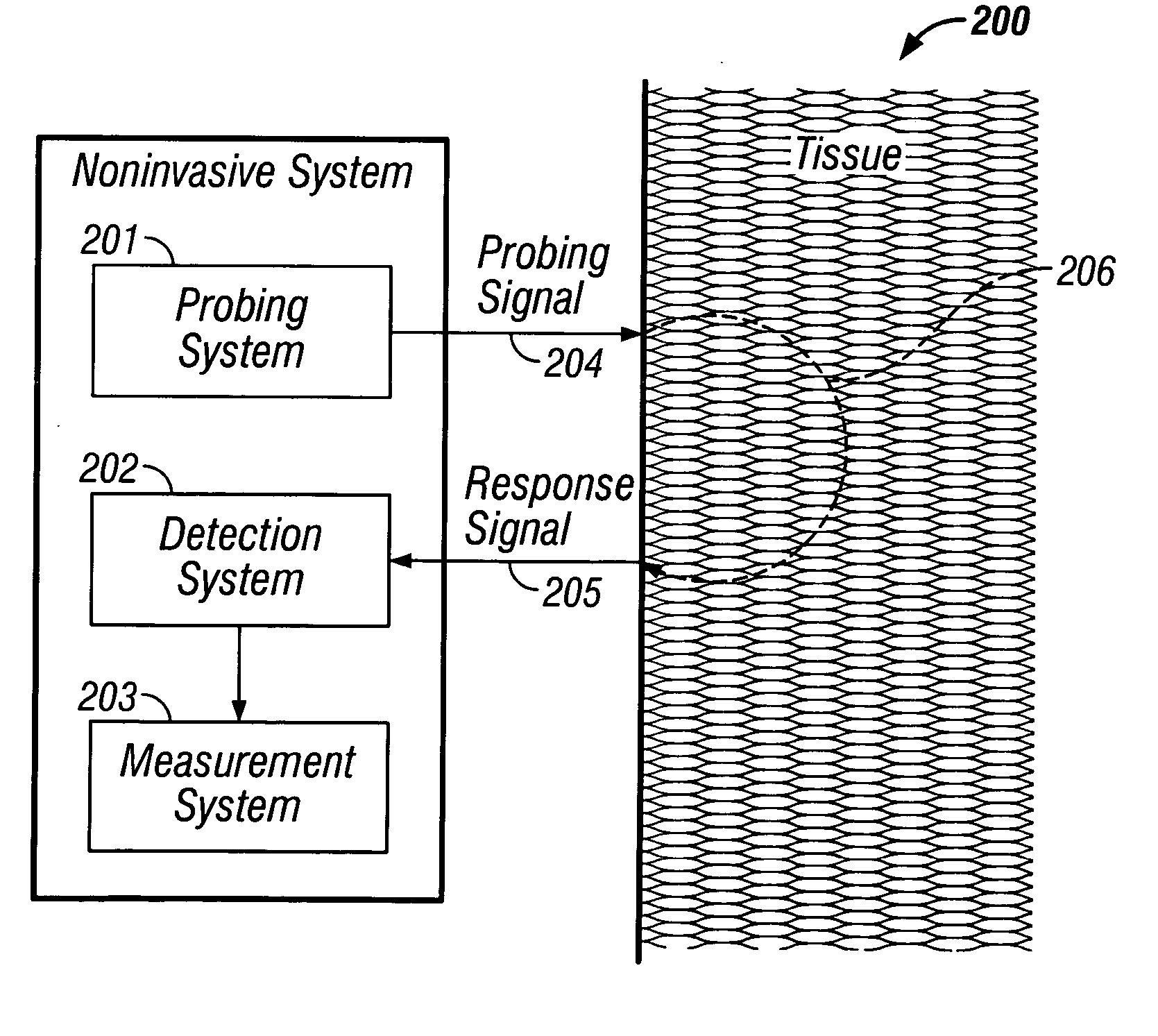
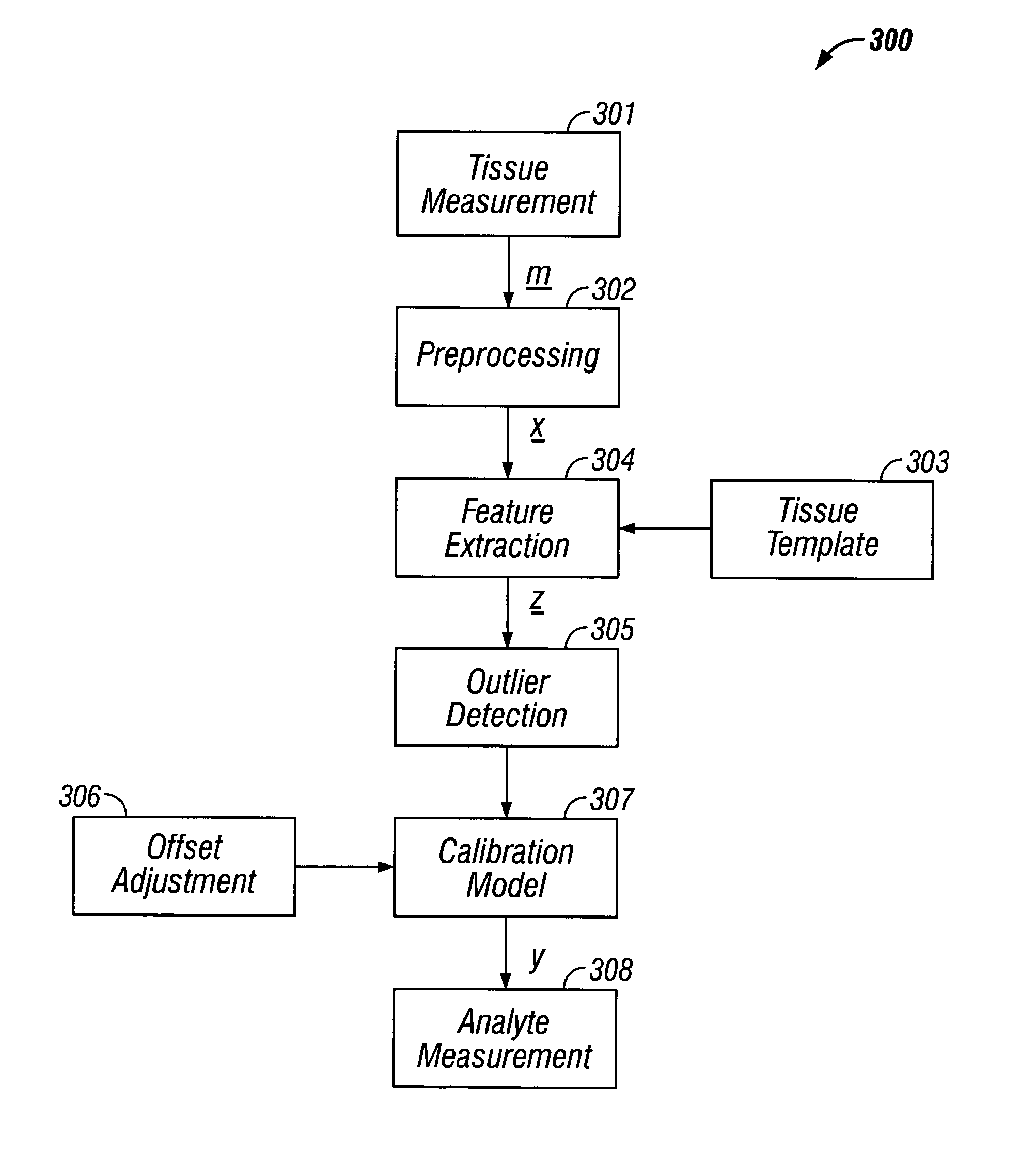
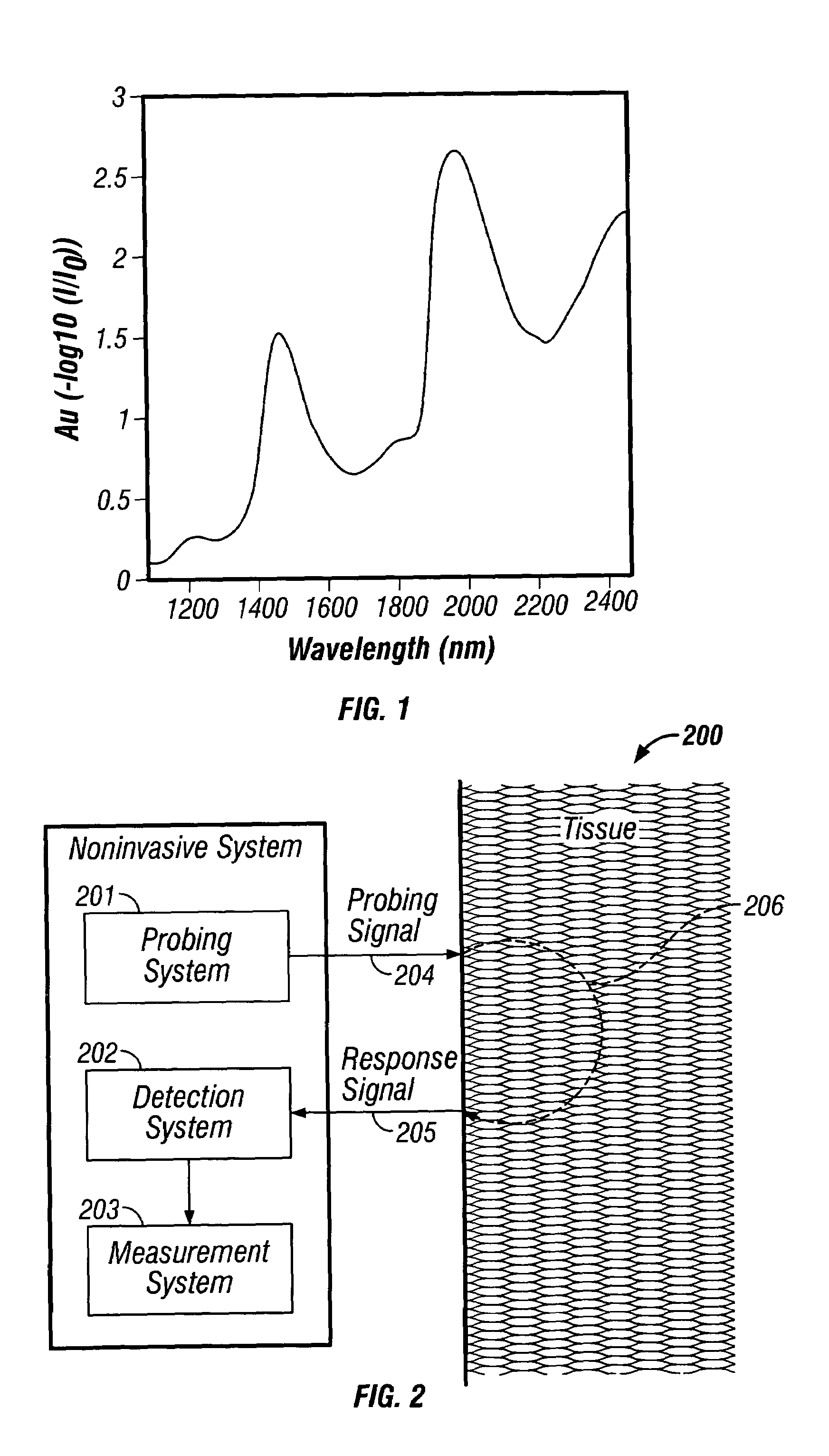
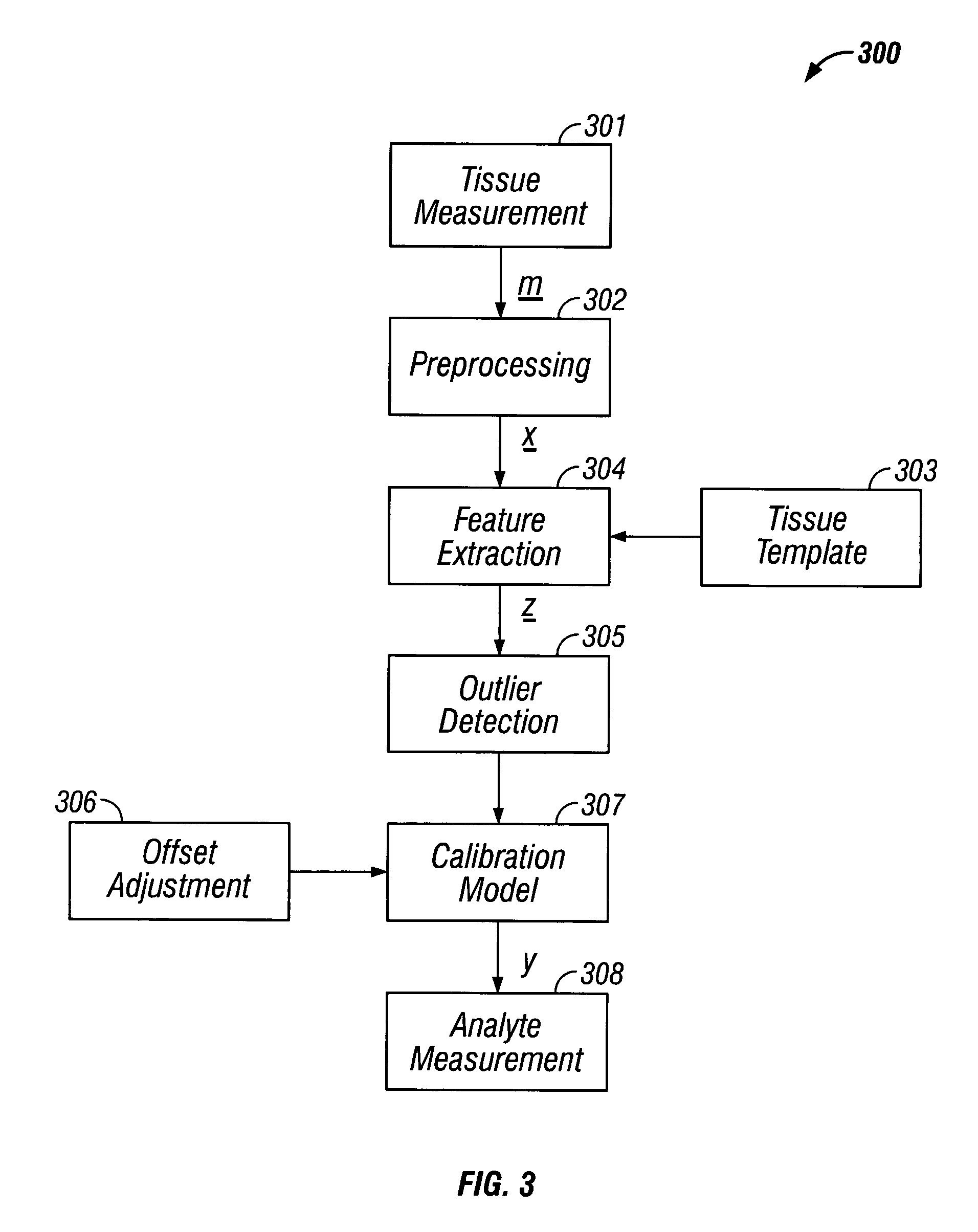
Indirect measurement of tissue analytes through tissue properties
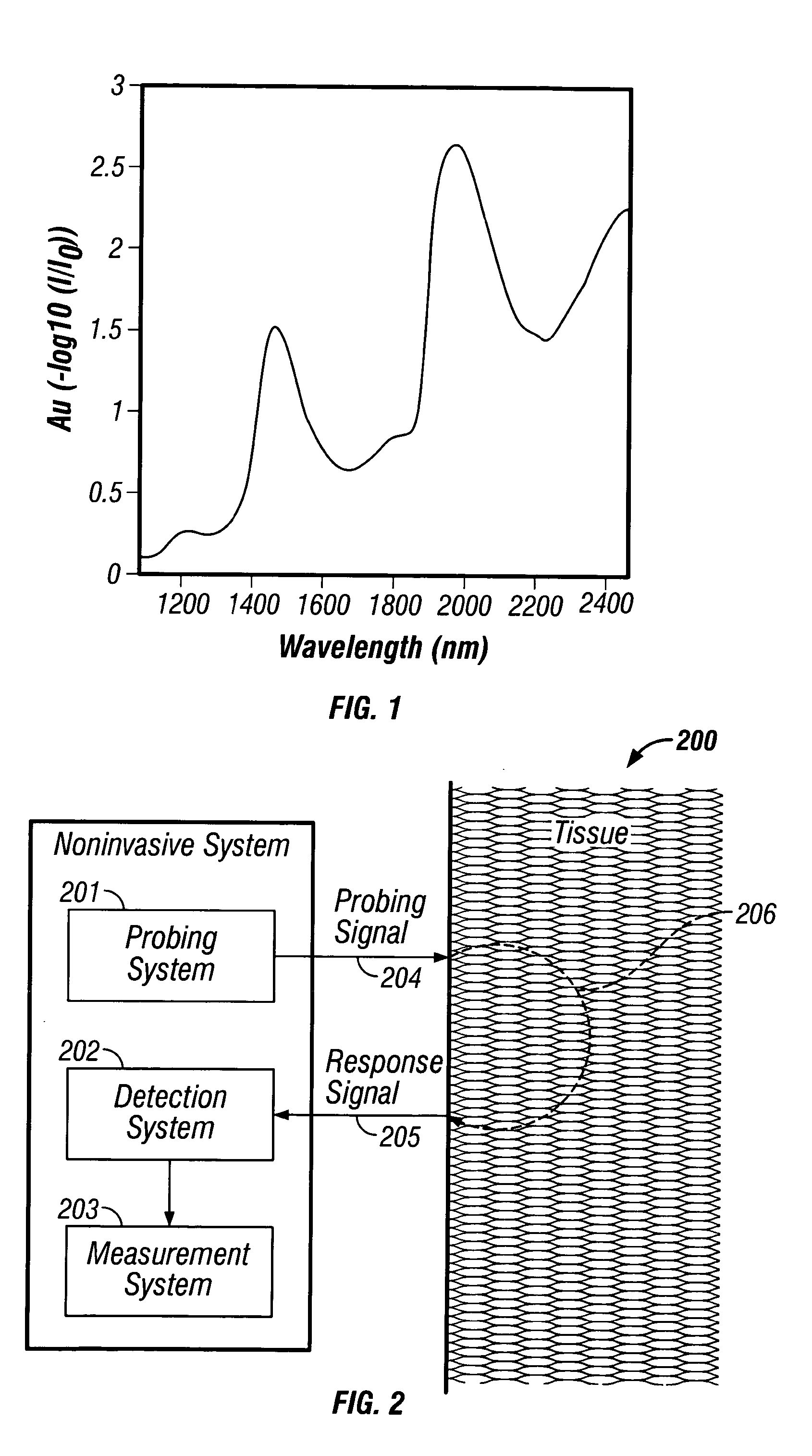
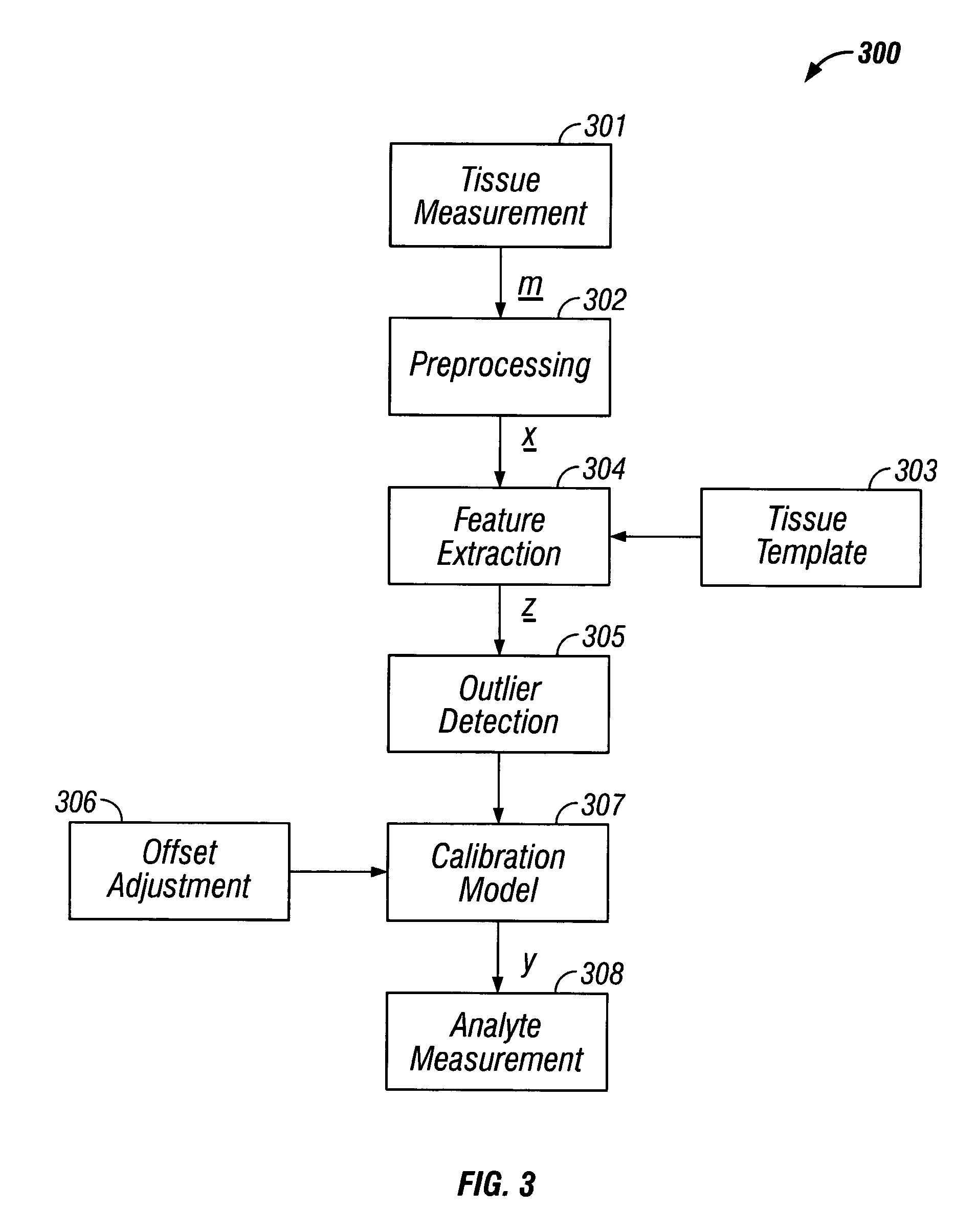
InactiveUS20040127777A1Improve accuracy and precisionLimited stateDiagnostics using spectroscopyRaman scatteringAnalyteMedicine
Methods and system for noninvasive determination of tissue analytes utilize tissue properties as reflected in key features of an analytical signal to improve measurement accuracy and precision. Physiological conditions such as changes in water distribution among tissue compartments lead to complex alterations in the measured analytical signal of skin, leading to a biased noninvasive analyte measurement. Changes in the tissue properties are detected by identifying key features in the analytical signal responsive to physiological variations. Conditions not conducive to the noninvasive measurement are detected. Noninvasive measurements that are biased by physiological changes in tissue are compensated. In an alternate embodiment, the analyte is measured indirectly based on natural physiological response of tissue to changes in analyte concentration. A system capable of such measurements is provided.
Owner:GLT ACQUISITION +1
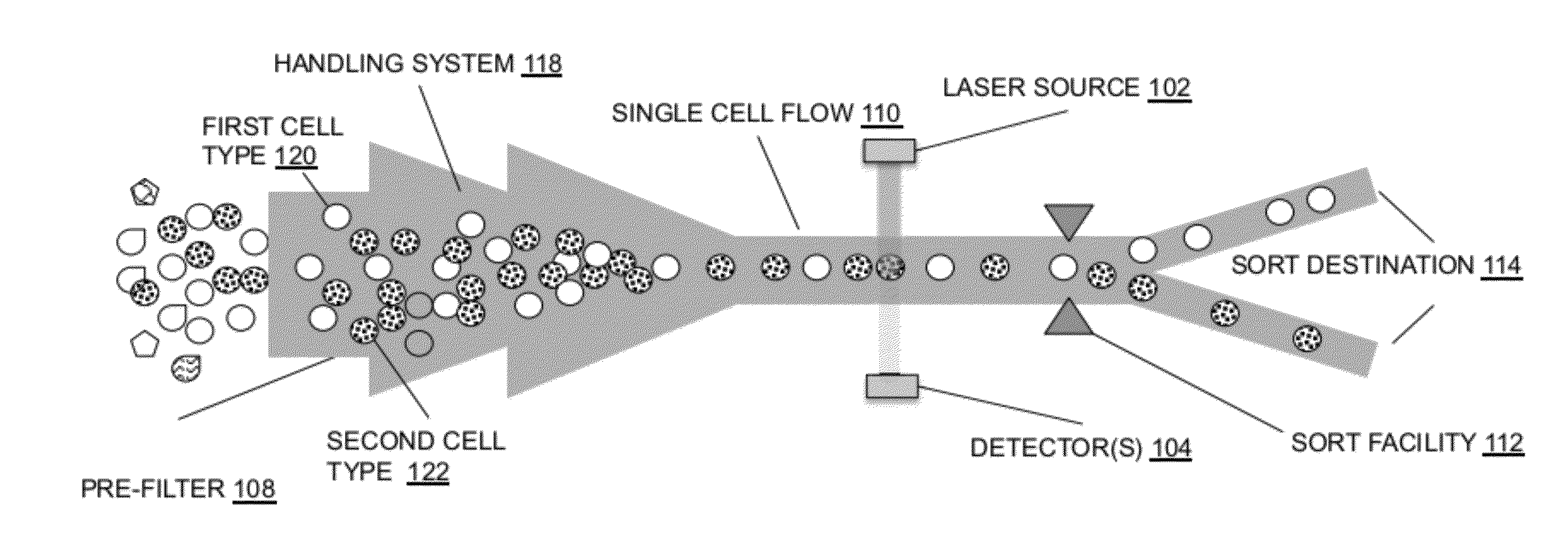
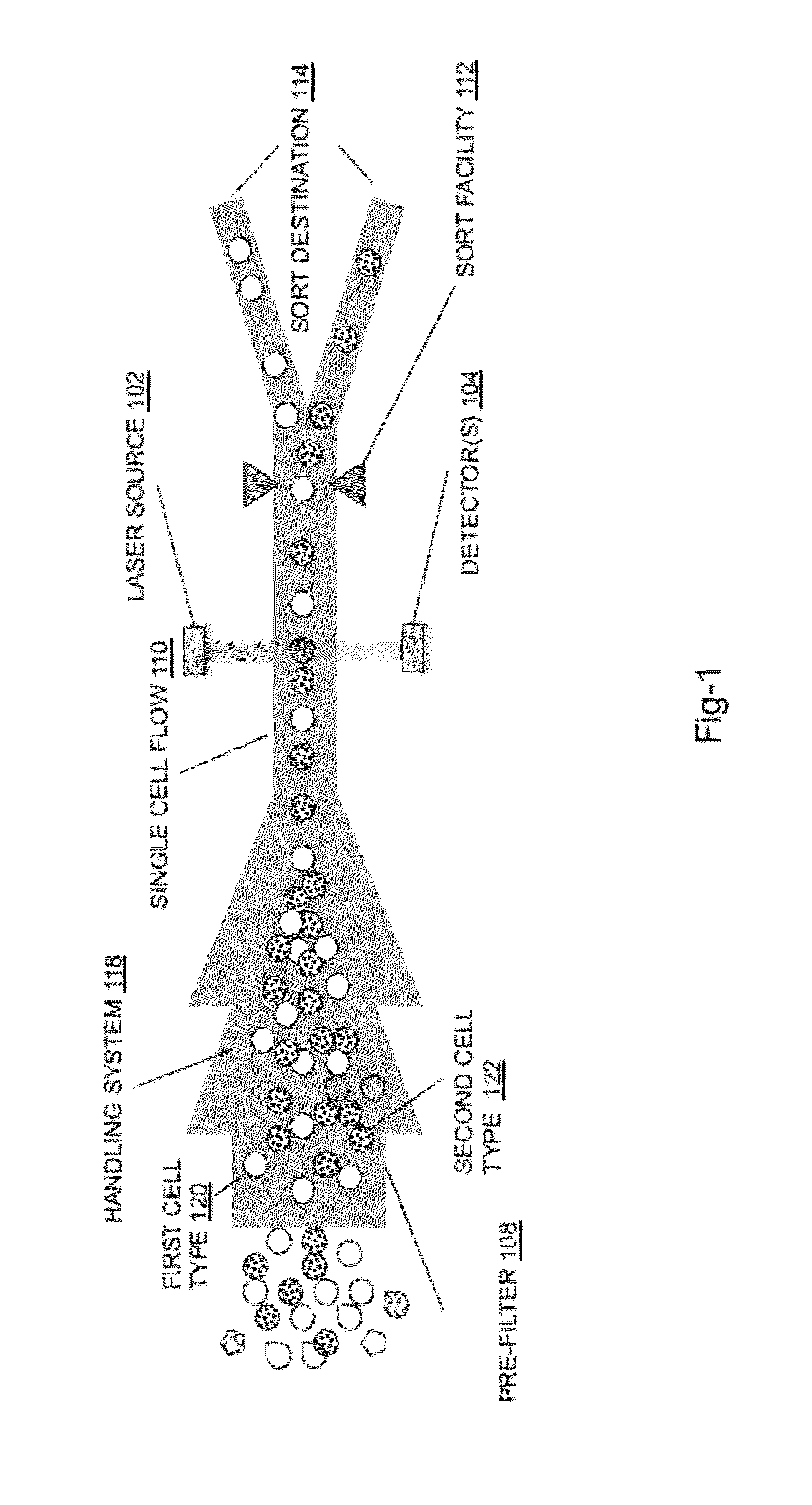
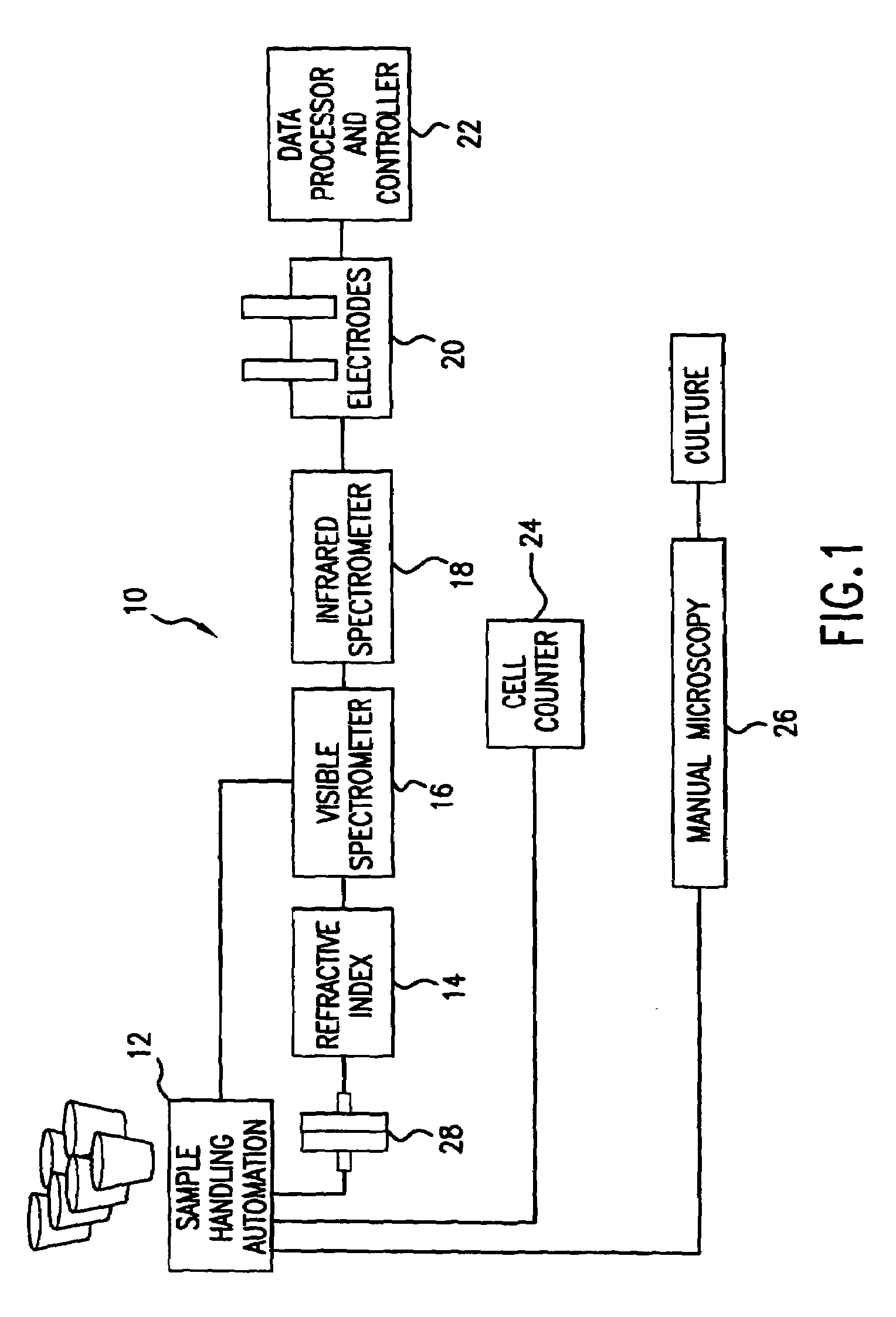
System for identifying and sorting living cells
ActiveUS20120122084A1Bioreactor/fermenter combinationsMaterial analysis using sonic/ultrasonic/infrasonic wavesFlow cellIr absorption
In embodiments of the present invention, a system and method of cytometry may include presenting a single sperm cell to at least one laser source configured to deliver light to the sperm cell in order to induce bond vibrations in the sperm cell DNA, and detecting the signature of the bond vibrations. The bond vibration signature is used to calculate a DNA content carried by the sperm cell which is used to identify the sperm cell as carrying an X-chromosome or Y-chromosome. Another system and method may include flowing cells past at least one QCL source one-by-one using a fluid handling system, delivering QCL light to a single cell to induce resonant mid-IR absorption by one or more analytes of the cell, and detecting, using a mid-infrared detection facility, the transmitted mid-infrared wavelength light, wherein the transmitted mid-infrared wavelength light is used to identify a cell characteristic.
Owner:1087 SYST
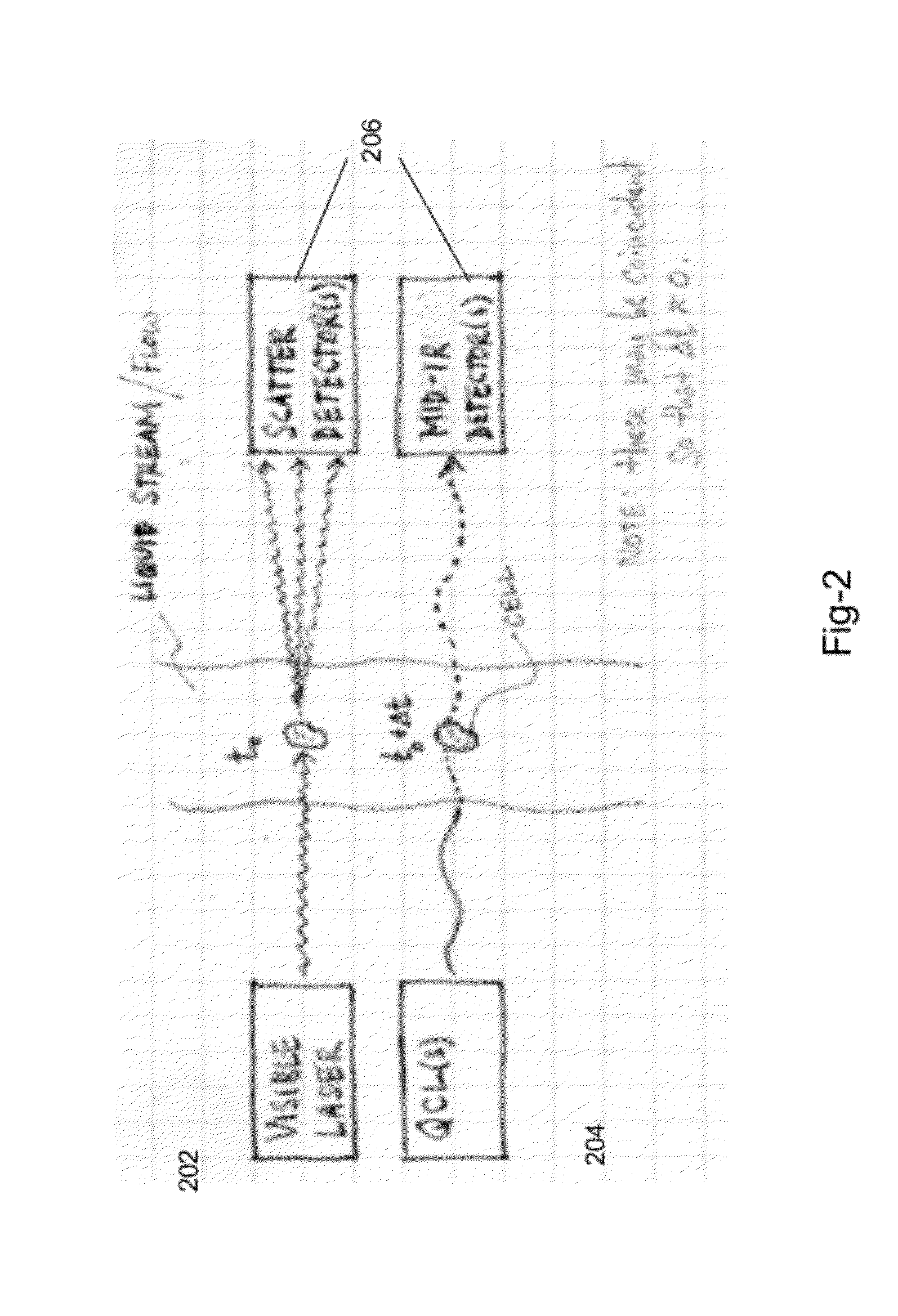
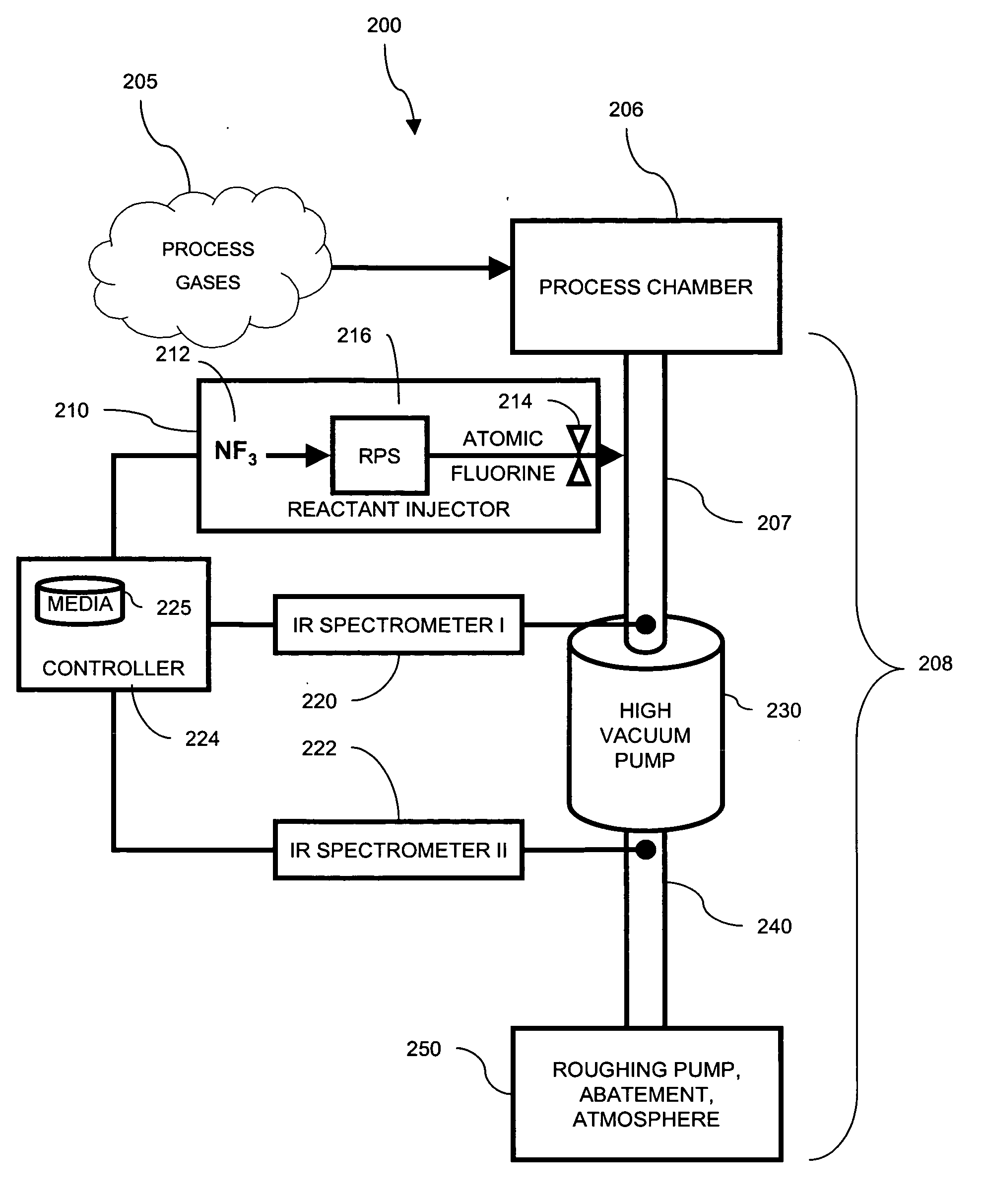
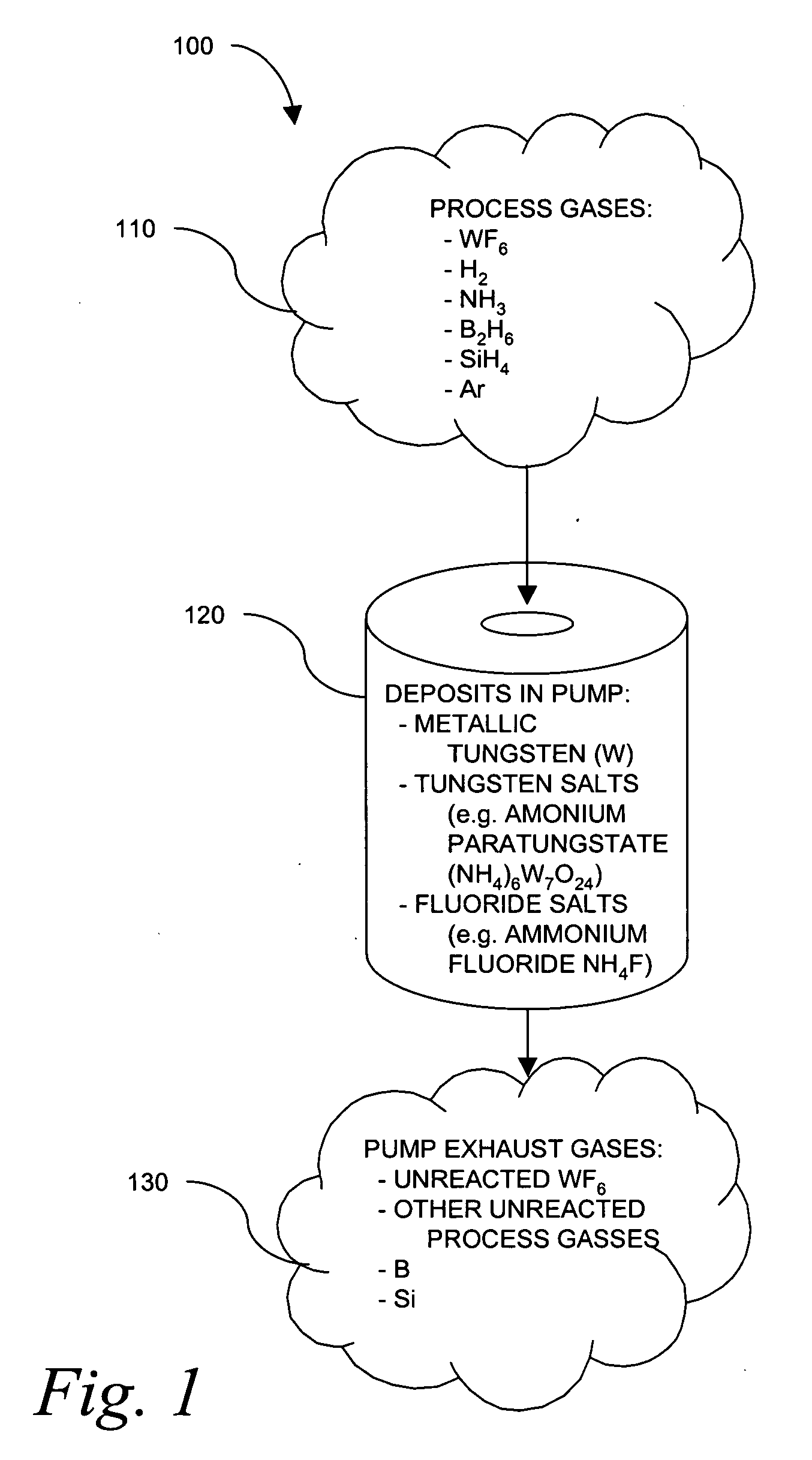
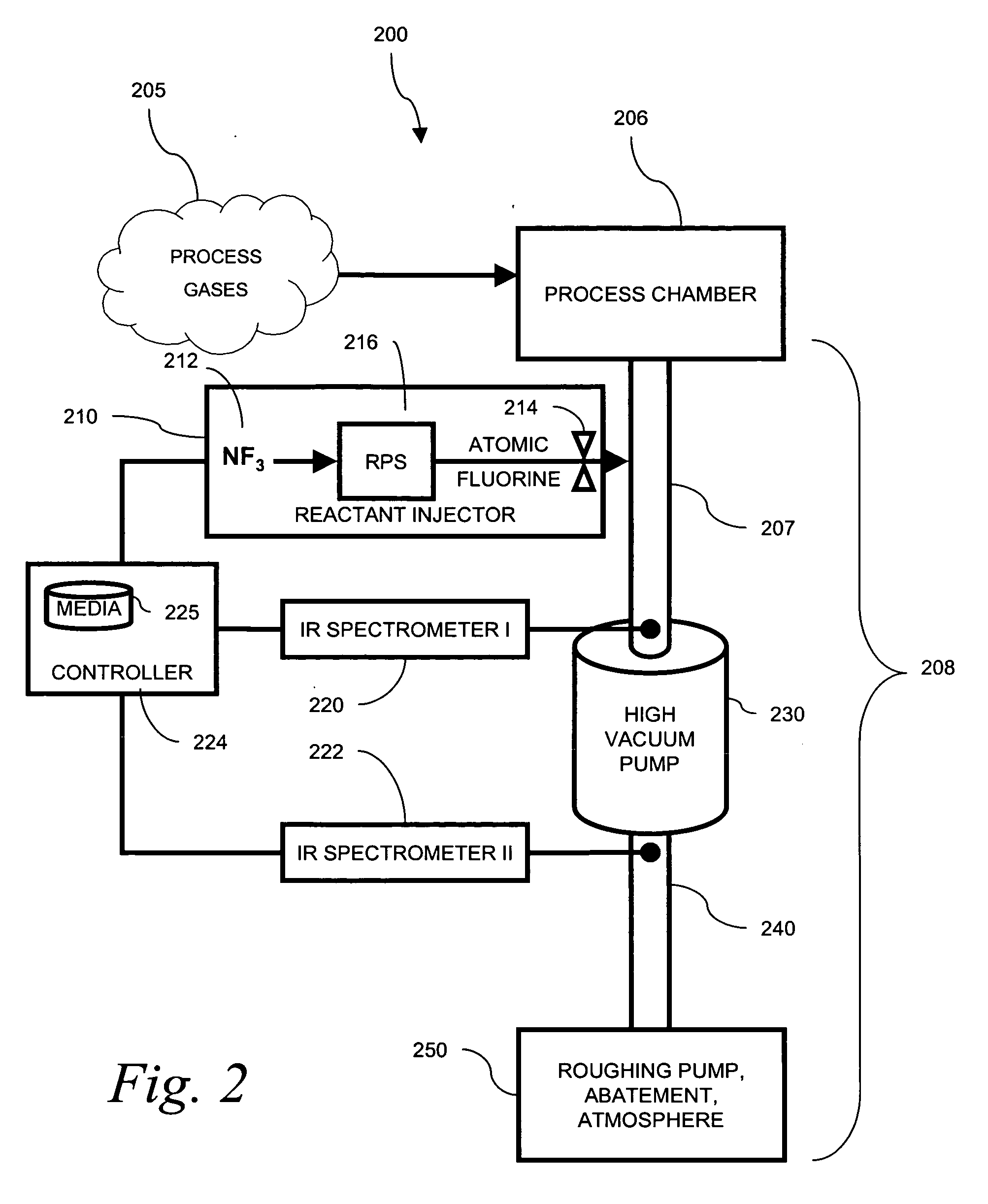
Use of spectroscopic techniques to monitor and control reactant gas input into a pre-pump reactive gas injection system
ActiveUS20090320881A1Produce some attenuationTrend downRotary/oscillating piston combinations for elastic fluidsHollow article cleaningGas phaseReactive gas
The present invention relates to vacuum processing systems in which process gases are introduced in a process chamber and are exhausted through a vacuum processing system exhaust path. Deposits made by the exhausted gas are reduced or eliminated by introducing a reactive gas upstream of the device affected by deposits. The amount of introduced reactive gas is controlled by measuring gas phase concentrations of exhausted gas components upstream and downstream of the affected device, and, from those measurements, determining whether the components are being consumed in deposits on the affected device.
Owner:EDWARDS VACUUM LLC
Raman-active taggants and their recognition
InactiveUS6610351B2Easy to useQuality improvementMaterial nanotechnologyRadiation applicationsMaximum dimensionActive component
An organic or organoelement, linear or branched, monomeric or polymeric composition of matter having a Raman-active component in the form of particles. The particles having a maximum dimension of 50 mum. The Raman-active compound is applied to a substrate. When the Raman-active compound is exposed to a laser light wavelength which is batochromically well beyond a spectral region of maximum absorbance of said Raman-active compound, Raman scattering can be detected.
Owner:QUANTAG SYST
Optical sensor with layered plasmon structure for enhanced detection of chemical groups by SERS
InactiveUS20060034729A1Produced in advanceRadiation pyrometryMicrobiological testing/measurementExcitation beamLight excitation
An optical sensor and method for use with a visible-light laser excitation beam and a Raman spectroscopy detector, for detecting the presence chemical groups in an analyte applied to the sensor are disclosed. The sensor includes a substrate, a plasmon resonance mirror formed on a sensor surface of the substrate, a plasmon resonance particle layer disposed over the mirror, and an optically transparent dielectric layer about 2-40 nm thick separating the mirror and particle layer. The particle layer is composed of a periodic array of plasmon resonance particles having (i) a coating effective to binding analyte molecules, (ii) substantially uniform particle sizes and shapes in a selected size range between 50-200 nm (ii) a regular periodic particle-to-particle spacing less than the wavelength of the laser excitation beam. The device is capable of detecting analyte with an amplification factor of up to 1012-1014, allowing detection of single analyte molecules.
Owner:POPONIN VLADIMIR
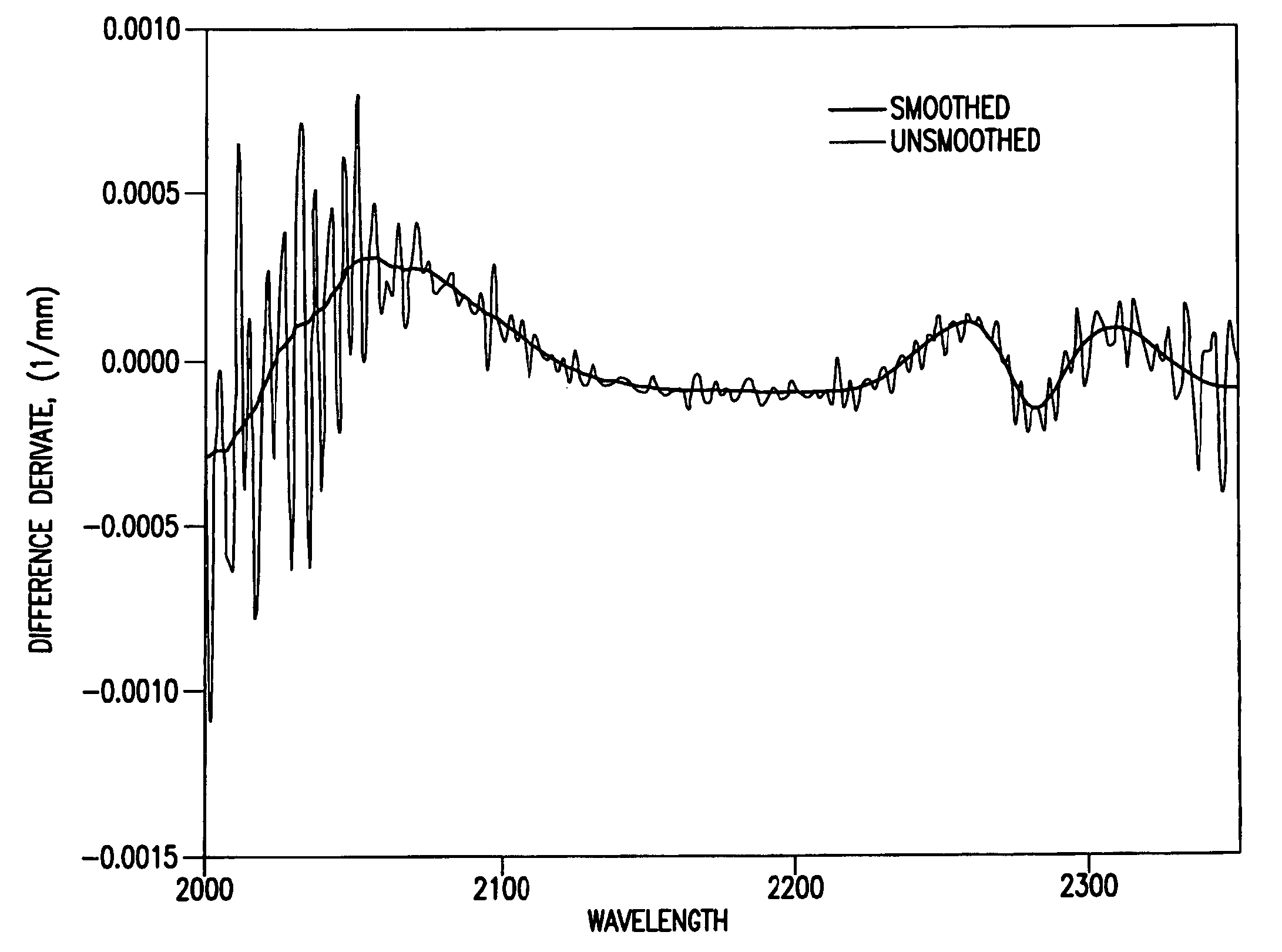
Reagentless analysis of biological samples by applying mathematical algorithms to smoothed spectra
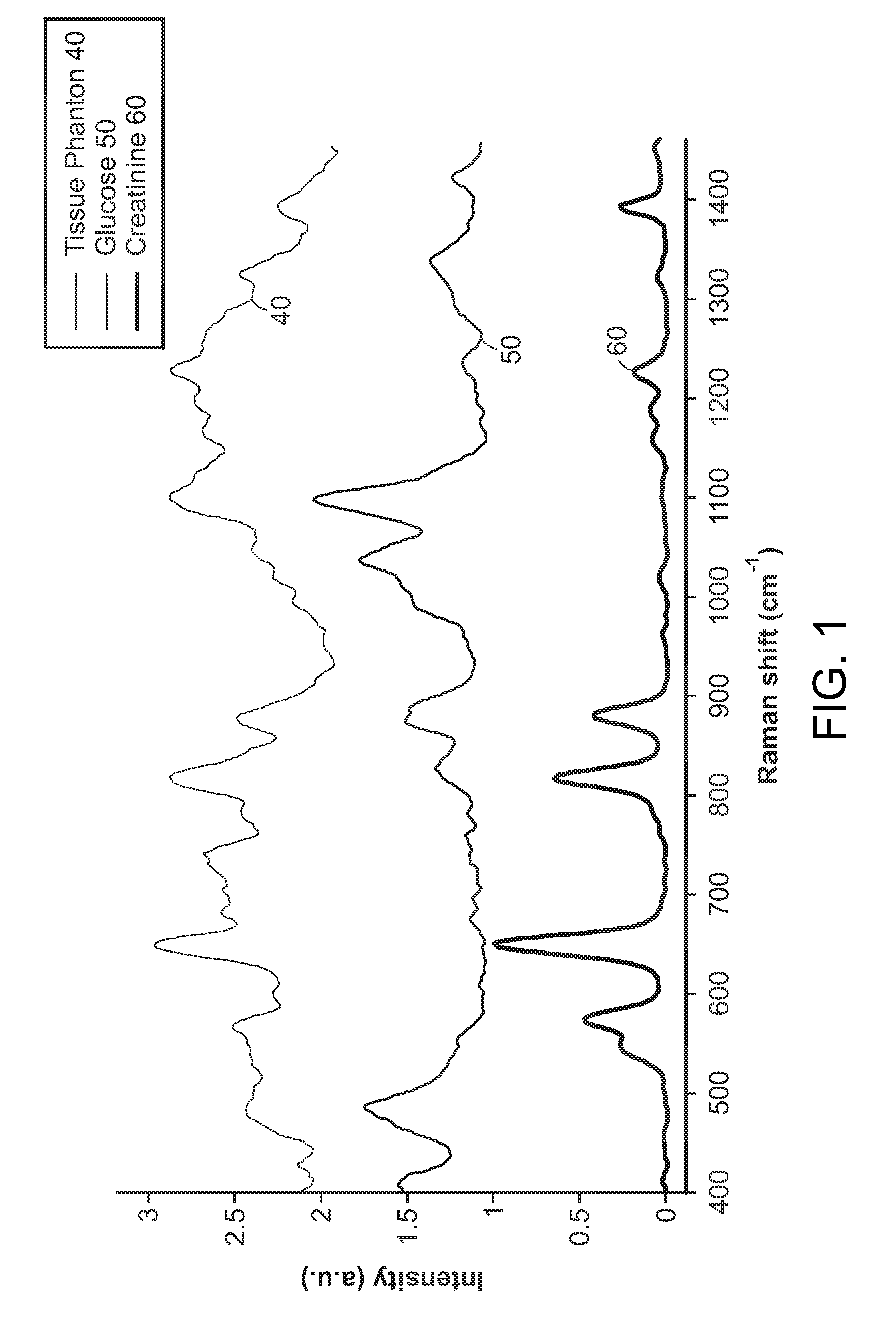
InactiveUS7303922B2Improve accuracyImprove automationPhase-affecting property measurementsRaman scatteringCreatinine riseRefractive index
Apparatus and method for determining at least one parameter, e. g., concentration, of at least one analyte, e. g., urea, of a biological sample, e. g., urine. A biological sample particularly suitable for the apparatus and method of this invention is urine. In general, spectroscopic measurements can be used to quantify the concentrations of one or more analytes in a biological sample. In order to obtain concentration values of certain analytes, such as hemoglobin and bilirubin, visible light absorption spectroscopy can be used. In order to obtain concentration values of other analytes, such as urea, creatinine, glucose, ketones, and protein, infrared light absorption spectroscopy can be used. The apparatus and method of this invention utilize one or more mathematical techniques to improve the accuracy of measurement of parameters of analytes in a biological sample. The invention also provides an apparatus and method for measuring the refractive index of a sample of biological fluid while making spectroscopic measurements substantially simultaneously.
Owner:ABBOTT LAB INC
Color translating UV microscope
A color translating UV microscope for research and clinical applications involving imaging of living or dynamic samples in real time and providing several novel techniques for image creation, optical sectioning, dynamic motion tracking and contrast enhancement comprises a light source emitting UV light, and visible and IR light if desired. This light is directed to the condenser via a means of selecting monochromatic, bandpass, shortpass, longpass or notch limited light. The condenser can be a brightfield, darkfield, phase contrast or DIC. The slide is mounted in a stage capable of high speed movements in the X, Y and Z dimensions. The microscope uses broadband, narrowband or monochromat optimized objectives to direct the image of the sample to an image intensifier or UV sensitive video system. When an image intensifier is used it is either followed by a video camera, or in the simple version, by a synchronized set of filters which translate the image to a color image and deliver it to an eyepiece for viewing by the microscopist. Between the objective and the image intensifier there can be a selection of static or dynamic switchable filters. The video camera, if used, produces an image which is digitized by an image capture board in a computer. The image is then reassembled by an overlay process called color translation and the computer uses a combination of feedback from the information in the image and operator control to perform various tasks such as optical sectioning and three dimensional reconstruction, coordination of the monochromater while collecting multiple images sets called image planes, tracking dynamic sample elements in three space, control of the environment of the slide including electric, magnetic, acoustic, temperature, pressure and light levels, color filters and optics, control for microscope mode switching between transmitted, reflected, fluorescent, Raman, scanning, confocal, area limited, autofluorescent, acousto-optical and other modes.
Owner:RICHARDSON TECH
Indirect measurement of tissue analytes through tissue properties
InactiveUS7039446B2Improve accuracy and precisionLimited stateDiagnostics using spectroscopyRaman scatteringAnalyteMedicine
Owner:GLT ACQUISITION +1
System and method for high throughput screening of droplets
InactiveUS20030119193A1Sequential/parallel process reactionsComponent separationHigh-Throughput Screening MethodsHigh flux
A system and method for high throughput screening of fluid samples. A reduced pressure is applied, via an injection valve, to a sample aspiration tube. A first fluid and a second fluid are alternatively aspirated, via the sample aspiration tube, the first fluid for filling a sample loop with samples, the second fluid for flushing the sample aspiration tube. Excess fluid aspirated from the first fluid source and all fluid aspirated from the second fluid source is captured in an inline trap.
Owner:BIOCIUS LIFE SCI
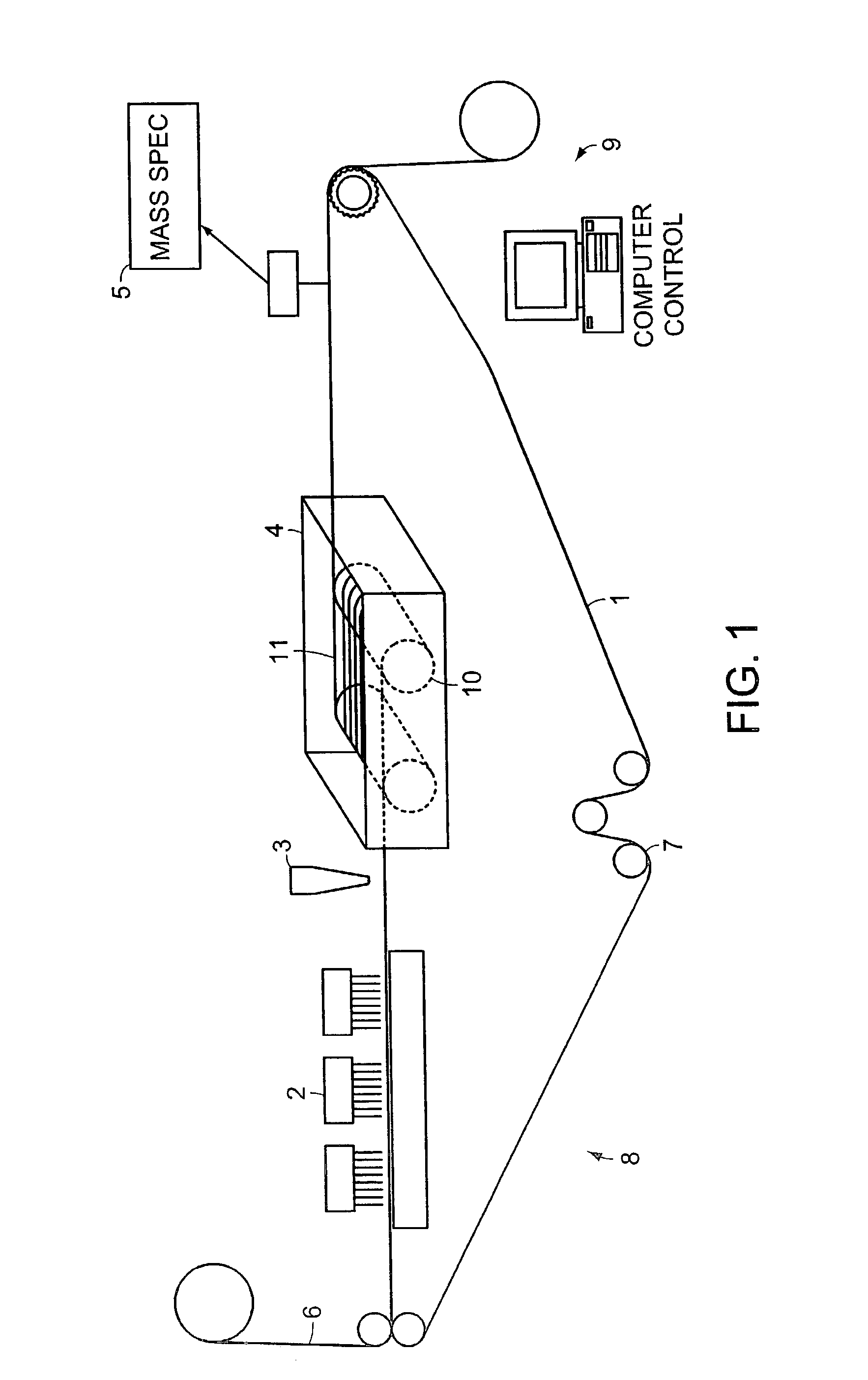
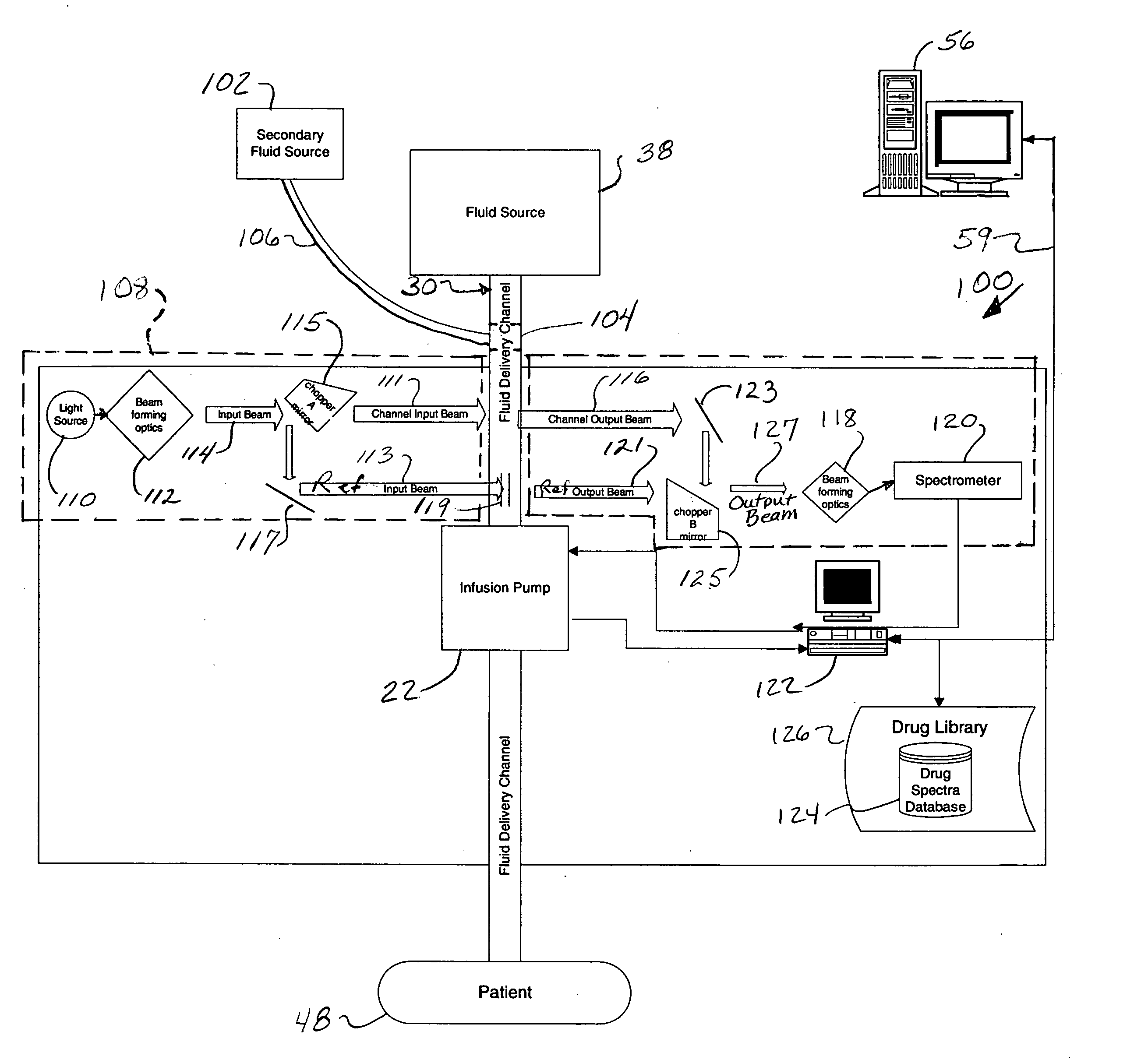
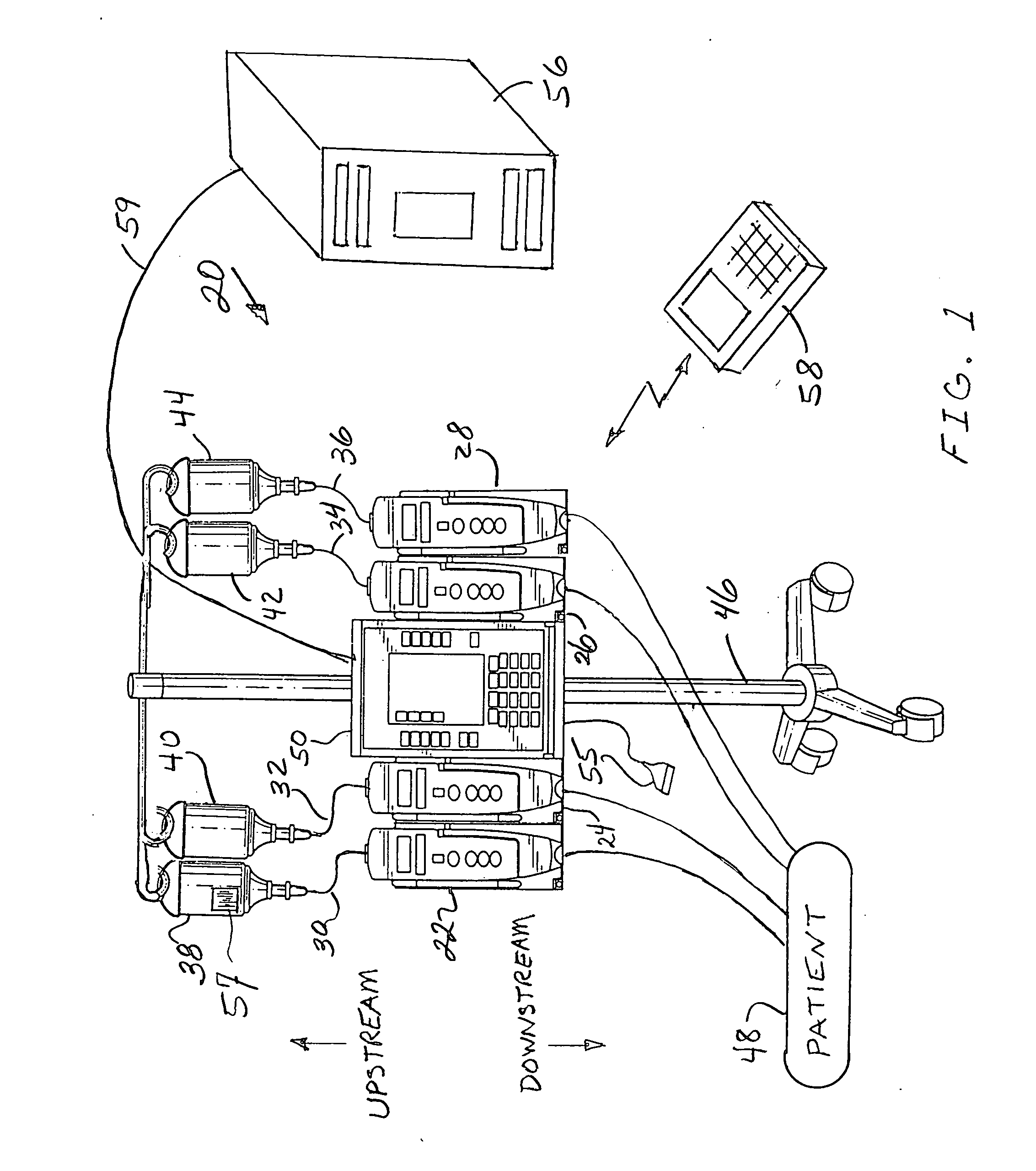
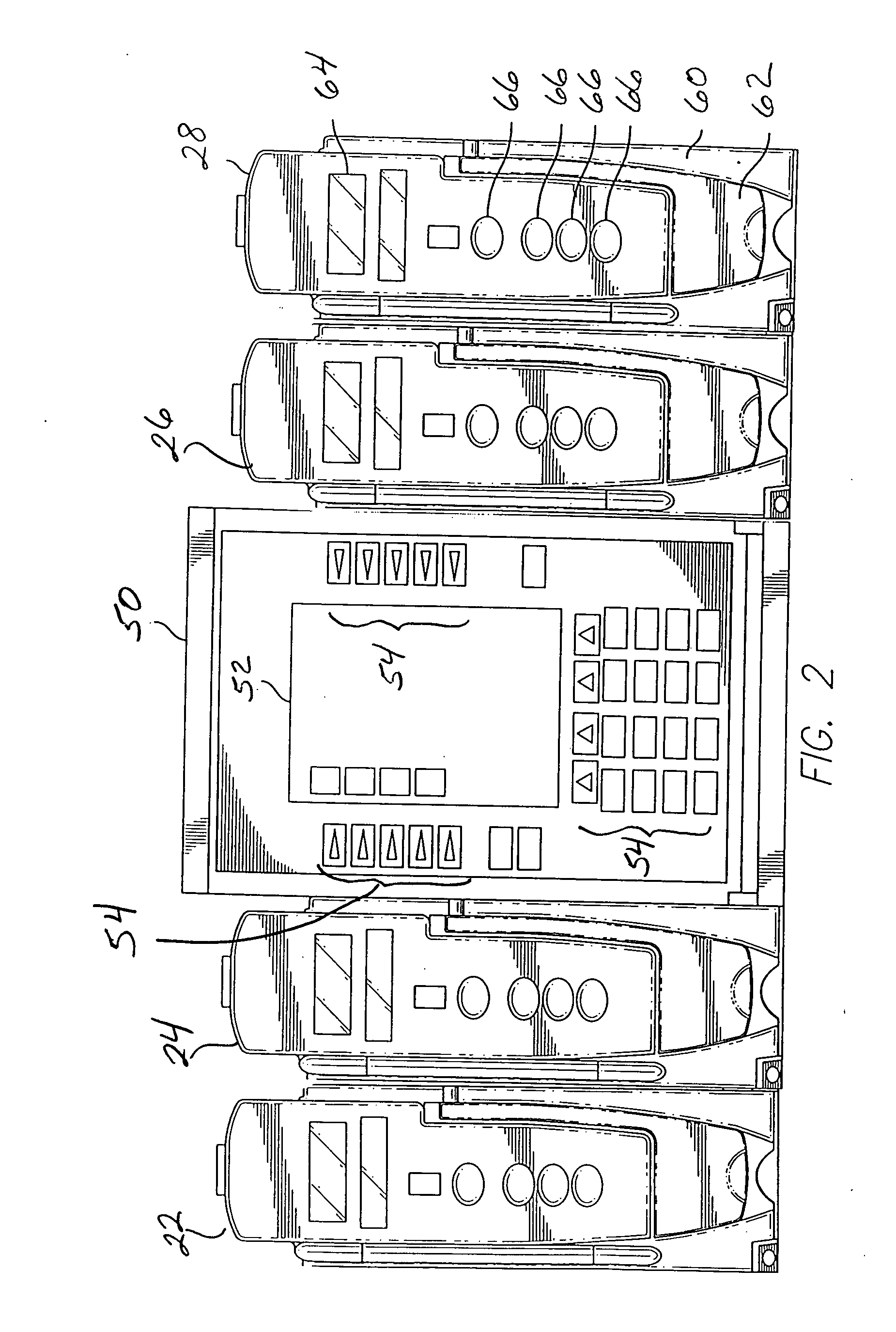
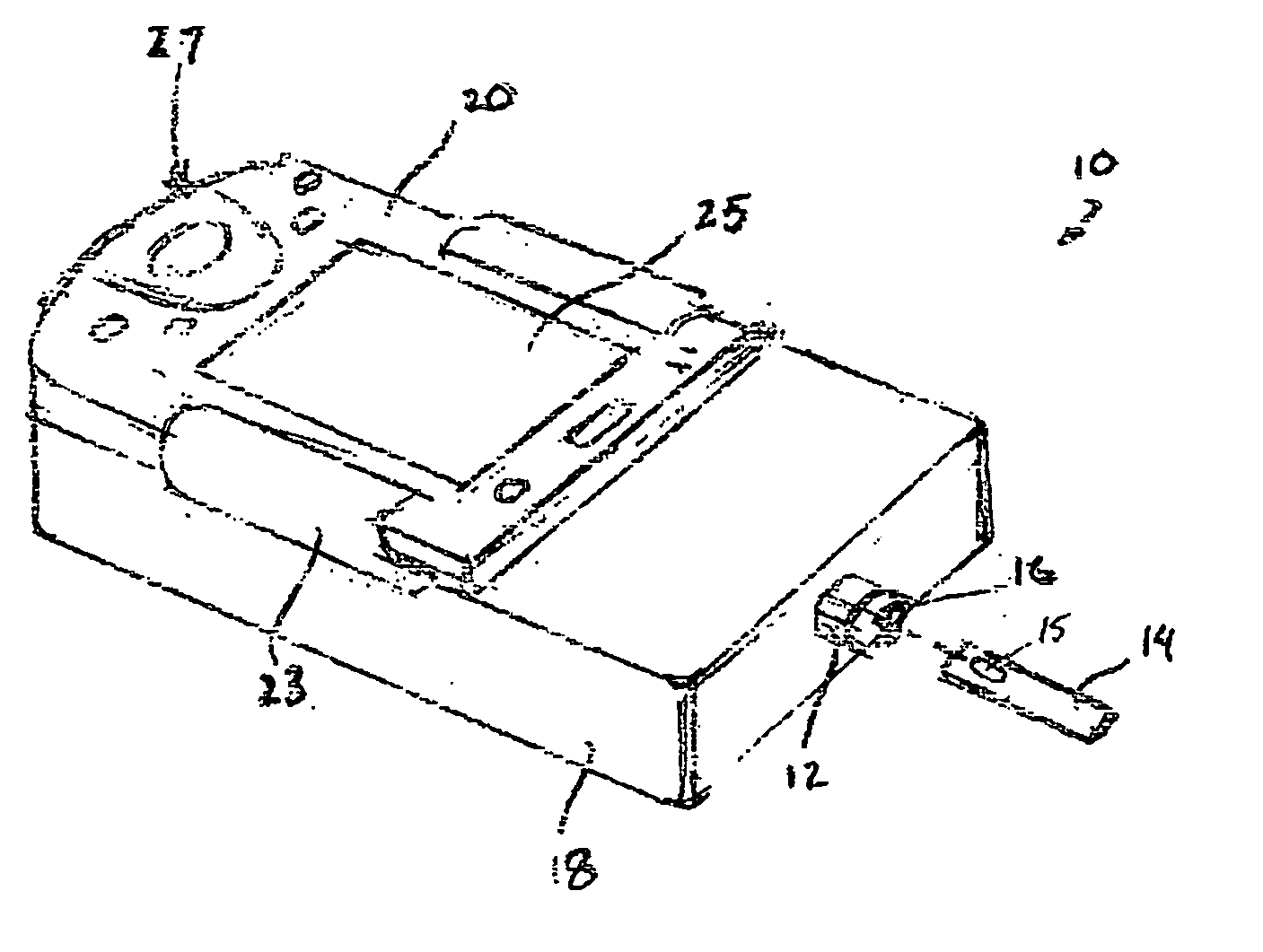
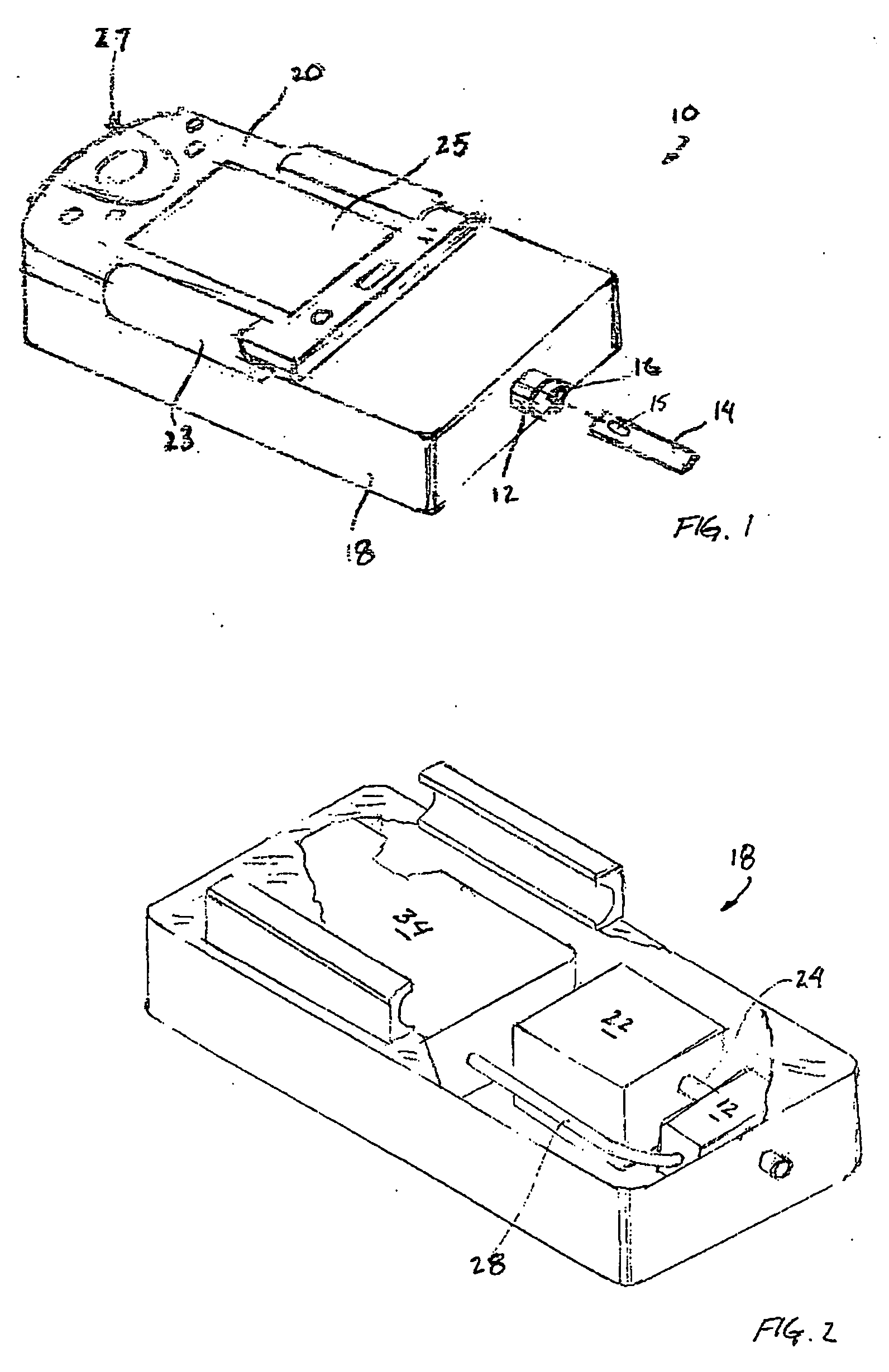
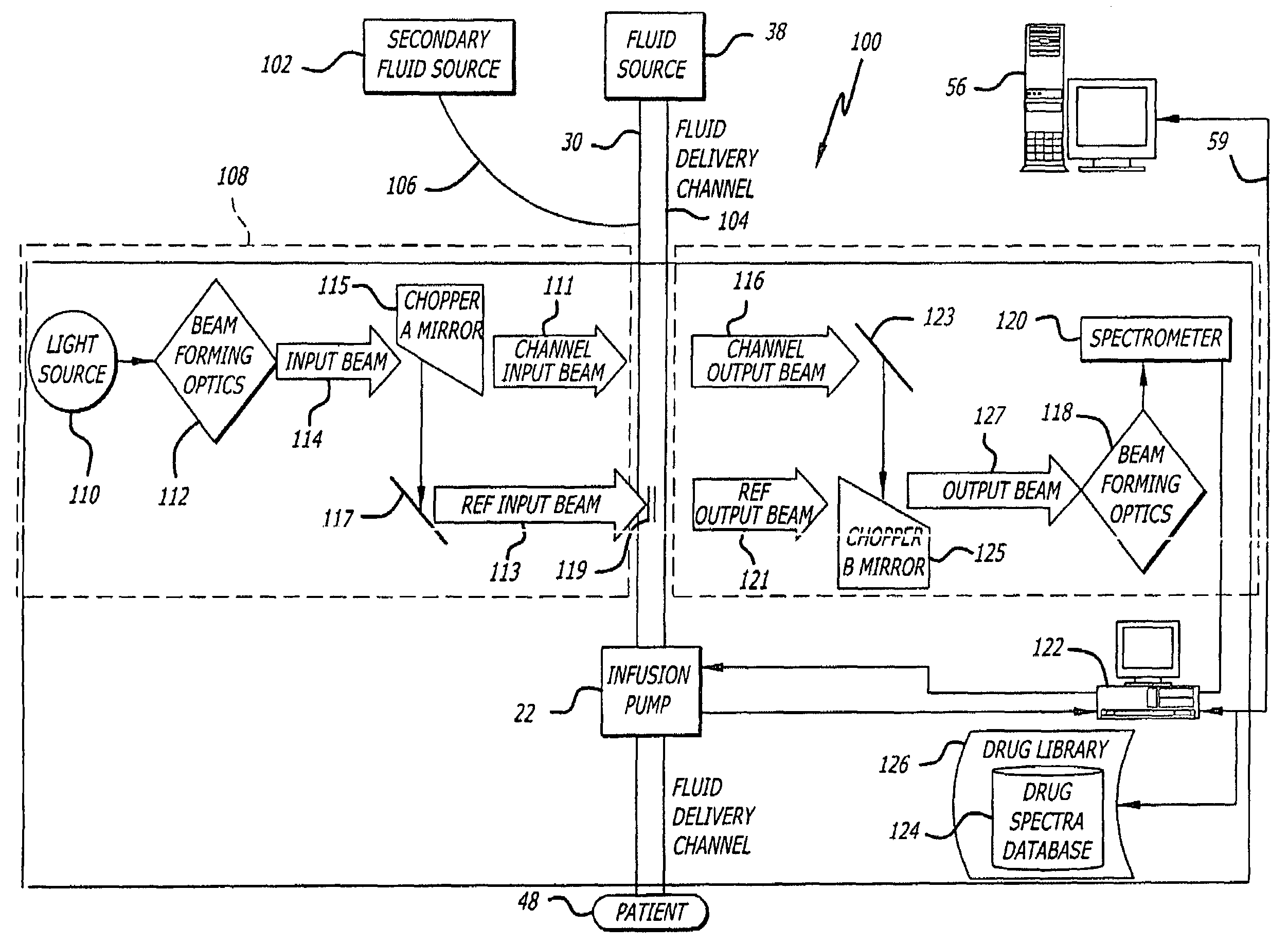
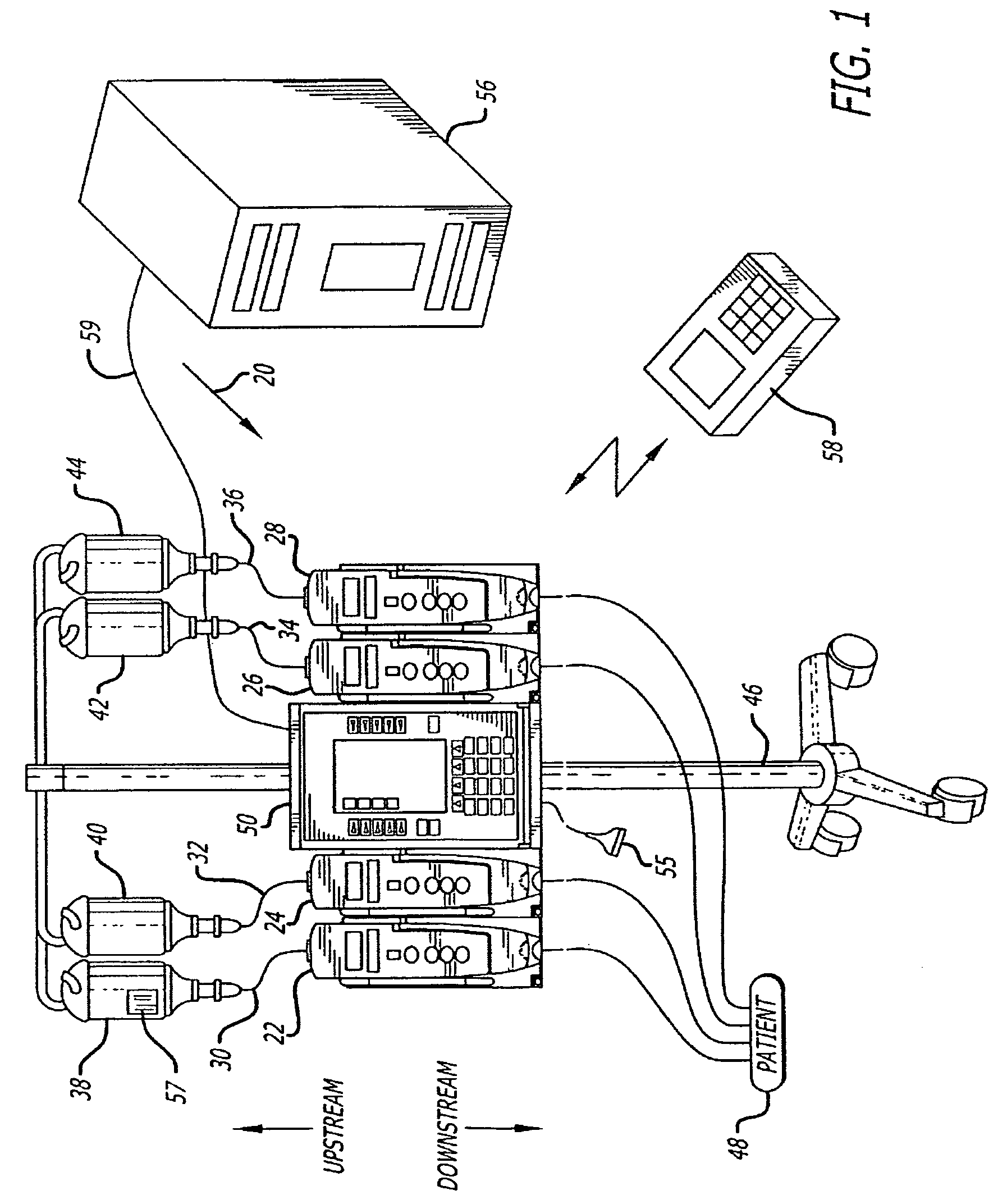
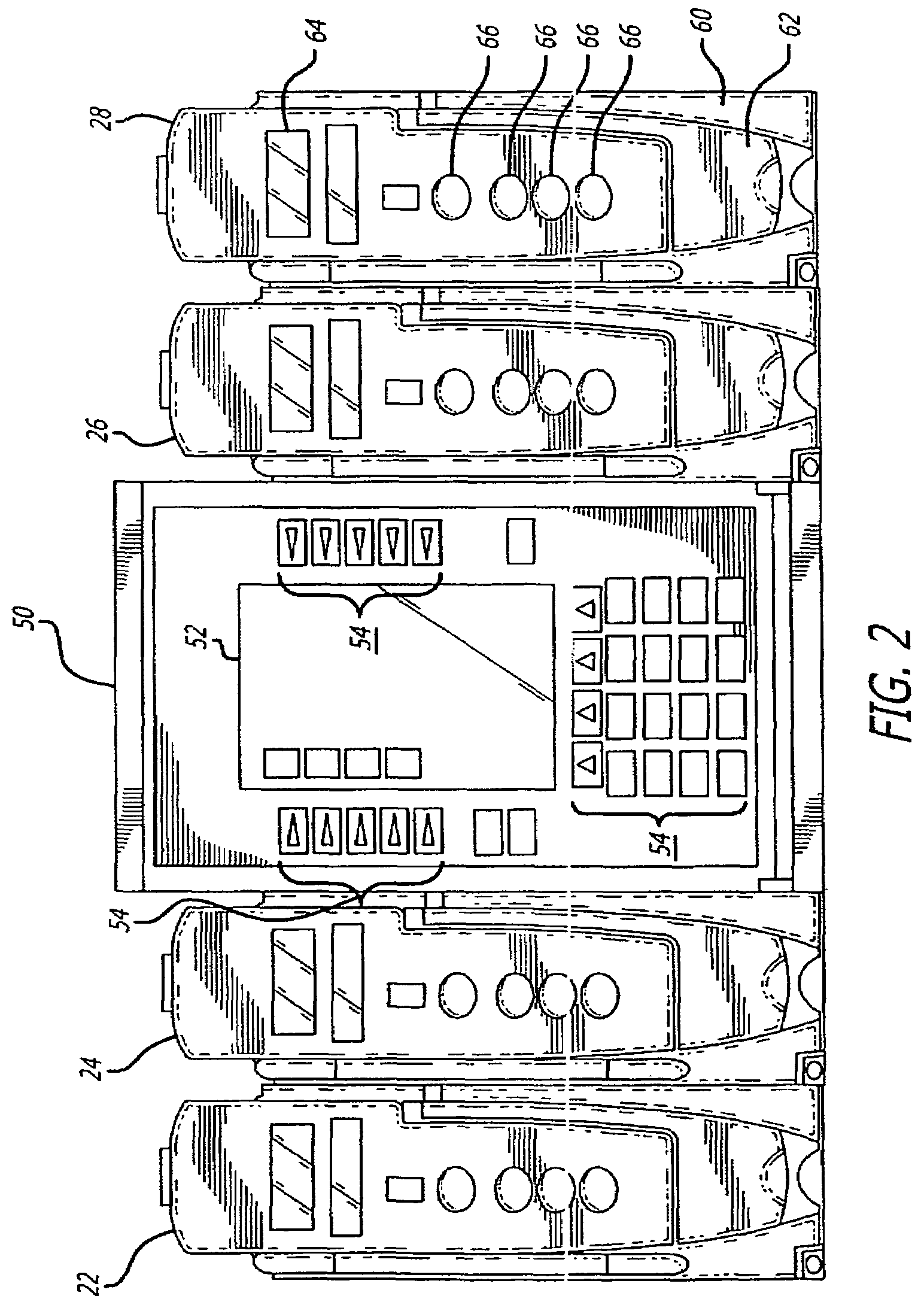
Fluid verification system and method for infusions
An apparatus and method are provided to verify the composition of a medical fluid in a fluid infusion channel by comparison with clinician-entered input. Light is transmitted through the channel and detected by a sensor that generates signals representative of the spectral data of the light detected. A processor compares the spectral data of the light detected to the spectral data associated with the expected contents of the channel to verify that the correct fluid is being infused.
Owner:CAREFUSION 303 INC
Raman-active taggants and their recognition
InactiveUS20020025490A1Easy to useQuality improvementOptical radiation measurementMaterial nanotechnologyLaser lightLasing wavelength
An organic or organoelement, linear or branched, monomeric or polymeric composition of matter having a Raman-active component in the form of particles. The particles having a maximum dimension of 50 mum. The Raman-active compound is applied to a substrate. When the Raman-active compound is exposed to a laser light wavelength which is batochromically well beyond a spectral region of maximum absorbance of said Raman-active compound, Raman scattering can be detected.
Owner:QUANTAG SYST
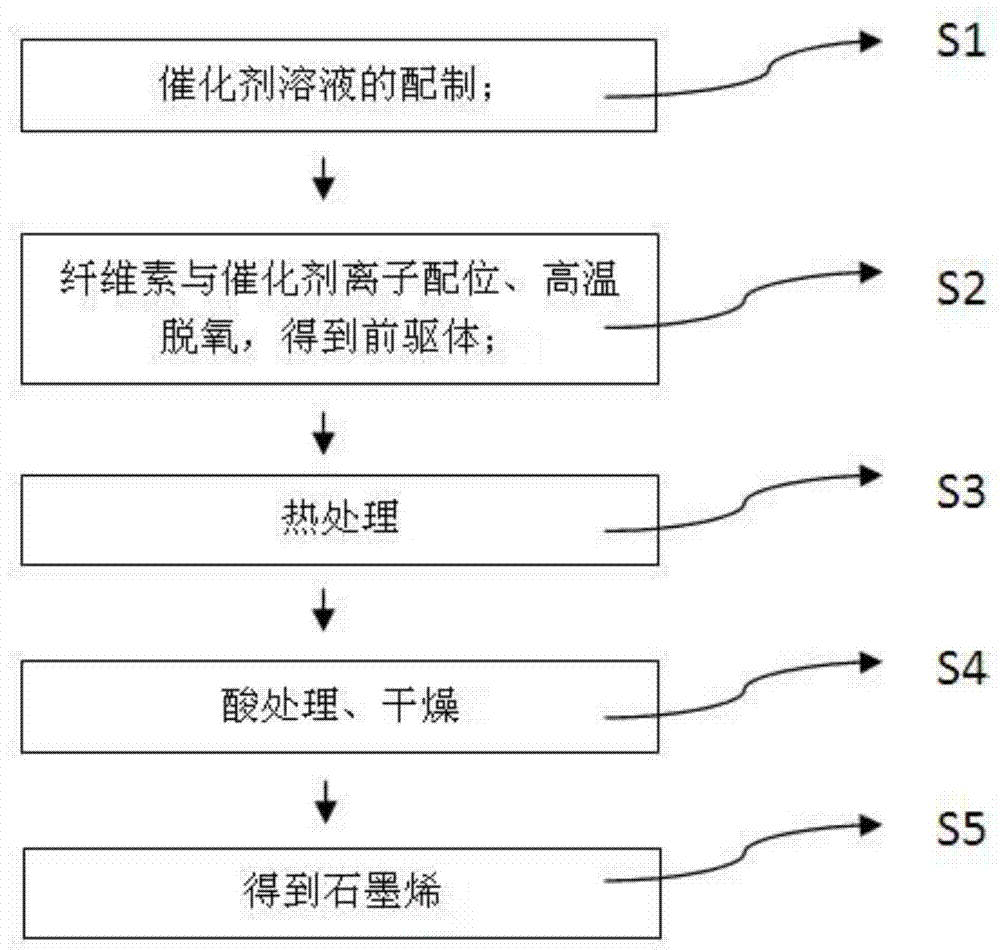
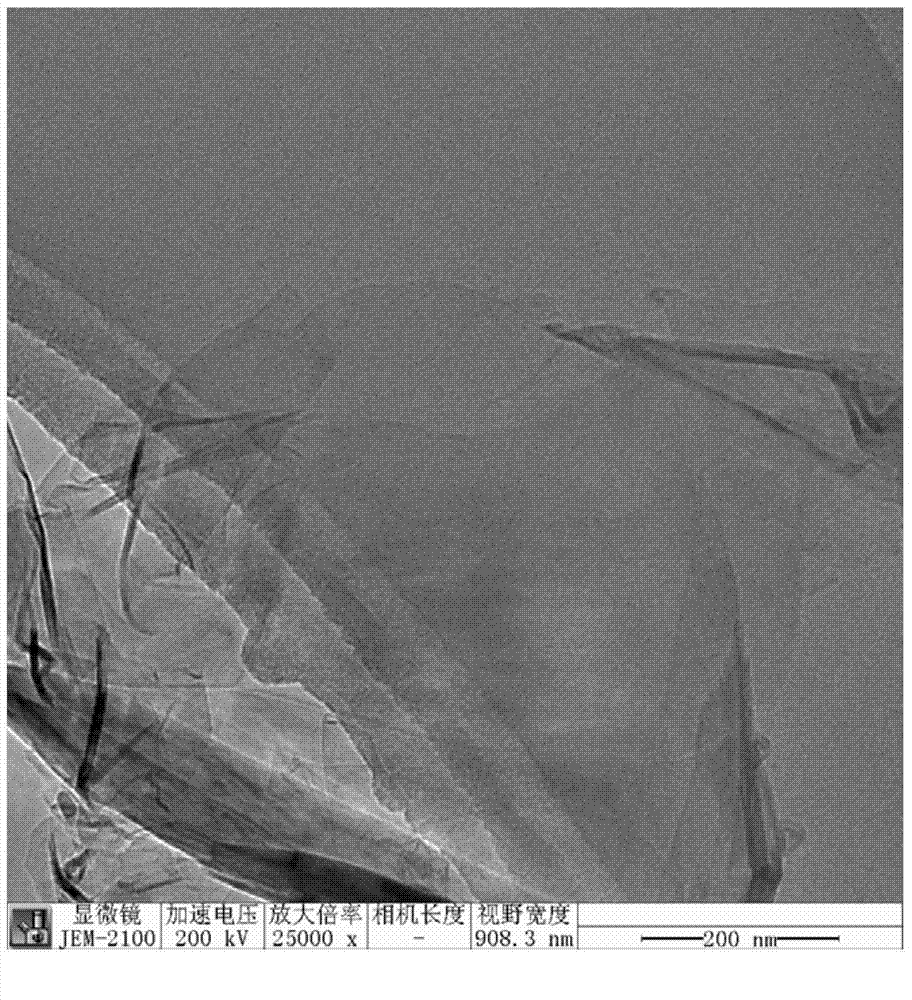
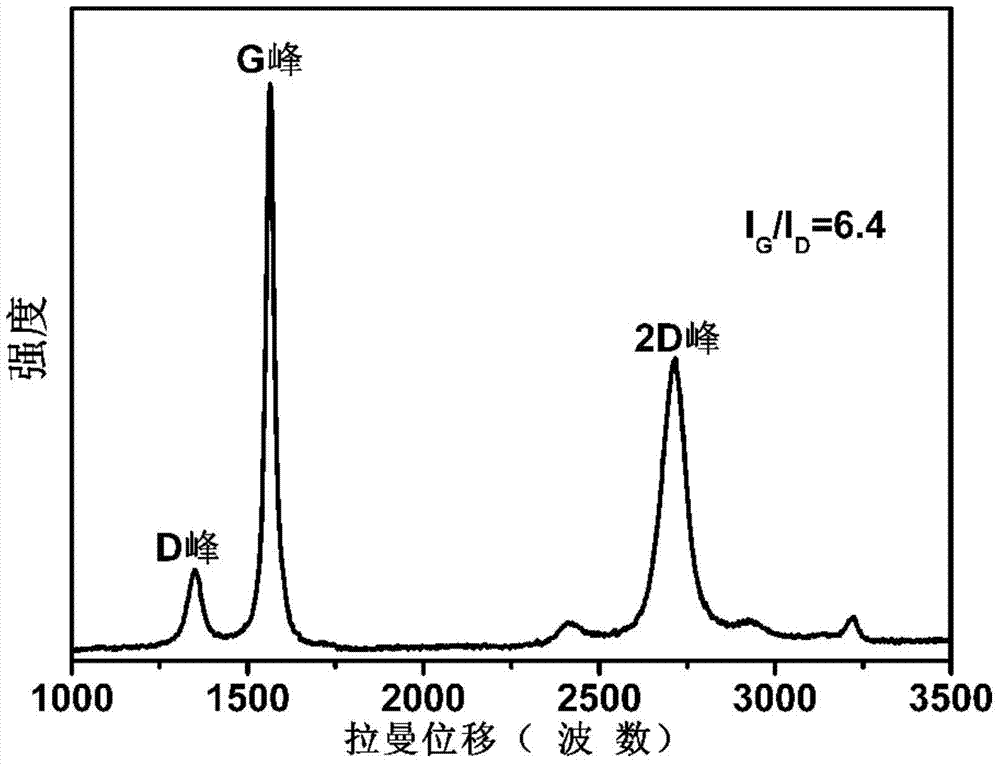
Method for preparing biomass graphene employing cellulose as raw material
ActiveCN104724699AUniform sizeIncrease productionPhysical/chemical process catalystsGrapheneCellulosePolymer science
The invention provides a preparation method of graphene, and particularly relates to a method for preparing biomass graphene employing cellulose as a raw material. The specific preparation method comprises the following steps: 1, preparing a catalyst solution; 2, carrying out ionic coordination and high-temperature deoxidization on cellulose and a catalyst, so as to obtain a precursor; 3, carrying out thermal treatment; 4, carrying out acid treatment, and drying to obtain the graphene, wherein the prepared graphene is uniform in morphology, has a single-layer or multi-layer two-dimensional layered structure; the dimension is 0.5-2 microns; and the electrical conductivity is 25,000-45,000S / m. The preparation method is simple in preparation technology, low in cost, high in yield, high in production safety, and controllable in product dimension and physical property; industrialized production can be realized; the graphene prepared by the method can be applied to electrode materials of super capacitors and lithium ion batteries, and can also be added to resin and rubber as an additive; and the physical property of the material can be improved.
Owner:HEILONGJIANG UNIV +1
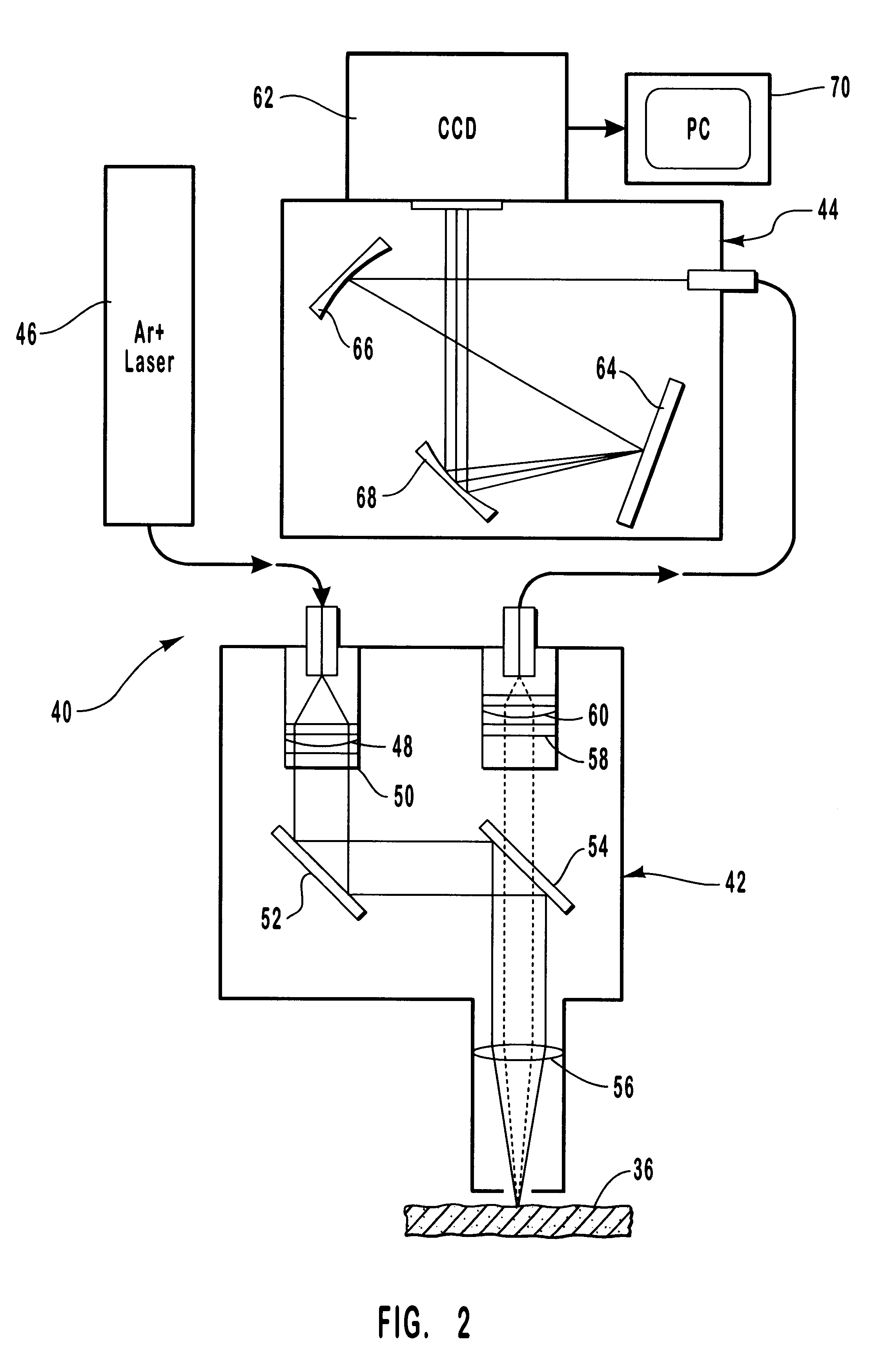
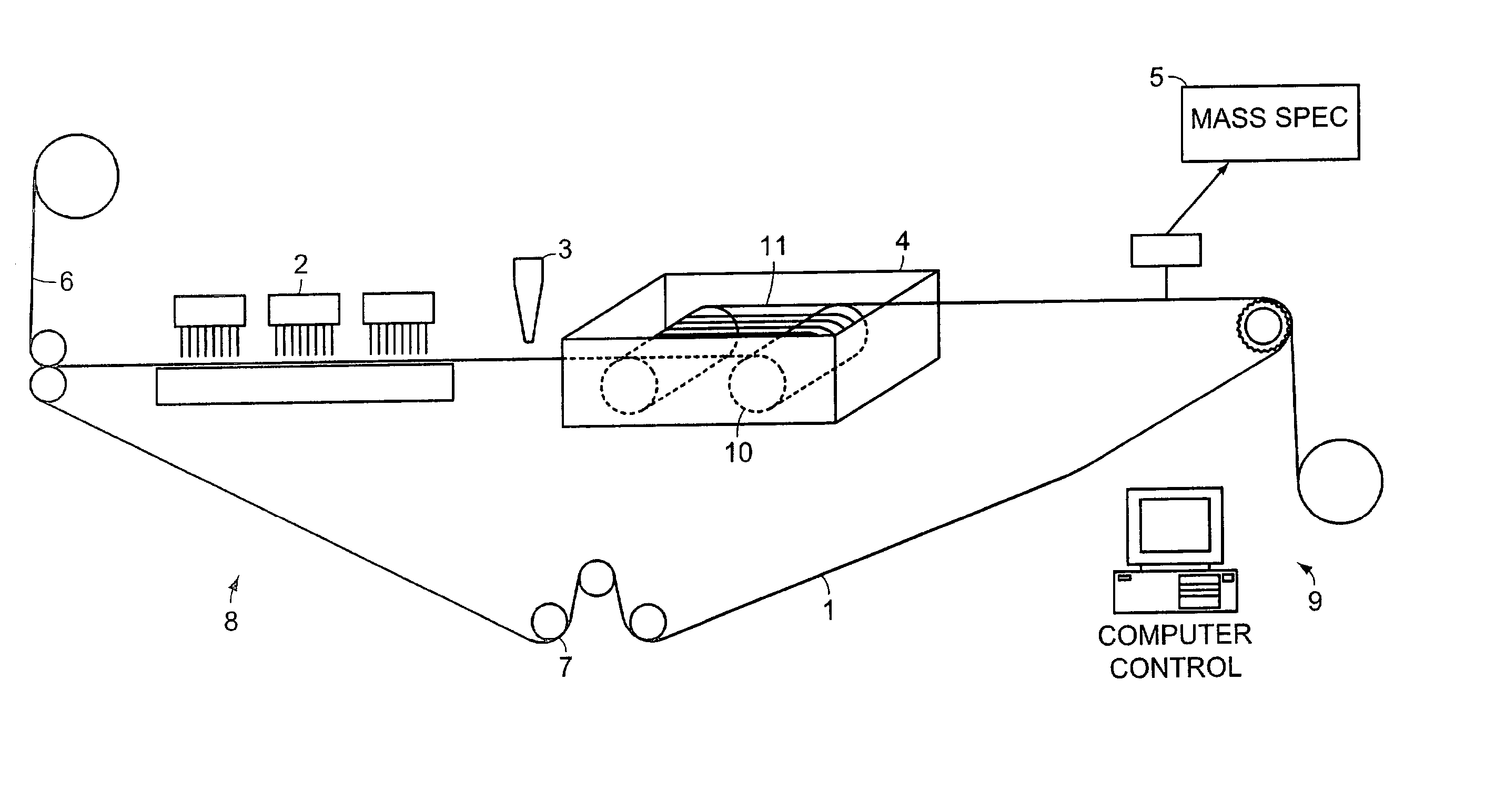
Method and apparatus for non-invasive measurement of blood analytes
InactiveUS20060063993A1Shorten the timeReduce the amount requiredDiagnostics using spectroscopyRaman scatteringBlood levelAnalyte
The present invention discloses a method and apparatus and method for achieving non-invasive measurement of analytes from human and animal blood through the skin using Raman lightwave technology. The apparatus includes a hydraulic tissue permeation unit, which controls the amount of blood in the laser tissue interaction region. Two or more spectra are obtained at different blood levels. These spectra are used to improve the measurements.
Owner:YU DEJIN +1
Raman-active taggants and thier recognition
InactiveUS20040058058A1Easy to useQuality improvementMaterial nanotechnologyRadiation pyrometryLasing wavelengthLaser light
An organic or organoelement, linear or branched, monomeric or polymeric composition of matter having a Raman-active component in the form of particles. The particles having a maximum dimension of 50 mum. The Raman-active compound is applied to a substrate. When the Raman-active compound is exposed to a laser light wavelength which is batochromically well beyond a spectral region of maximum absorbance of said Raman-active compound, Raman scattering can be detected.
Owner:SHCHEGOLIKHIN ALEXANDER NIKITOVICH +4
Safety shut-off device for laser surgical instruments employing blackbody emitters
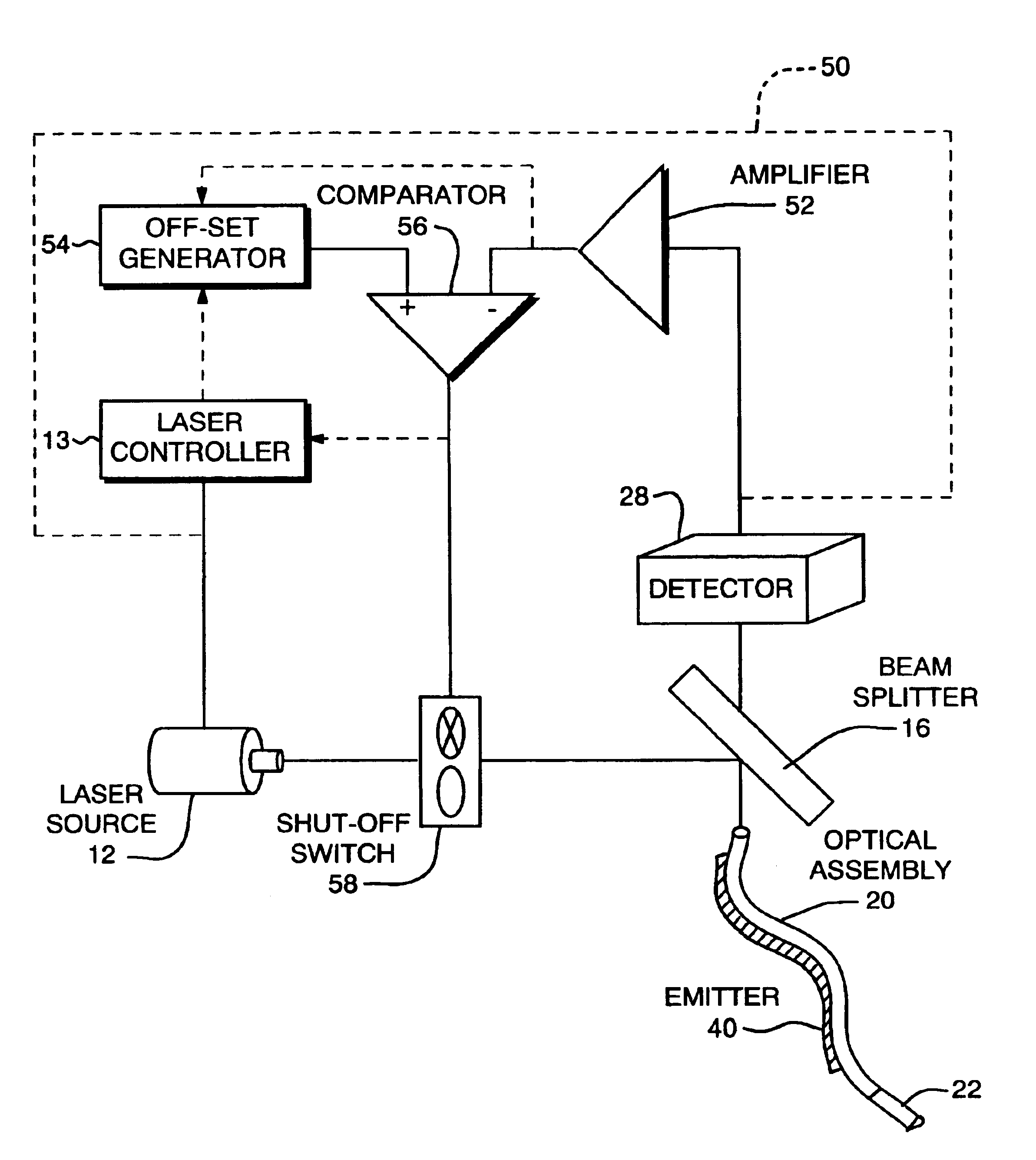
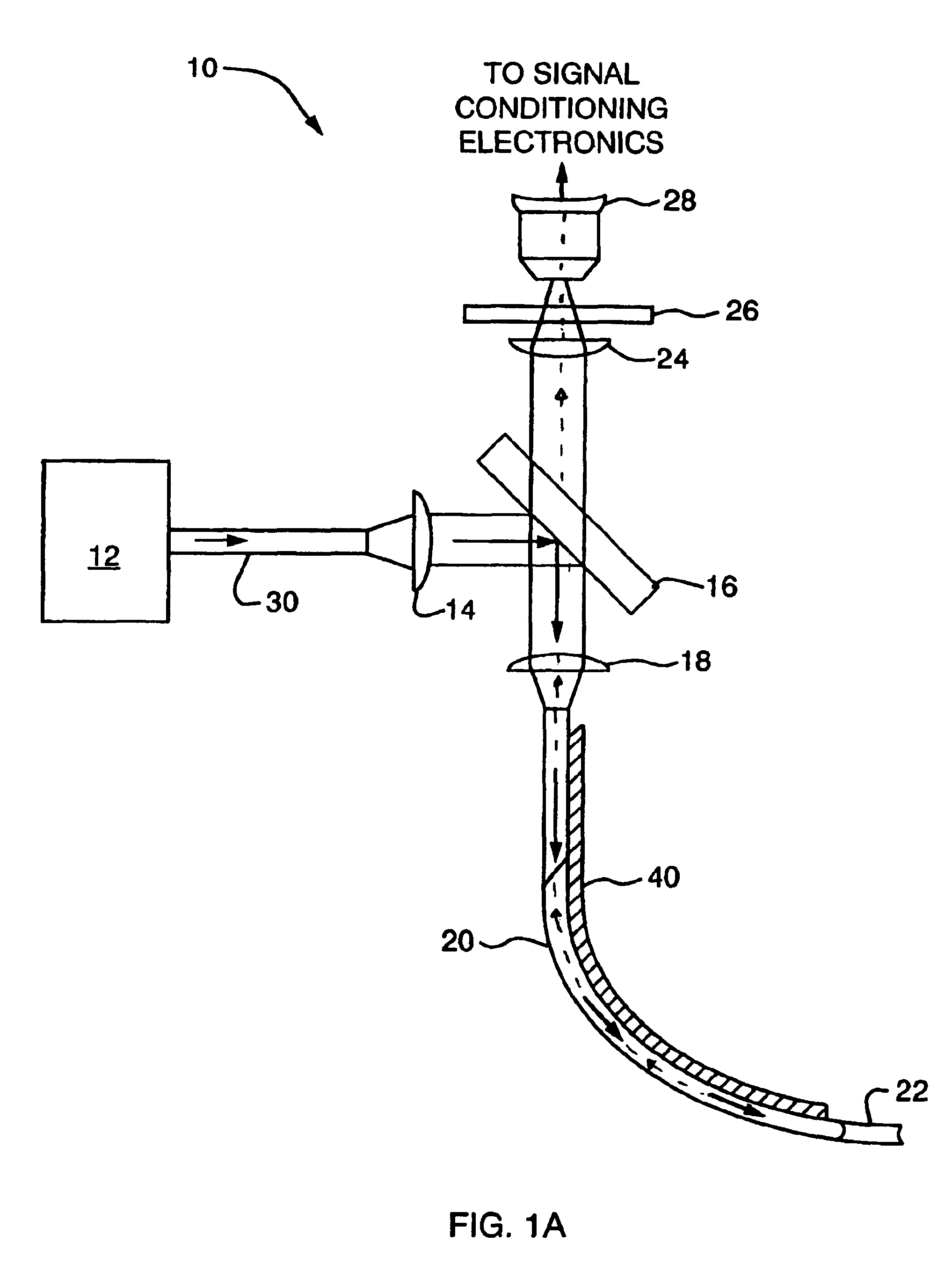
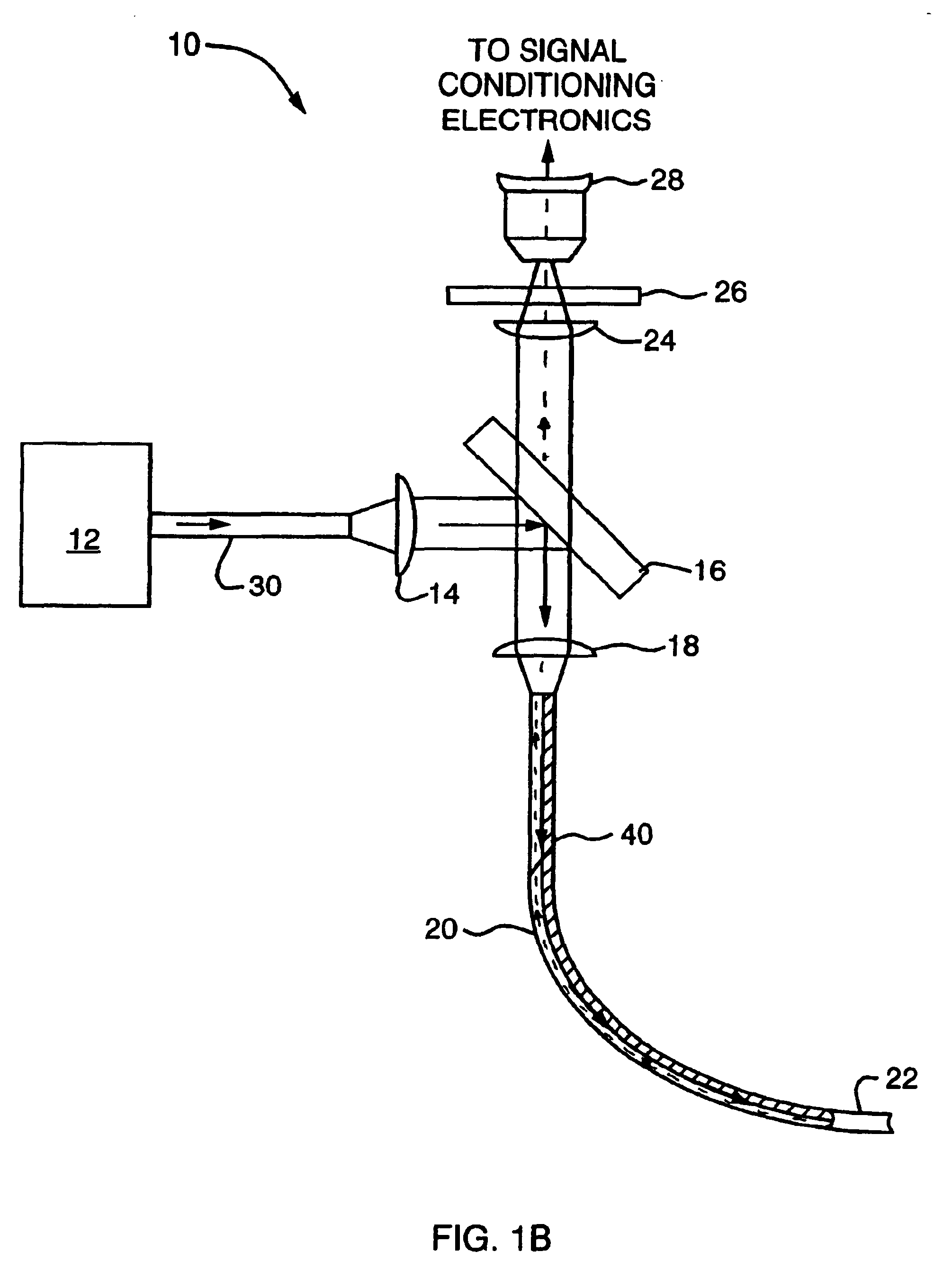
InactiveUS6932809B2High detection sensitivityHigh strengthThermometer detailsPhotometry using reference valueInfraredTherapeutic radiation
Methods and systems are disclosed for detecting overheating in an optical device before harmful consequences, such as severe local heating, can result. In one embodiment of the invention, a blackbody emitter is disposed in close proximity to a therapeutic optical fiber to absorb therapeutic radiation at a fault and re-emit blackbody (infrared) radiation. The emitter can be coupled to the fiber but, during normal operation, lies outside the optical path between the output of the laser radiation and the site of treatment. Systems and catheters incorporating such emitters are also described for effective monitoring of the laser power transmitted along the optical fiber within the phototherapy device.
Owner:CARDIOFOCUS INC
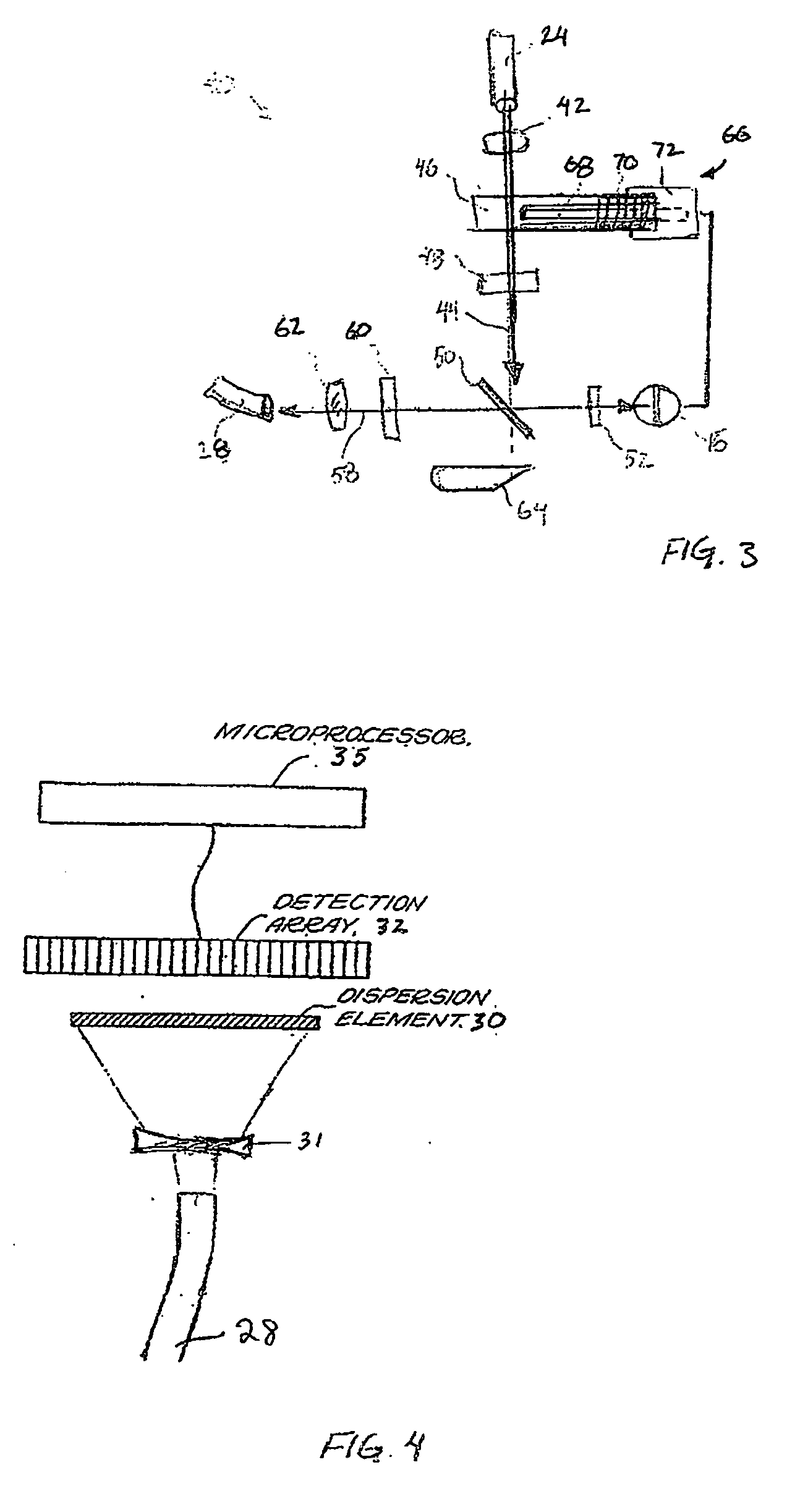
Handheld raman blood analyzer
InactiveUS20060166302A1Bioreactor/fermenter combinationsBiological substance pretreatmentsAnalyteImage resolution
Methods and apparatus for in vitro detection of an analyte in a blood sample using low resolution Raman spectroscopy are disclosed. The blood analyzer includes a disposable strip for receiving a sample of blood on a target region, the target region including gold sol-gel to provide surface enhanced Raman scattering. A light source irradiates the target region to produce a Raman spectrum consisting of scattered electromagnetic radiation that is separated into different wavelength components by a dispersion element. A detection array detects a least some of the wavelength components of the scattered light and provides data to a processor for processing the data. The results of the processed data are displayed on a screen to inform a user about an analyte within the blood sample.
Owner:PRESCIENT MEDICAL
Fluid verification system and method for infusions
An apparatus and method are provided to verify the composition of a medical fluid in a fluid infusion channel by comparison with clinician-entered input. Light is transmitted through the channel and detected by a sensor that generates signals representative of the spectral data of the light detected. A processor compares the spectral data of the light detected to the spectral data associated with the expected contents of the channel to verify that the correct fluid is being infused.
Owner:CAREFUSION 303 INC
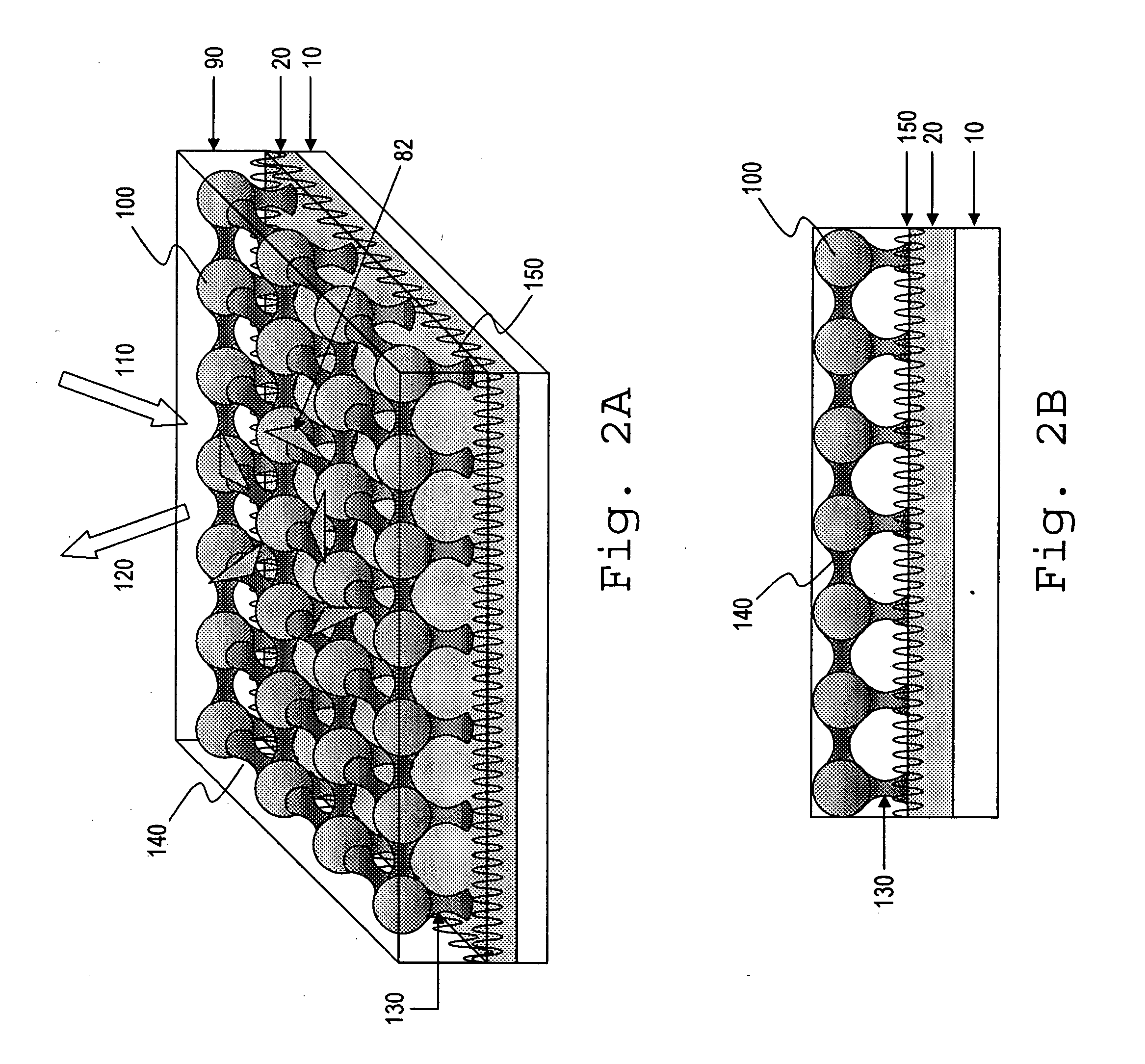
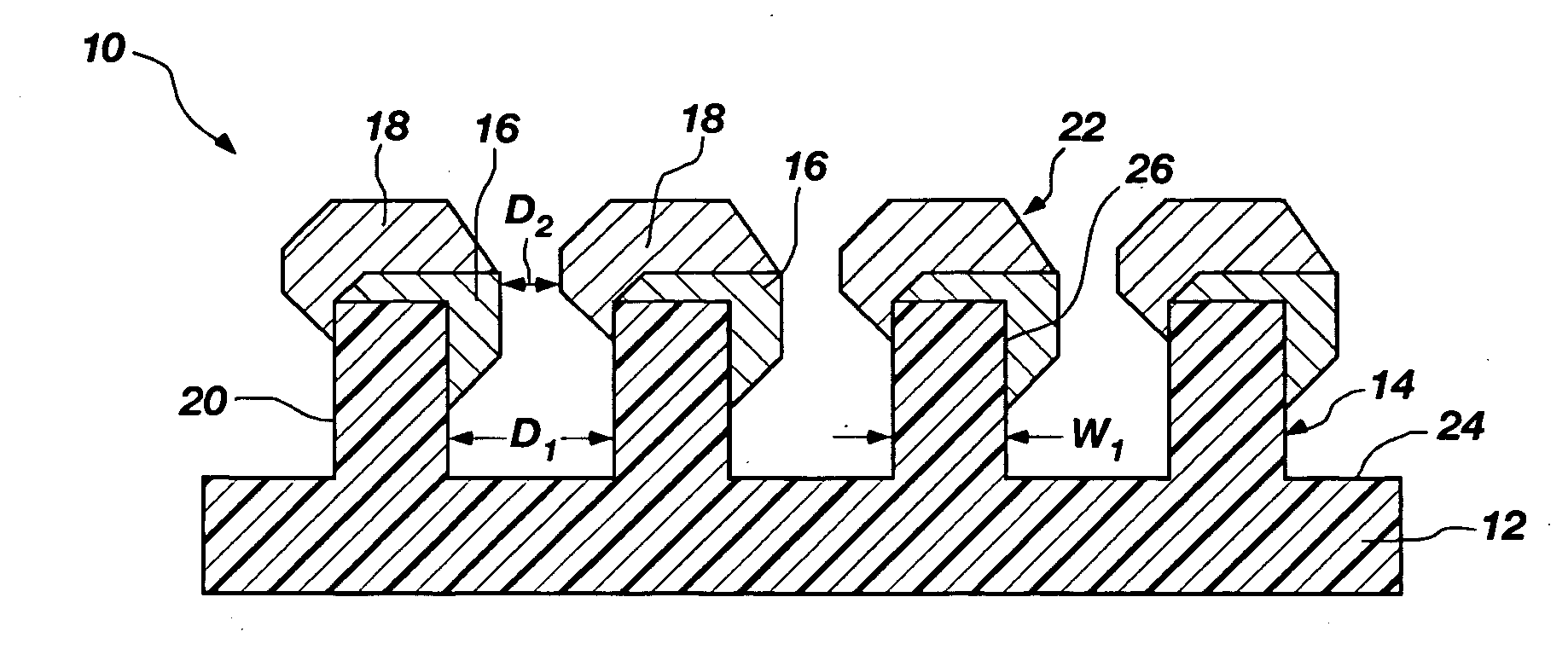
Nanoscale structures, systems, and methods for use in nano-enhanced raman spectroscopy (NERS)
NERS-active structures for use in Raman spectroscopy include protrusions extending from a surface of a substrate. A Raman signal-enhancing material is disposed on at least one surface of a first protrusion and at least one surface of a second protrusion. The Raman signal-enhancing material disposed on the first protrusion projects laterally in a direction generally towards the second protrusion, and the Raman signal-enhancing material disposed on the second protrusion projects laterally in a direction generally towards the first protrusion. At least a portion of the Raman signal-enhancing projecting from the first protrusion and at least a portion of the Raman signal-enhancing material projecting from the second protrusion may be separated by a distance of less than about 10 nanometers. Raman spectroscopy systems include such NERS-active structures, and methods for performing Raman spectroscopy include irradiating an analyte proximate such a NERS-active structure and detecting Raman-scattered radiation scattered by the analyte.
Owner:HEWLETT PACKARD DEV CO LP
Optical analysis technique and sensors for use therein
InactiveUS6020207AMaterial analysis by observing effect on chemical indicatorChemiluminescene/bioluminescenceLiquid coreChemical species
The detection of a chemical specie of interest is accomplished by immobilizing sensing molecules on the inner wall of a liquid core optical waveguide, the waveguide comprising a capillary tube and the sensing molecules being selected to interact with the specie of interest carried by the liquid which forms the waveguide core. The interaction produces a change in an optical characteristic of the waveguide which may be detected by illuminating the waveguide with analysis light.
Owner:WPI TAG ACQUISITION LLC
Surface-enhanced spectroscopy-active sandwich nanoparticles
Surface-enhanced Raman spectroscopy (SERS) uses nanoscale metal particles (SERS-active particles) or surface roughness to enhance the Raman signal of Raman-active analytes contacting the surface. SERS sandwich particles contain SERS-active particles sandwiching a Raman-active substance and serve as optical tags. Preferably, the particles are rod-shaped, with each layer (SERS-active and Raman-active) formed as a distinct stripe of the particle. These freestanding particles can be derivatized with surface ligands capable of associating with analytes of interest in, for example, a biological sample. The acquired Raman spectrum of the particle encodes the identity of the ligand. Because of the simplicity and intensity of Raman spectra, highly multiplexed assays are capable using SERS particles with different Raman-active species.
Owner:BECTON DICKINSON & CO +1
Portable raman diagnostic system
ActiveUS20120035442A1Small sizeReduce weightRaman scatteringDiagnostic recording/measuringSpectral bandsRaman Optical Activity Spectroscopy
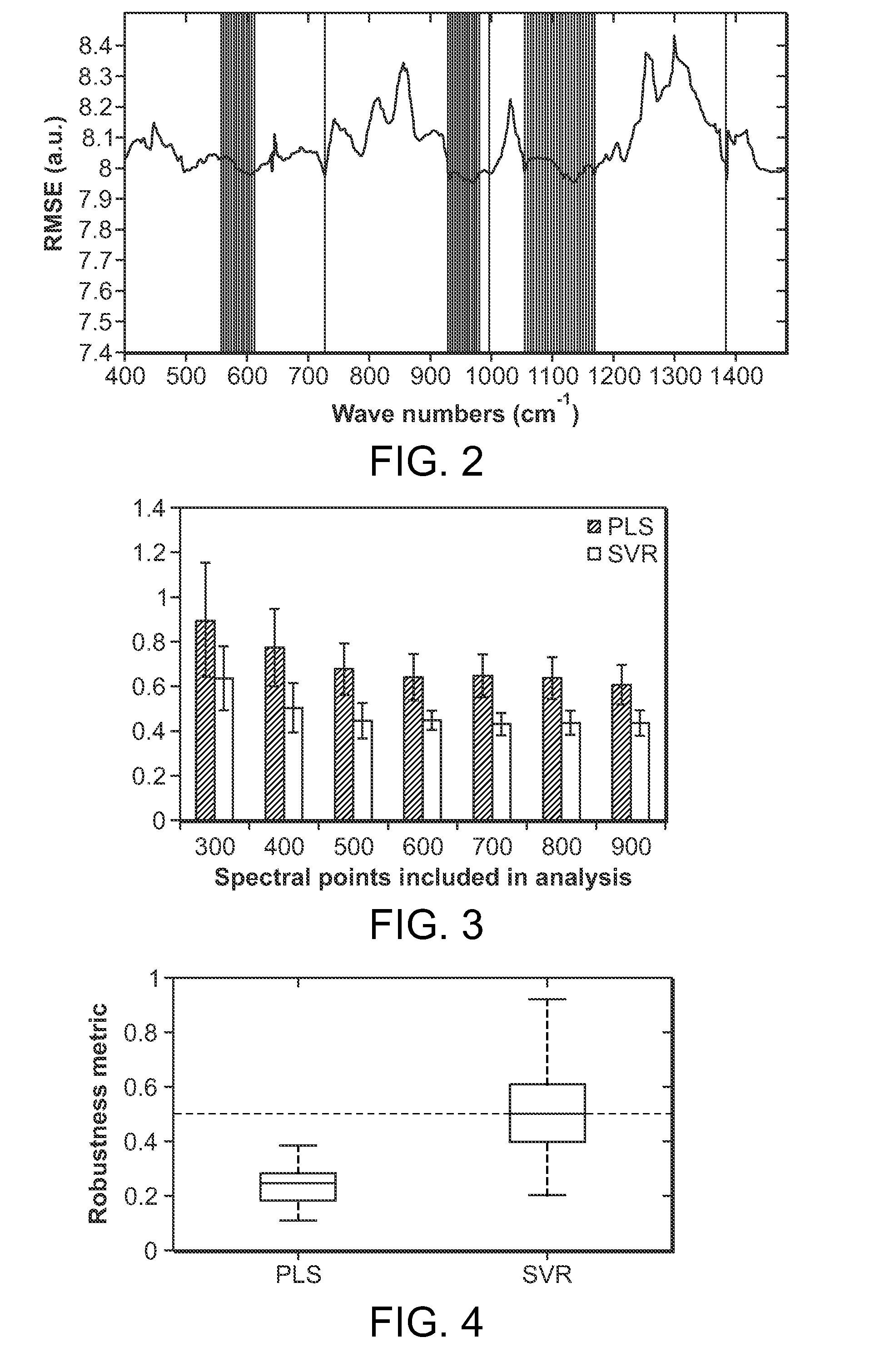
The present invention further relates to the selection of the specific filter combinations, which can provide sufficient information for multivariate calibration to extract accurate analyte concentrations in complex biological systems. The present invention also describes wavelength interval selection methods that give rise to the miniaturized designs. Finally, this invention presents a plurality of wavelength selection methods and miniaturized spectroscopic apparatus designs and the necessary tools to map from one domain (wavelength selection) to the other (design parameters). Such selection of informative spectral bands has a broad scope in miniaturizing any clinical diagnostic instruments which employ Raman spectroscopy in particular and other spectroscopic techniques in general.
Owner:MASSACHUSETTS INST OF TECH
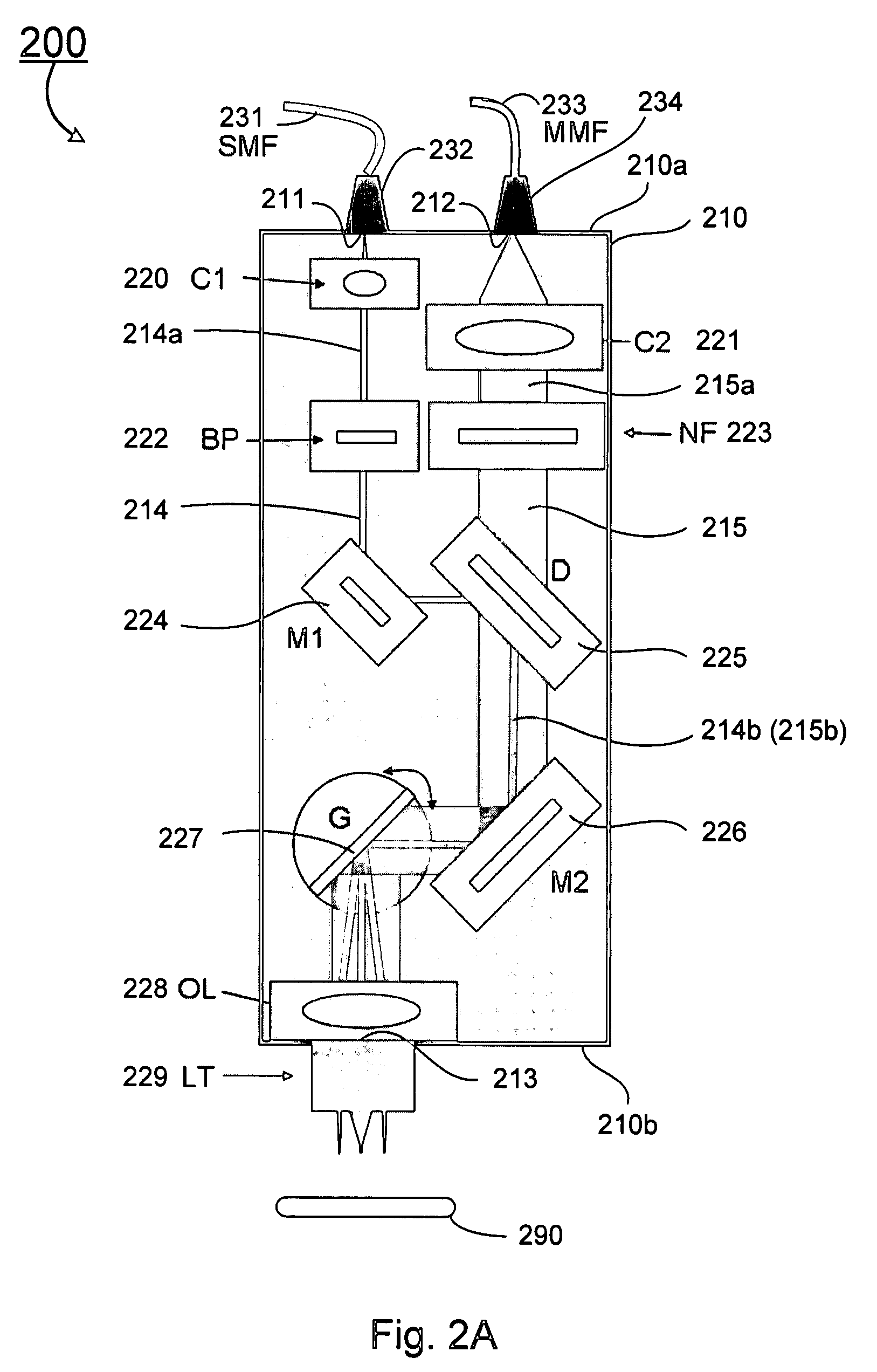
Double-clad fiber scanning microscope
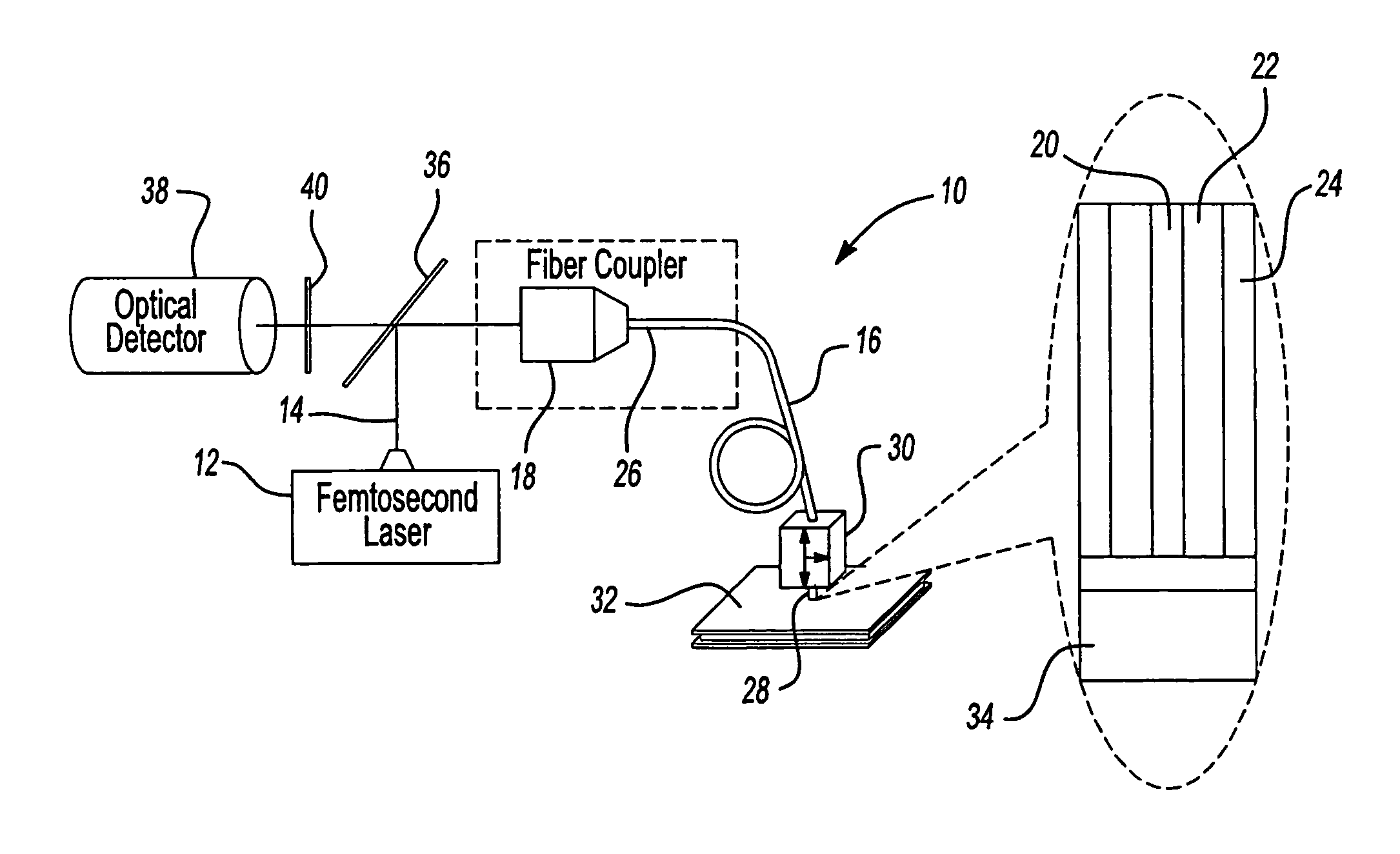
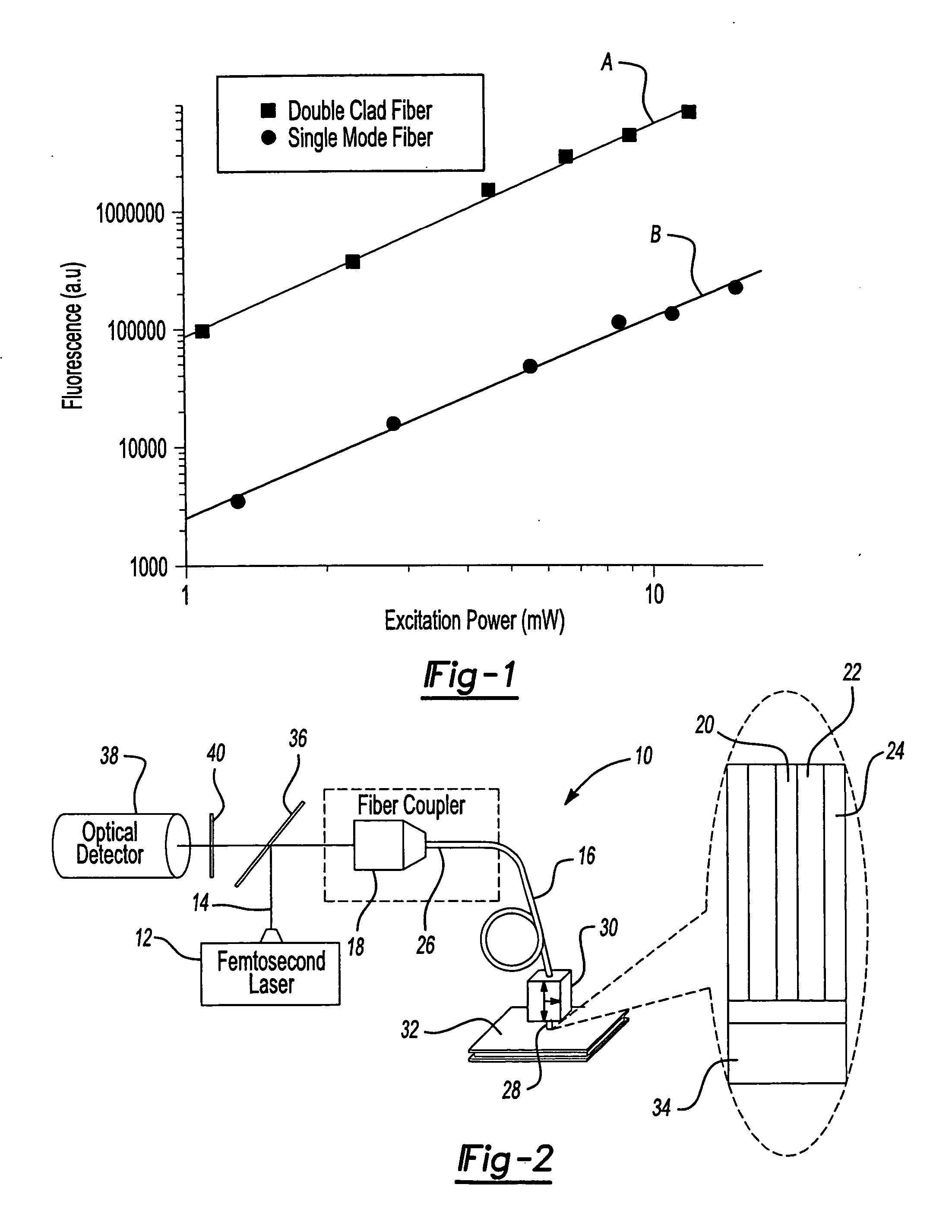
ActiveUS20070002435A1High resolutionImprove detection efficiencyRaman scatteringMicroscopesFiberDouble-clad fiber
A scanning microscope having a laser outputting an excitation laser beam and a fiber member having a first core and a second core. The second core is generally disposed within the first core and is operable to receive the excitation laser beam from the laser and transmit the excitation laser beam to a sample to be tested. A moveable stage supports an end of the fiber member and / or a sample to be tested and is operable to move the end of the fiber member and the sample to be tested relative to each other.
Owner:RGT UNIV OF MICHIGAN
Analyzing and correlating spectra, identifying samples and their ingredients, and displaying related personalized information
Obtaining two spectra from the same sample under two different conditions at about the same time for comparison, where at least one of the spectra measures magnitudes of electromagnetic radiation on at least four different ranges or weightings of wavelengths or frequencies. Classifying a sample using these spectra obtained by a user, and using spectra obtained from different samples by different users to identify the sample. Computing correlations between data related to food and ingredient consumption by one or more users over time, and data related to passive personal log data, user entered feedback, user interaction data or personal information related to those users, and detecting: foods or ingredients to which a user may be allergic or intolerant; a possible medical condition of a user; a possible link between food and ingredient consumption and a medical or health condition; or a similarity between at least two such users.
Owner:TELLSPEC
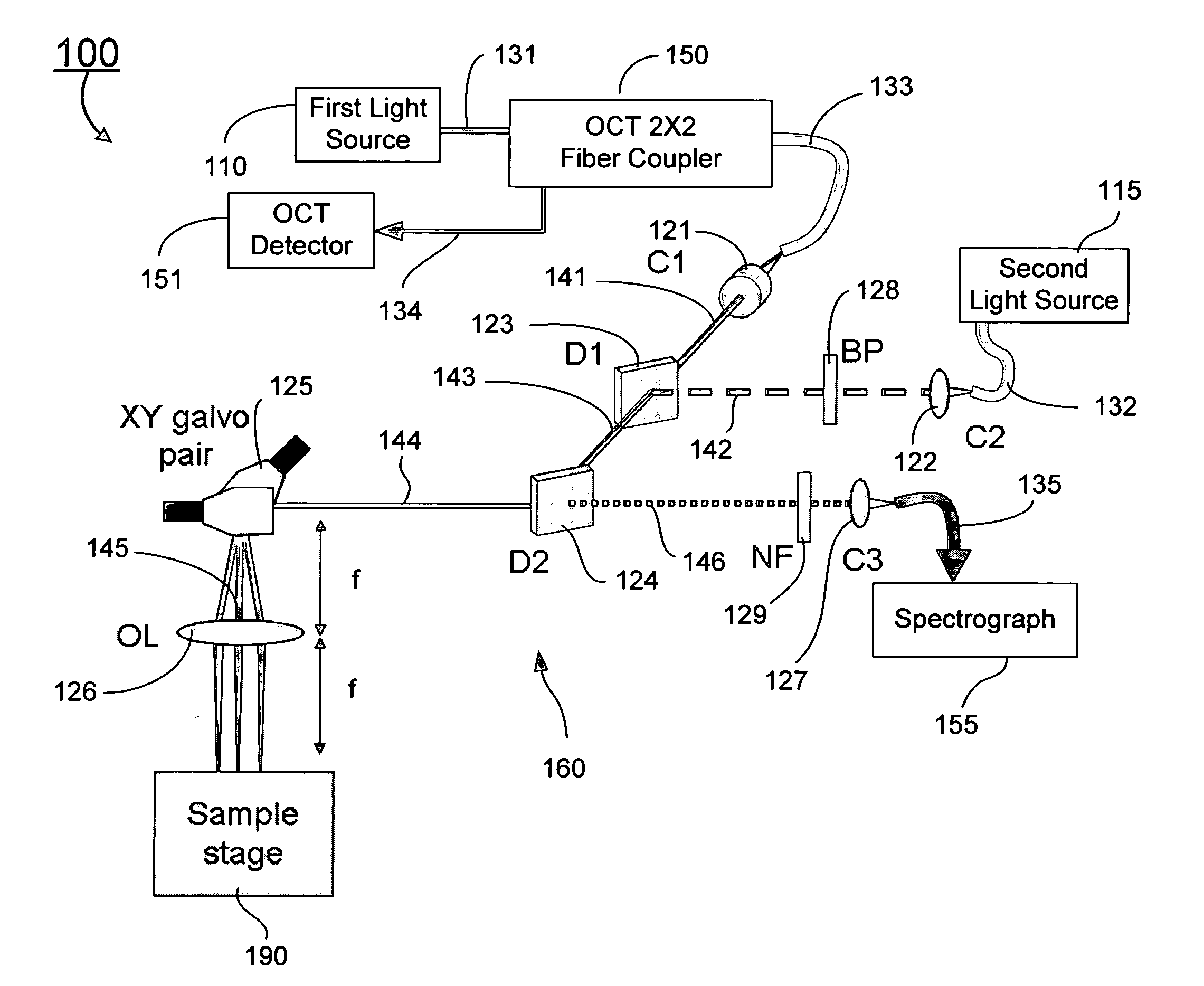
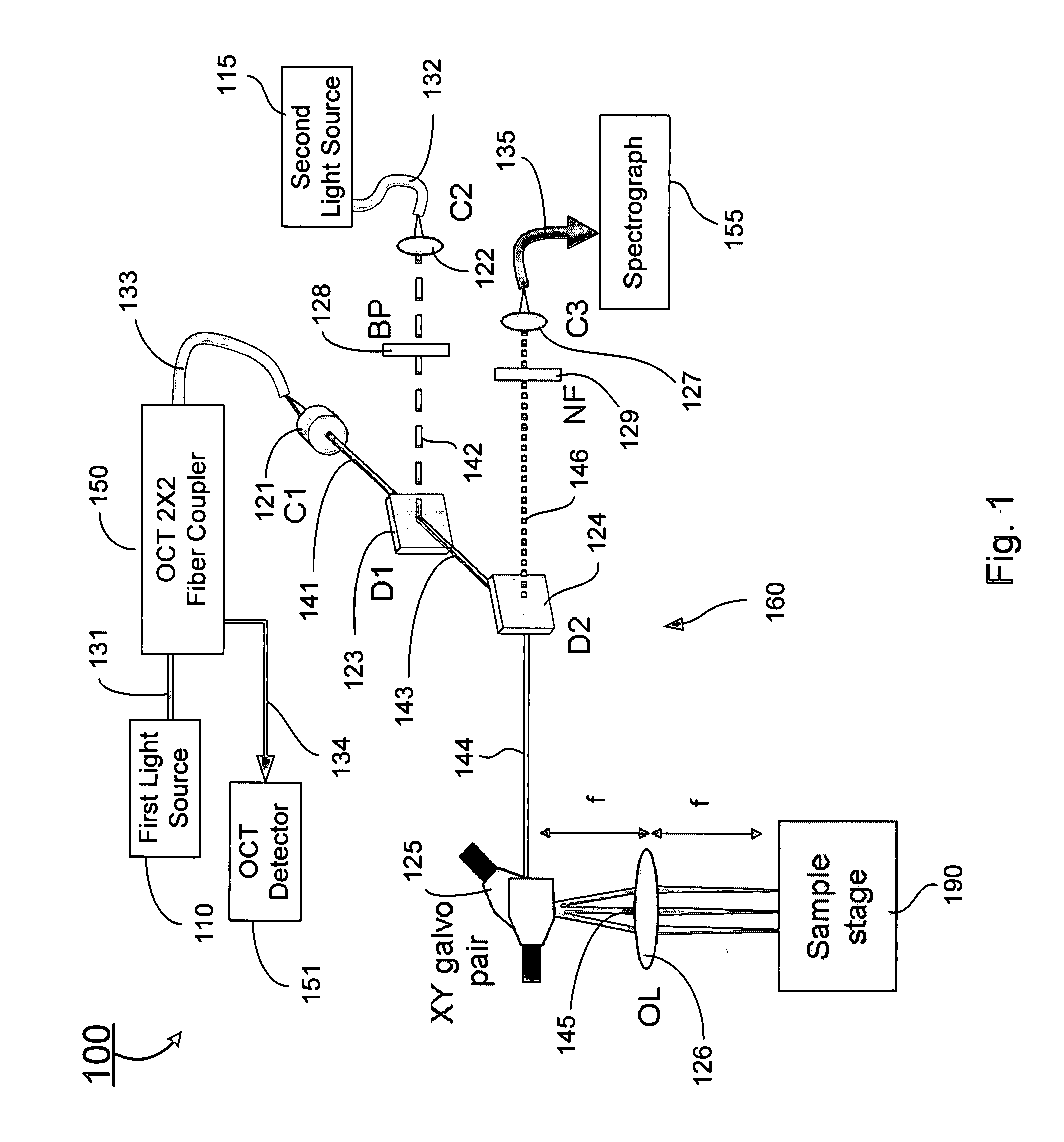
Combined raman spectroscopy-optical coherence tomography (rs-oct) system and applications of the same
ActiveUS20090021724A1Eliminate the problemRadiation pyrometryInterferometersBiological bodyMonochromatic color
An apparatus for evaluating a target of interest of a living subject. In one embodiment, the apparatus has a first light source for generating a broadband light, a second light source for generating a monochromatic light, a beamsplitter optically coupled to the first light source for receiving the broadband light and splitting it into a reference light and a sample light, a reference arm optically coupled to the beamsplitter for receiving the reference light and returning it into the beamsplitter, and a probe having a working end placed proximal to a target of interest of a living subject, optically coupled to the beamsplitter and the second light source for receiving the sample light and the monochromatic light, delivering them from the working end to the target of interest, collecting from the working end a backscattering light and a Raman scattering light that are obtained from interaction of the sample light and the monochromatic light with the target of interest, respectively, and returning the backscattering light into the beamsplitter so as to generate an interference signal between the returned backscattering light and the returned reference light in the beamsplitter.
Owner:VANDERBILT UNIV
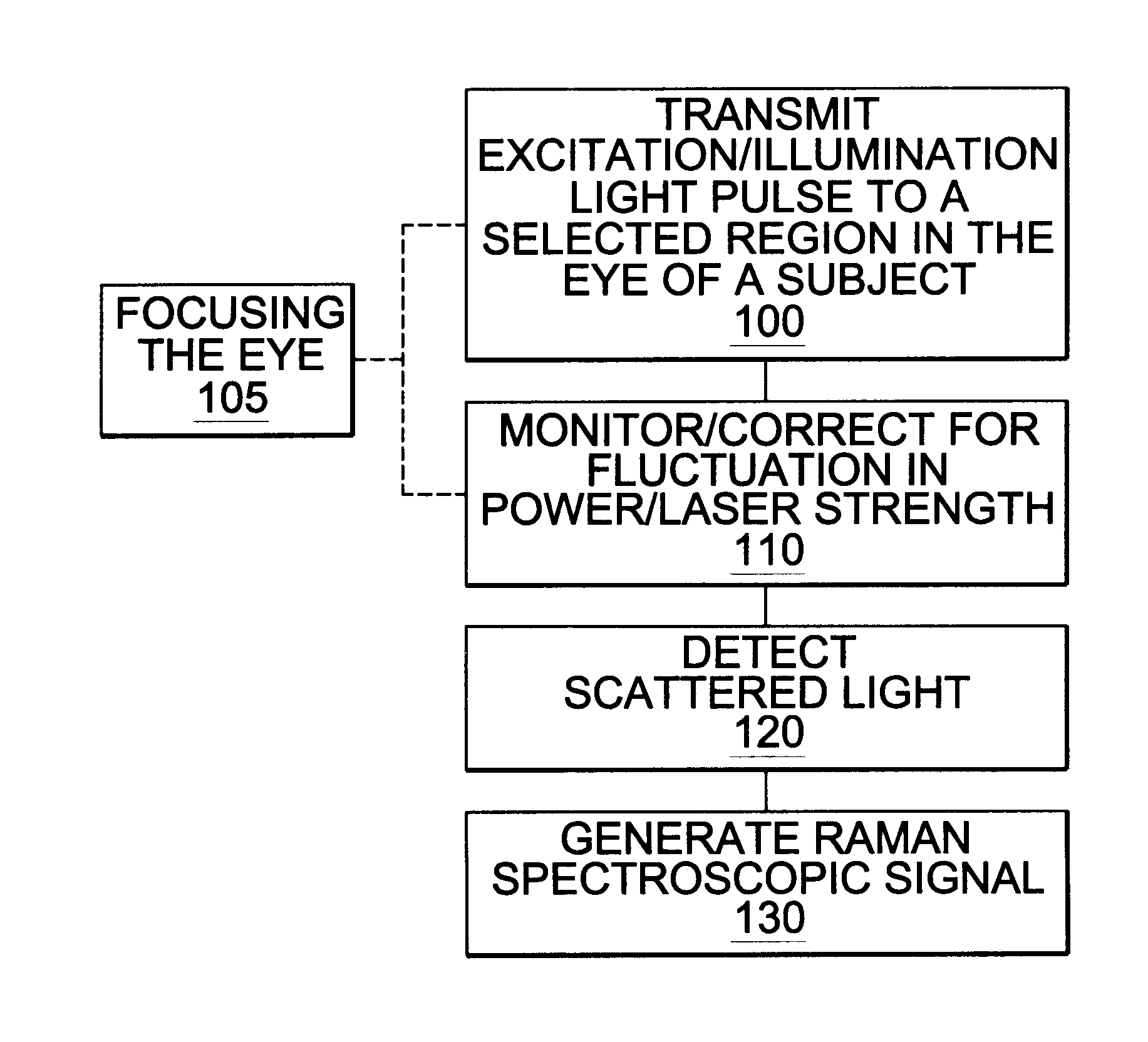
Assessing blood brain barrier dynamics or identifying or measuring selected substances or toxins in a subject by analyzing Raman spectrum signals of selected regions in the eye
InactiveUS6574501B2Reduced energy/density exposure ratingImproved margin of safetyRaman scatteringDiagnostic recording/measuringConjunctivaNon invasive
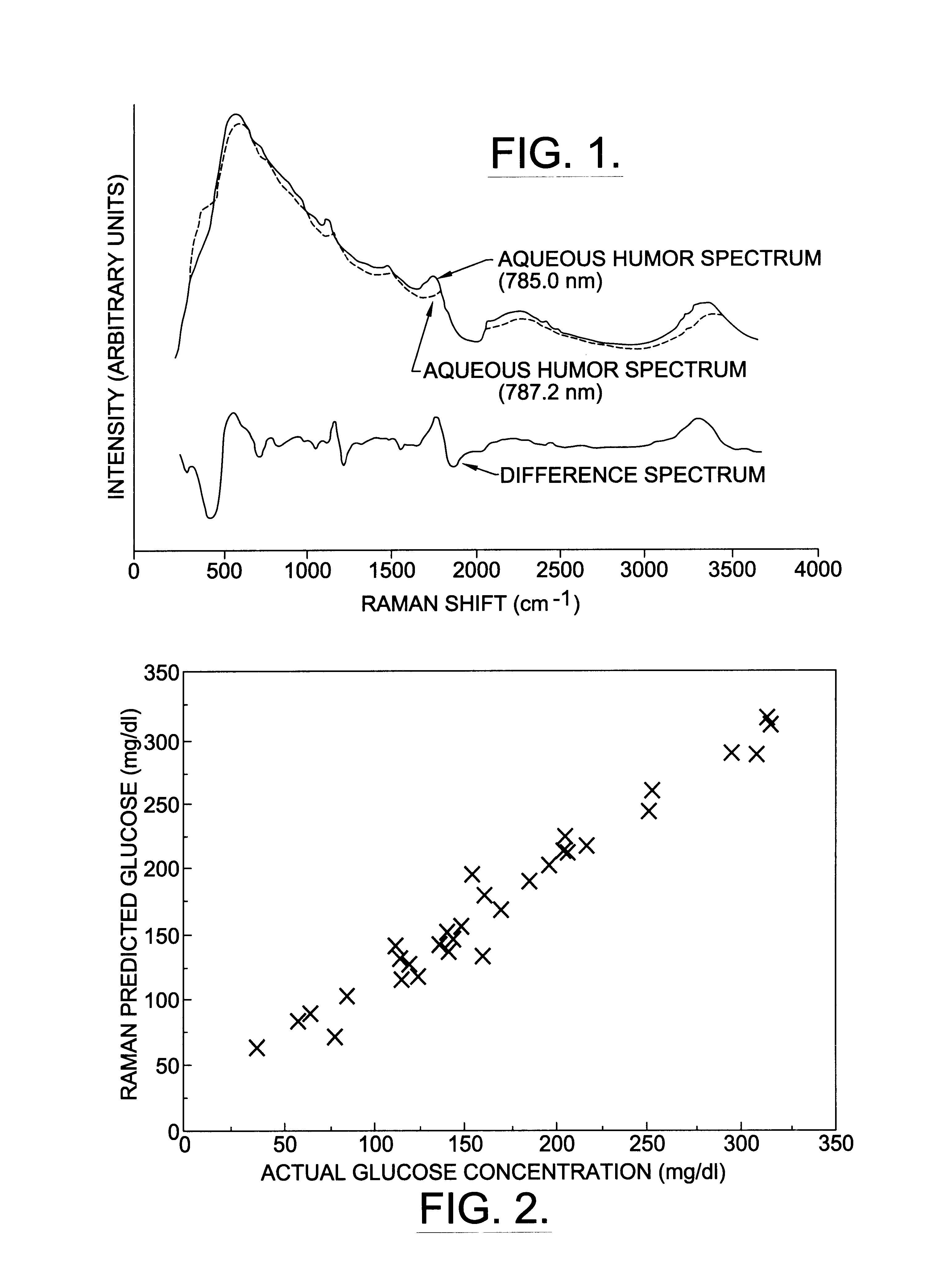
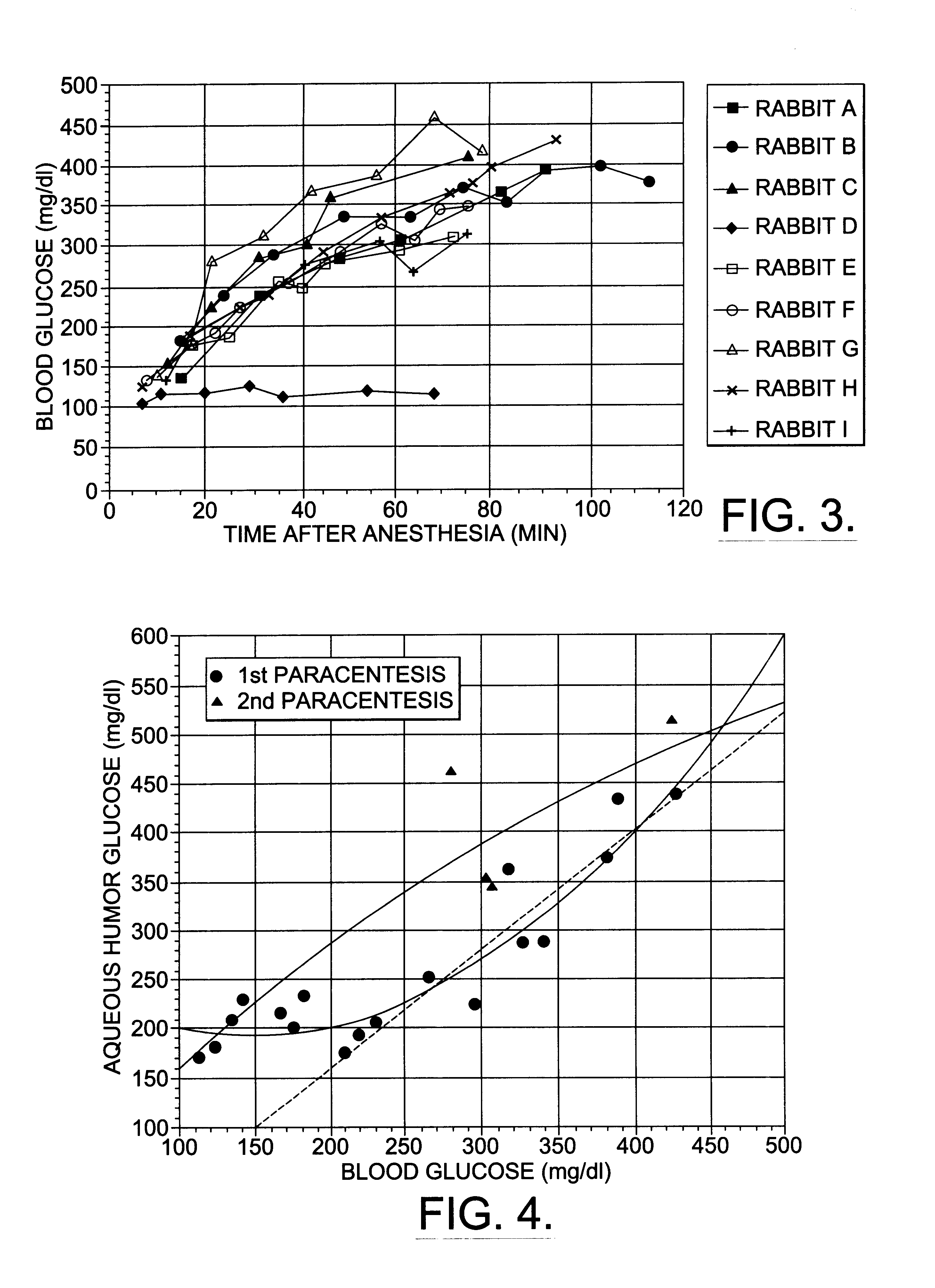
A non-invasive method for analyzing the blood-brain barrier includes obtaining a Raman spectrum of a selected portion of the eye and monitoring the Raman spectrum to ascertain a change to the dynamics of the blood brain barrier. Also, non-invasive methods for determining the brain or blood level of an analyte of interest, such as glucose, drugs, alcohol, poisons, and the like, comprises: generating an excitation laser beam (e.g., at a wavelength of 600 to 900 nanometers); focusing the excitation laser beam into the anterior chamber of an eye of the subject so that aqueous humor, vitreous humor, or one or more conjunctiva vessels in the eye is illuminated; detecting (preferably confocally detecting) a Raman spectrum from the illuminated portion of the eye; and then determining the blood level or brain level (intracranial or cerebral spinal fluid level) of an analyte of interest for the subject from the Raman spectrum. In certain embodiments, the detecting step may be followed by the step of subtracting a confounding fluorescence spectrum from the Raman spectrum to produce a difference spectrum; and determining the blood level and / or brain level of the analyte of interest for the subject from that difference spectrum, preferably using linear or nonlinear multivariate analysis such as partial least squares analysis. Apparatus for carrying out the foregoing methods are also disclosed.
Owner:CHILDRENS HOSPITAL OF LOS ANGELES +1
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com