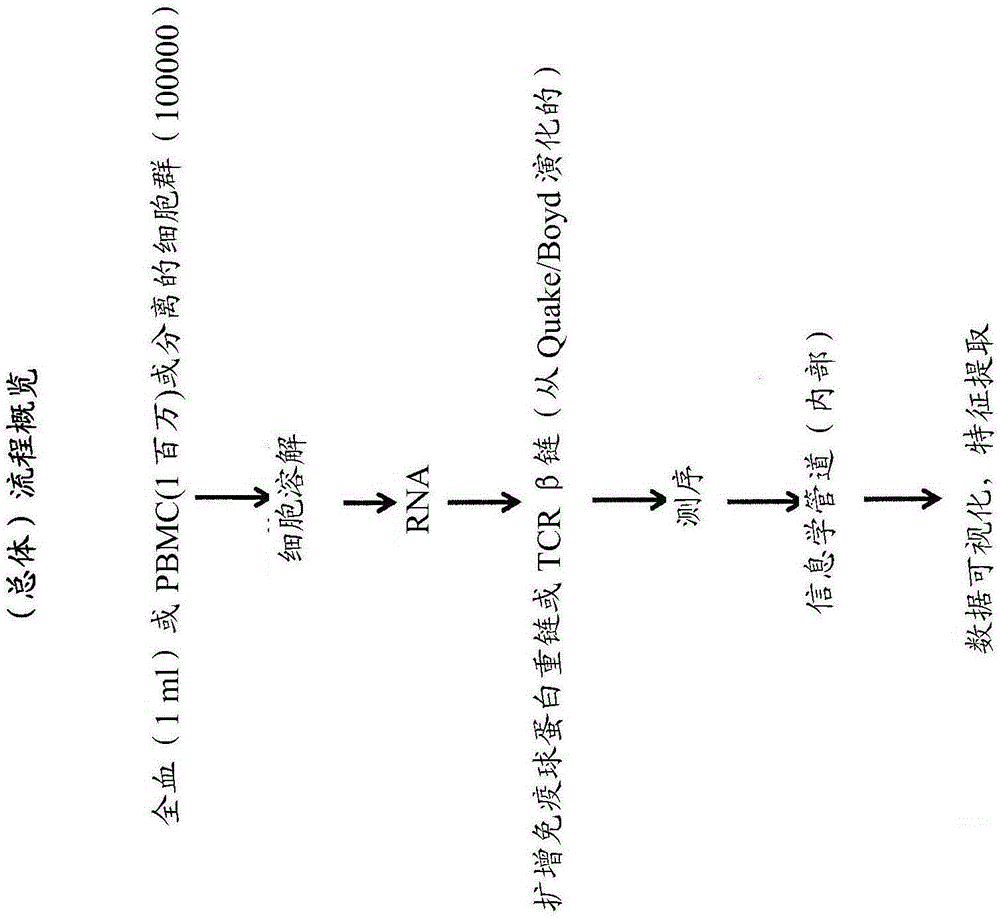Methods of sequencing the immune repertoire
A technology of immune group library and sequencing, which is applied in the fields of biochemical equipment and methods, recombinant DNA technology, microbial determination/testing, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
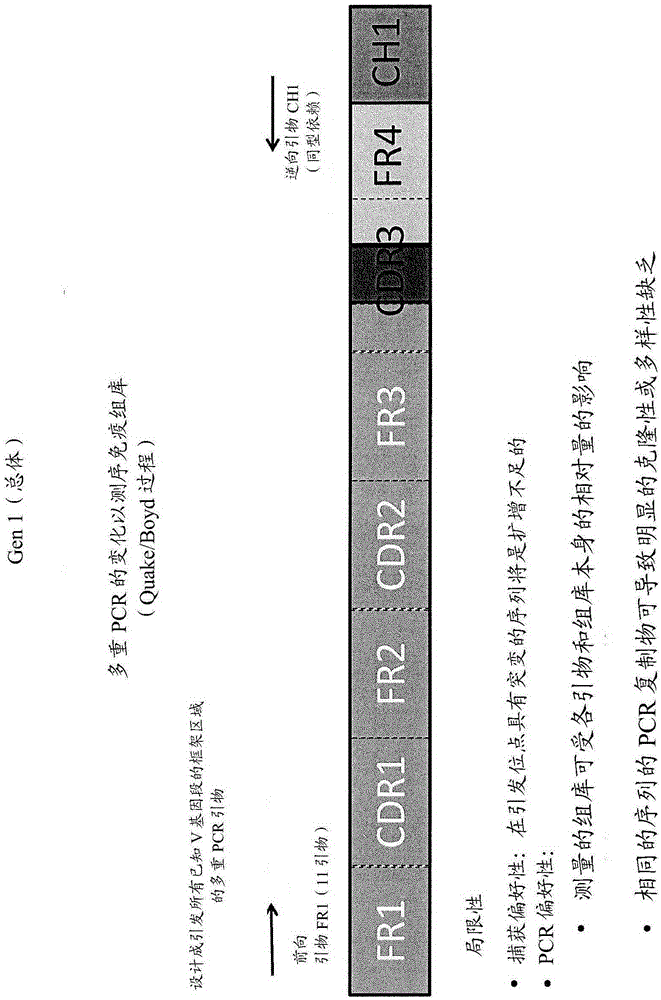
[0022] The present invention provides improved methods for sequencing immune repertoires. Previous methods of determining immune repertoires, such as those described in WO2011 / 140433 and WO2012 / 083069, are based on multiplex PCR. Multiplex PCR has a number of limitations that make it particularly unsuitable for precisely determining the immune repertoire. (See figure 2 ) These limitations include capture bias and amplification bias due to PCR. Multiplex PCR techniques for sequencing immune repertoires use primers designed to prime all framework regions of known V gene segments. Capture bias occurs when mutations occur at priming sites, and genes with mutations are underamplified. PCR bias arises due to uneven gene amplification due to the relative amounts of each primer and PCR replicates with the same sequence. Thus, PCR bias can lead to apparent lack of clonality or diversity. In general, the observed repertoire is either inaccurate, or it is a linear representation of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com