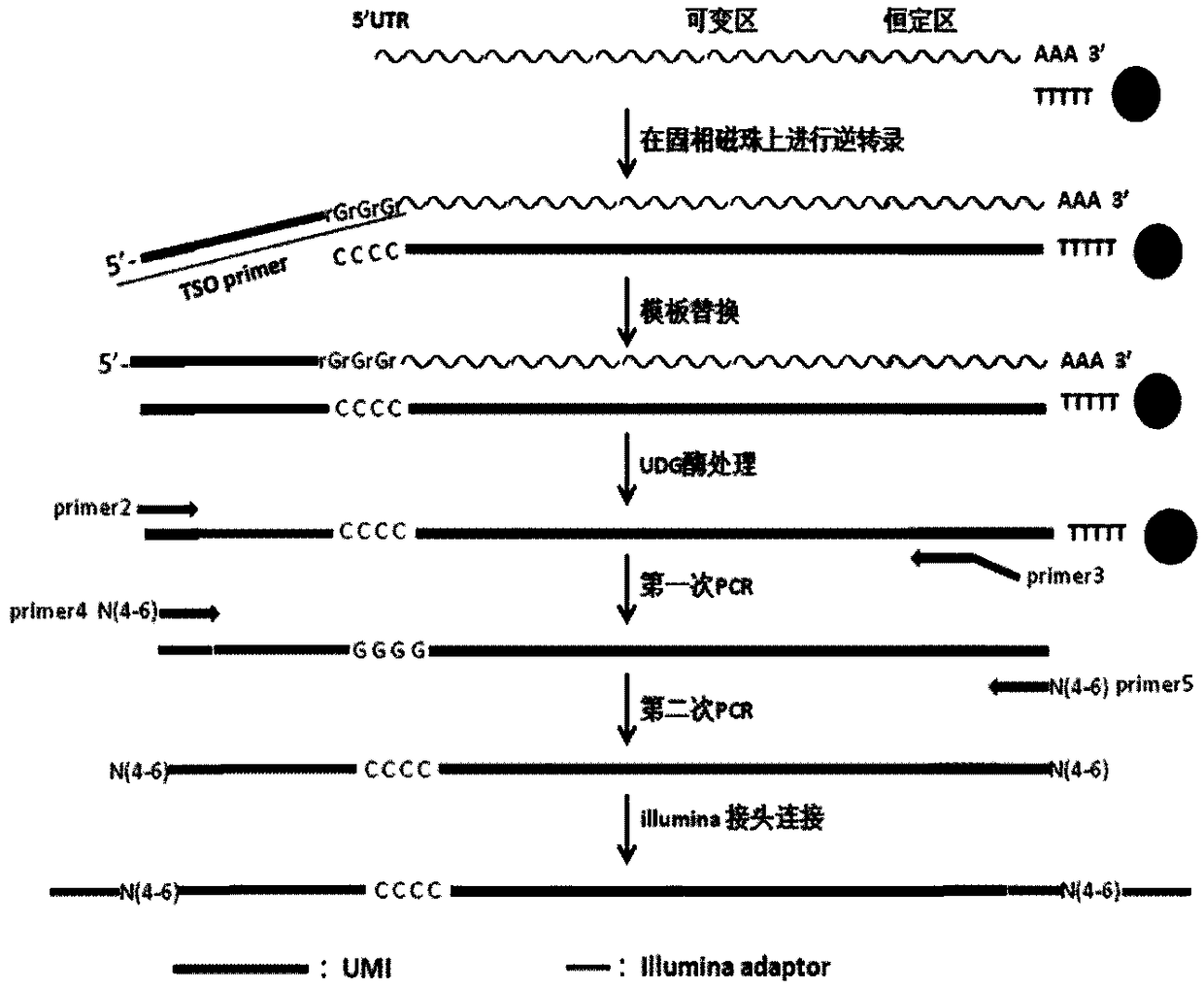
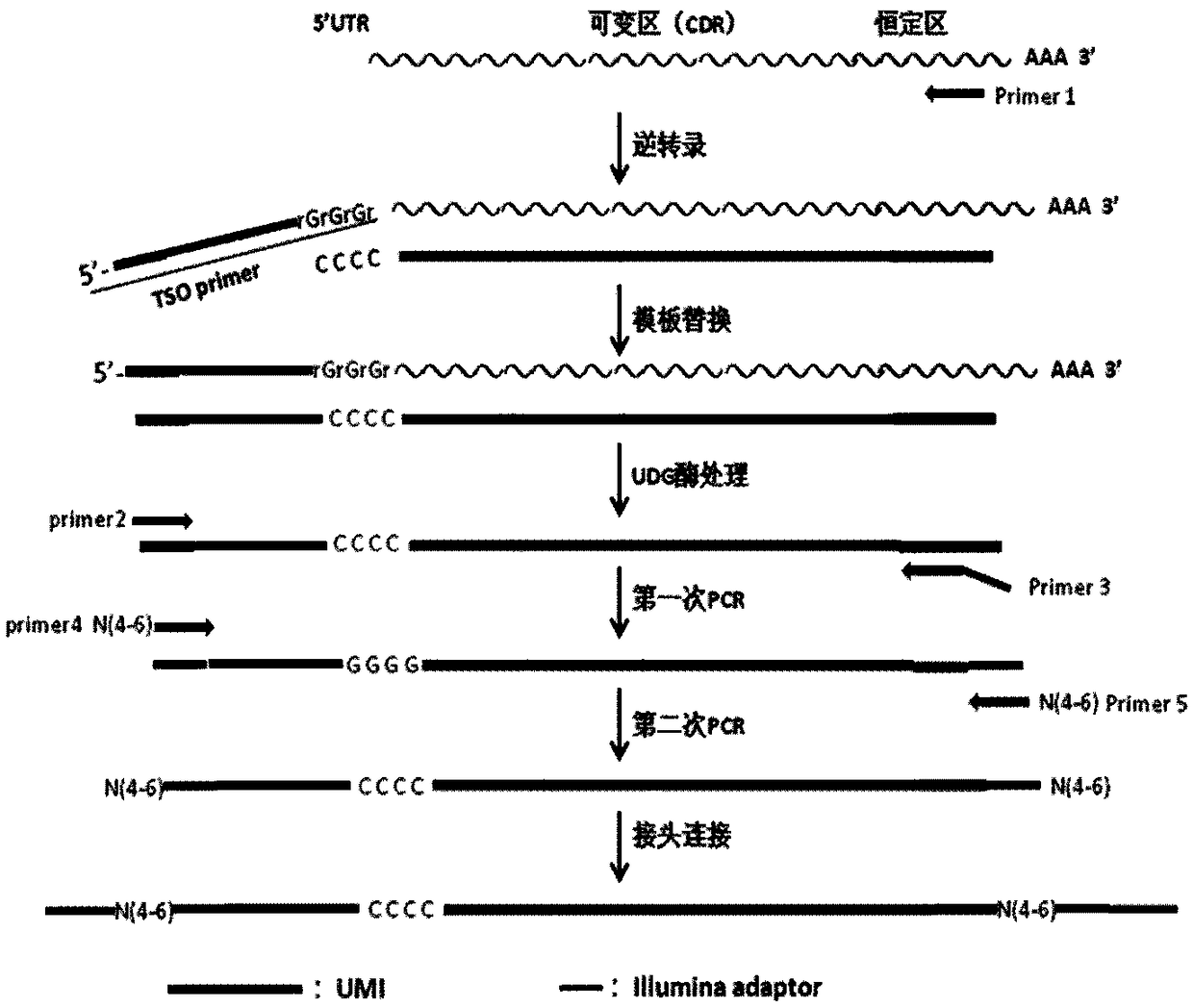
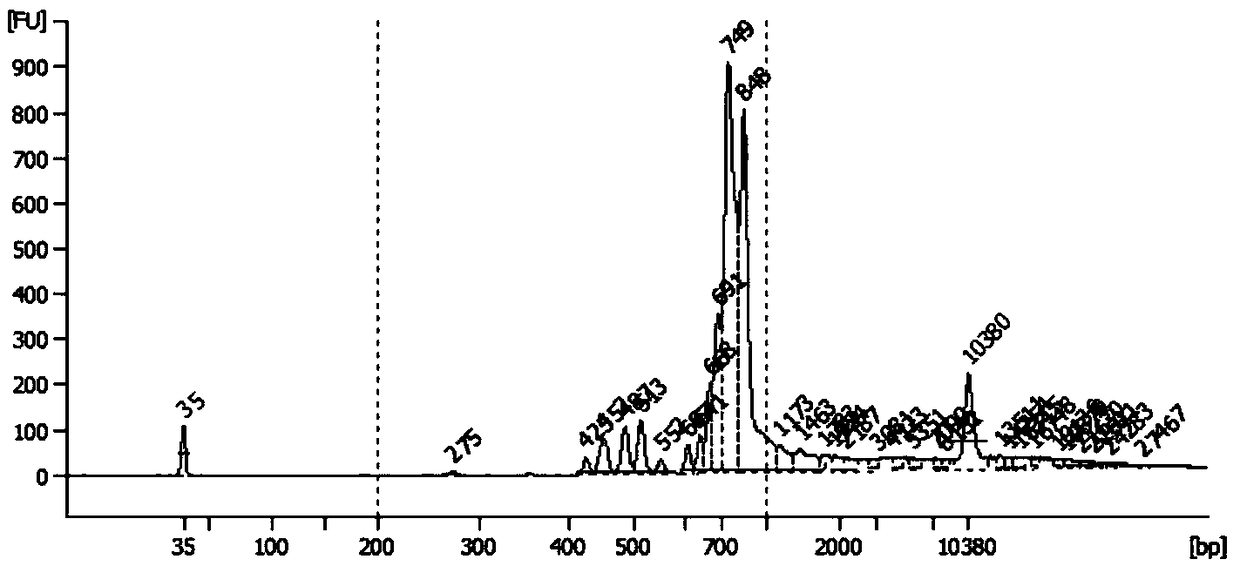
Method for constructing high-throughput sequencing library of immune repertoire
A technology of sequencing library and construction method, which is applied in the field of construction of high-throughput sequencing library of immune repertoire, and can solve problems such as errors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0132] Example 1 Construction method of immune repertoire high-throughput sequencing library of micro-initial samples
[0133] Sample processing: Use lymphocyte separation medium to separate healthy human peripheral blood mononuclear cells and count them (2×10 6cells / ml), 20,000 cells were taken for subsequent BCR library construction experiments;
[0134] 1. Preparation of cDNA template
[0136] Add 10000 lymphocytes to 100ul cell lysate for lysis to obtain lysate;
[0137] (2) mRNA isolation, reverse transcription, template replacement and UDG enzyme treatment
[0138] 1) Use the kit Dynabeads mRNA DIRECT Micro Kit (TermoFisher) to isolate mRNA from the lysed cell sample, and perform the operation according to the instructions;
[0139] 2) Configure the reverse transcription system as shown in Table 12, and immediately add it to the magnetic beads that have captured mRNA in the first step, resuspend the magnetic beads, and transfer them to PCR tub...
Embodiment 2
[0171] Example 2 The construction method of the high-throughput sequencing library of the immune repertoire of the conventional amount of initial samples
[0172] The sample in this embodiment takes the peripheral blood of a healthy person as a sample, and the specific implementation steps are as follows:
[0173] 1. Preparation of cDNA template
[0174] (1) Total RNA extraction
[0175] 1), lymphocyte separation;
[0176] 2), according to every 10 7 Add 1ml Trizol to each cell Add Trizol to the isolated lymphocytes to extract total RNA;
[0177] 3), the extracted RNA is subjected to agilent 2100 quality inspection;
[0178] (2) Reverse transcription, template replacement and UDG enzyme treatment (regular input sample)
[0179] 1), prepare the reaction system (mix1) shown in Table 14 in the PCR tube;
[0180] Table 18
[0181]
[0182] 2) Use a pipette or lightly flick the tube wall to mix the above reaction system, centrifuge briefly and place it on a PCR instrument...
Embodiment 3
[0217] By comparing the number of types of UMI before correction and the number of types of UMI after error correction to evaluate the impact on the sequencing accuracy, and multiple reads detected by the same UMI can be corrected each other to improve the accuracy of the reads sequence. The test uses 20 The data of each library is shown in Table 25:
[0218] Table 25
[0219]
[0220]
[0221] It can be seen that more than 70% of the UMI types have been corrected, indicating that the present invention has greatly improved the data accuracy. In addition, there are quite a few UMIs whose corresponding reads are greater than 2, and these reads can be mutually corrected , to remove PCR and sequencing errors and improve data accuracy.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com