Design method and detection method of specific primers for resistance gene of rifampicin antibiotic resistance caused by SNP
A resistance gene and design method technology, applied in the direction of microorganism-based methods, biochemical equipment and methods, and microorganism measurement/testing, can solve problems such as difficult detection and analysis, complicated operation, and long time consumption, and achieve an optimized reaction system , optimized reaction conditions, fast results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] 1. Primer design
[0036] (1) The resistance gene rpoB gene contains an open reading frame of 3552bp, using the reference sequence of rpoB of Staphylococcus aureus published by NCBI (GenBank NC_007795.1:522160-525711), which encodes 1183 amino acids.
[0037] (2) Use DNAman software to design primers based on the above reference sequence to generate candidate primers.
[0038] (3) Since the full-length sequence of the rpoB gene needs to be amplified, it must be taken at the beginning and end of the reference sequence, and the upstream primers start from the beginning of the reference sequence, and the downstream primers end from the end of the reference sequence. Therefore, some primers are designed The software cannot automatically generate specific primers. It is necessary to use the software and combine some long-range primer design principles to screen the primers. The design of long fragment primers must pay attention to the following issues:
[0039] a. Primer length: 25...
Embodiment 2
[0051] Specific primer pair amplification experiment
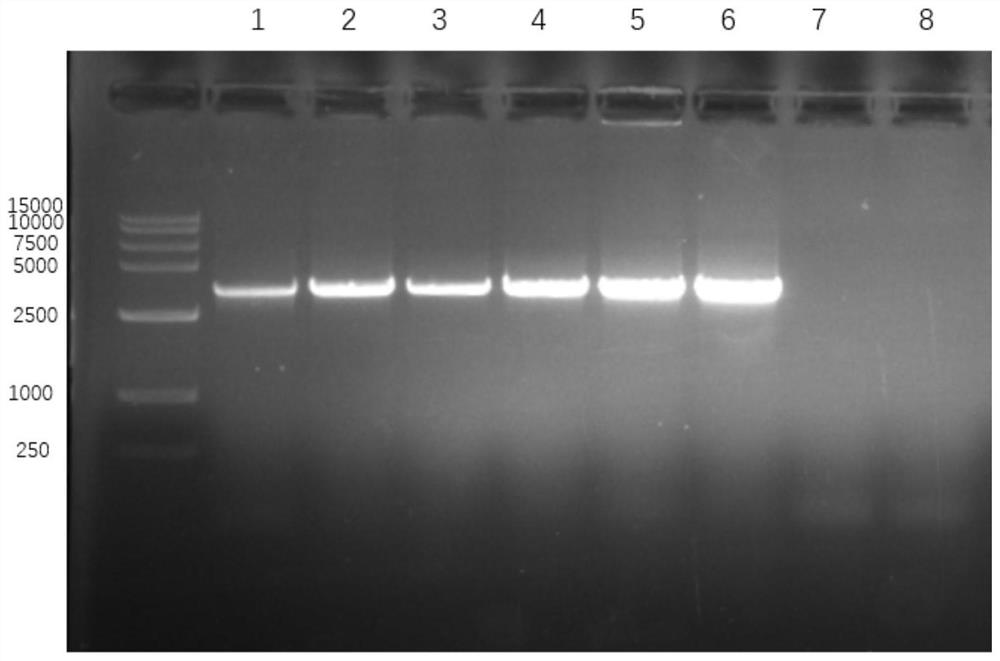
[0052] (1) Extract the genomic DNA of 6 strains of Rifampicin with different minimum inhibitory concentration (MIC) values of Staphylococcus aureus (denoted as Li 0~Li 5), 1 sample of activated sludge and 1 strain of E. coli ATCC25922.
[0053] (2) Using each genomic DNA extracted in step 1 as a template, perform PCR amplification with the specific primer pair screened in Example 1 to obtain the corresponding PCR product; perform agarose gel electrophoresis on the PCR product;
[0054] Among them, the PCR reaction system is:
[0055] The total reaction system is 50μL
[0056]
[0057] The PCR amplification program is: 94°C pre-denaturation 5min; 94°C, 2min, 97°C, 1min, 60°C, 1min, 35 cycles; 68°C, 5min, 68°C, 10min.
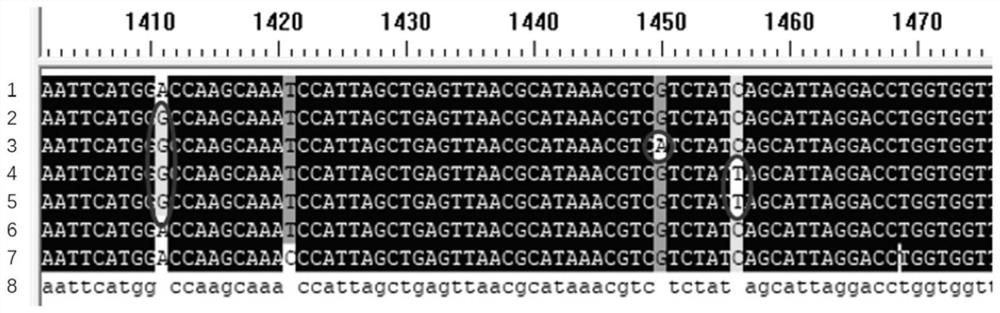
[0058] (3) Recover the PCR products and send them to BGI for sequencing.
[0059] Reference for agarose gel electrophoresis results figure 1 . Lanes 1 to 6 represent 6 strains of Staphylococcus aureus, lane 7 represen...
Embodiment 3
[0067] This embodiment provides a method for screening rifampicin-resistant strains, which includes the following steps:
[0068] (1) Obtain the genomic DNA of the strain to be screened;
[0069] (2) Use the specific primer pairs, DNA polymerase, nucleotides, etc. in Example 1 to extend the genomic DNA according to the amplification procedure in Example 2;
[0070] (3) Sequencing the amplified product after recovery, comparing it with the rpoB gene sequence of wild-type Staphylococcus aureus, obtaining the SNP locus information of the rpoB gene of the strain to be screened, and screening rifampicin based on its SNP locus information Resistant strains.
[0071] The specific primer pair used in this solution can accurately amplify the full-length sequence of the rpoB gene through conventional PCR, so that the strain screening can be effectively achieved.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com