Method and reagent for constructing nucleic acid single-stranded circular library
A single-stranded circular library and construction method technology, which is applied in chemical libraries, biochemical equipment and methods, and microbial measurement/testing, etc., can solve the problems of single-stranded molecular annealing, unfavorable anchor primer binding, etc., and achieve time-consuming The effect of less, flexible control of fragment length, and simple process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0048] The present invention will be described in further detail below through specific examples. Unless otherwise specified, the techniques used in the following examples are conventional techniques known to those skilled in the art; the instruments and reagents used, etc., are available to those skilled in the art through public channels such as commercially available and so on.
[0049] The terms used in the present invention are described as follows: the first joint is called the No. 1 joint in the specific embodiment; the second joint is called the No. 2 joint in the specific embodiment; the third joint is called the No. 3 joint in the specific embodiment connector.
[0050] In the present invention, concepts such as "first" and "second" used under any circumstances should not be interpreted as having sequential and technical meanings, and their function is only to distinguish them from other objects.
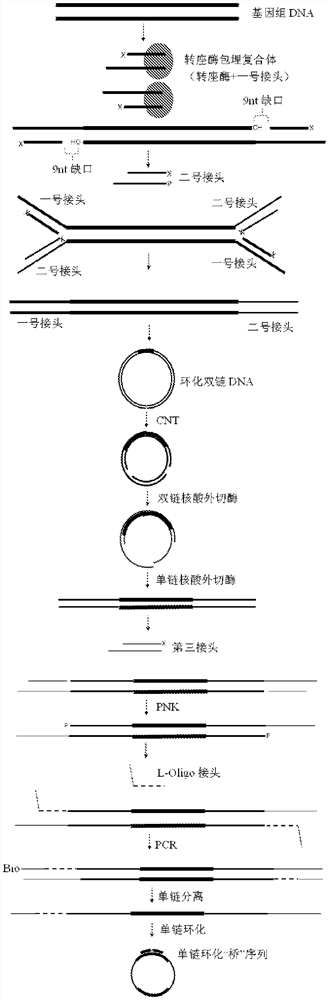
[0051] Please refer to figure 1 , a method for constructing a nucl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com