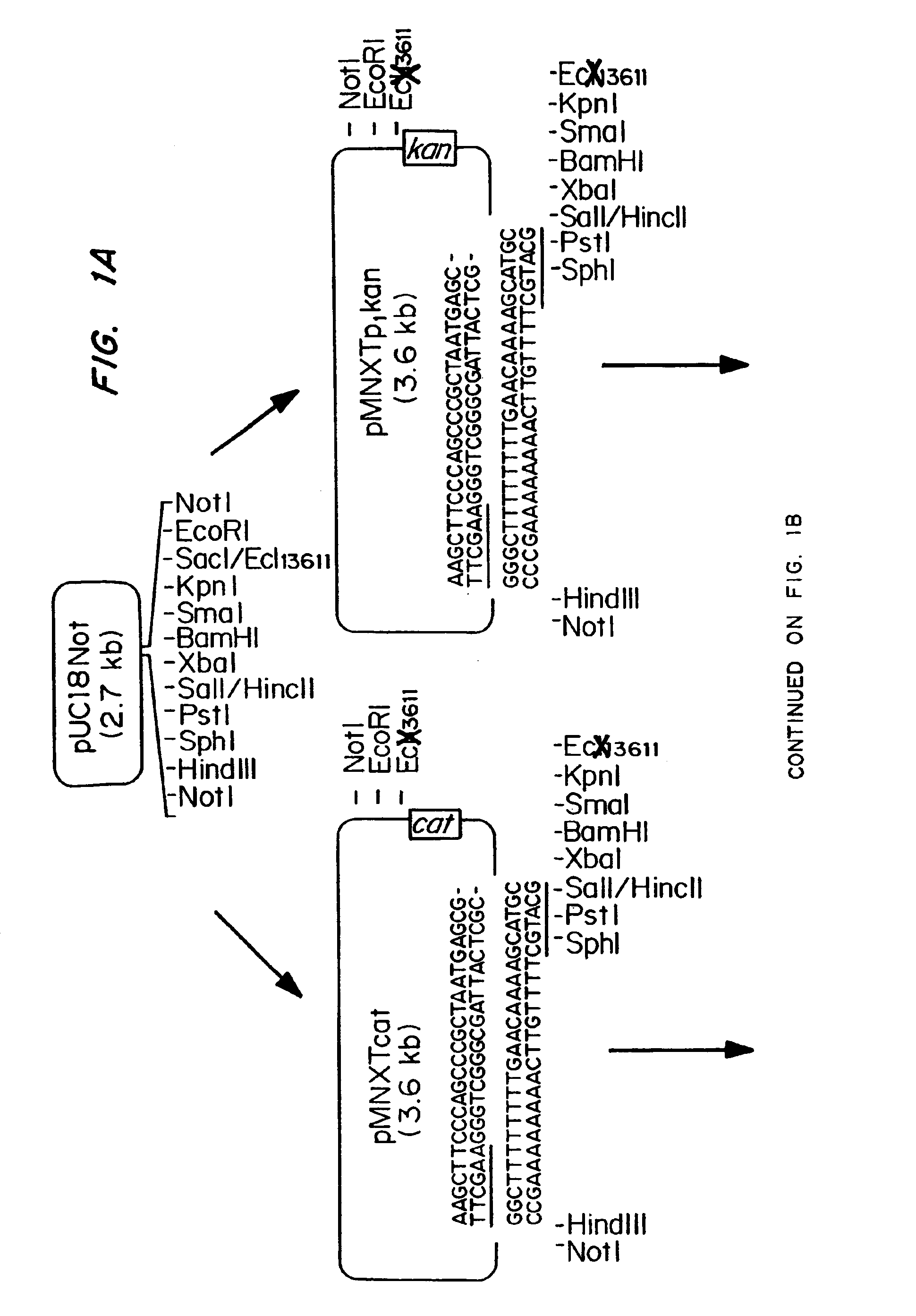
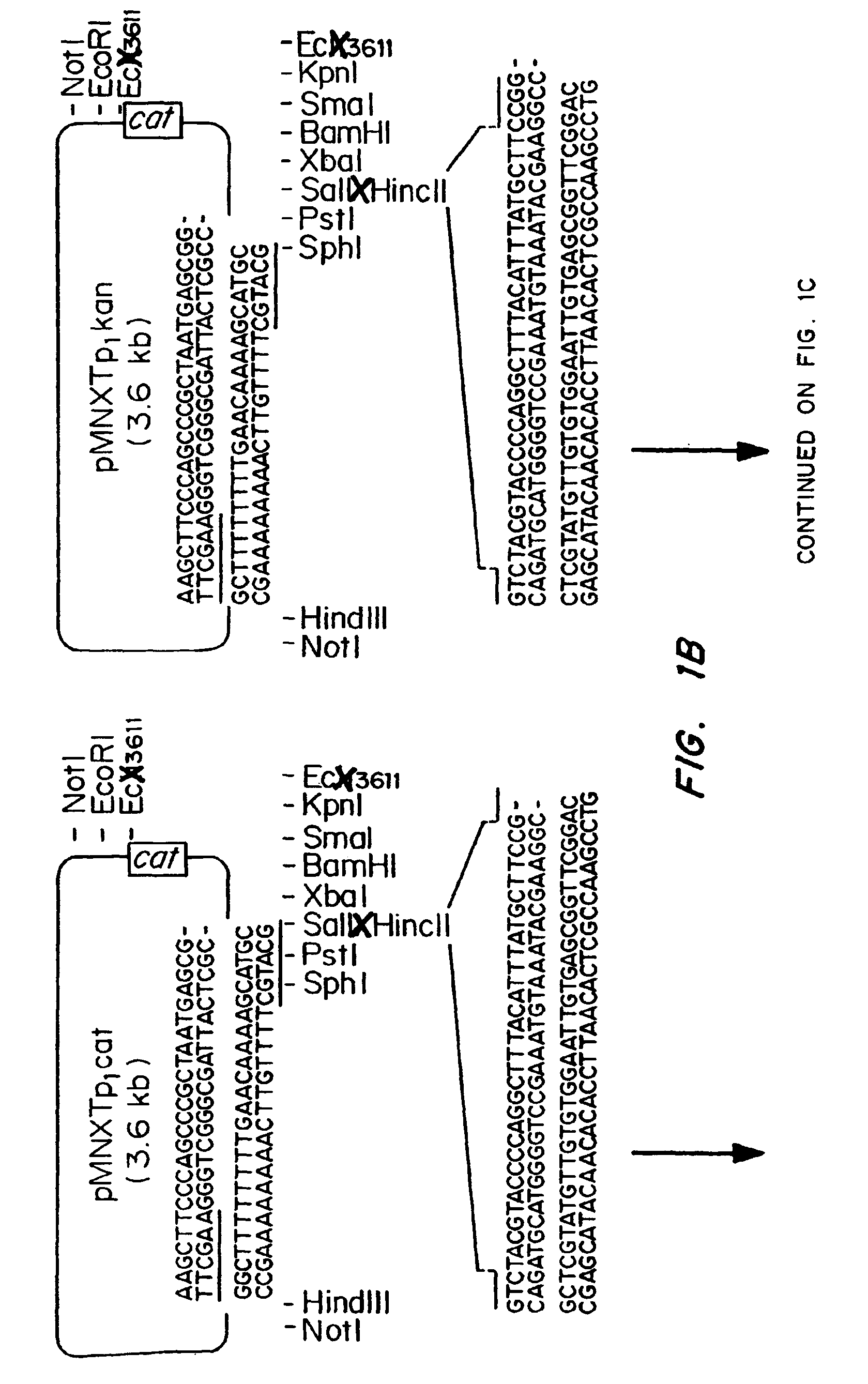
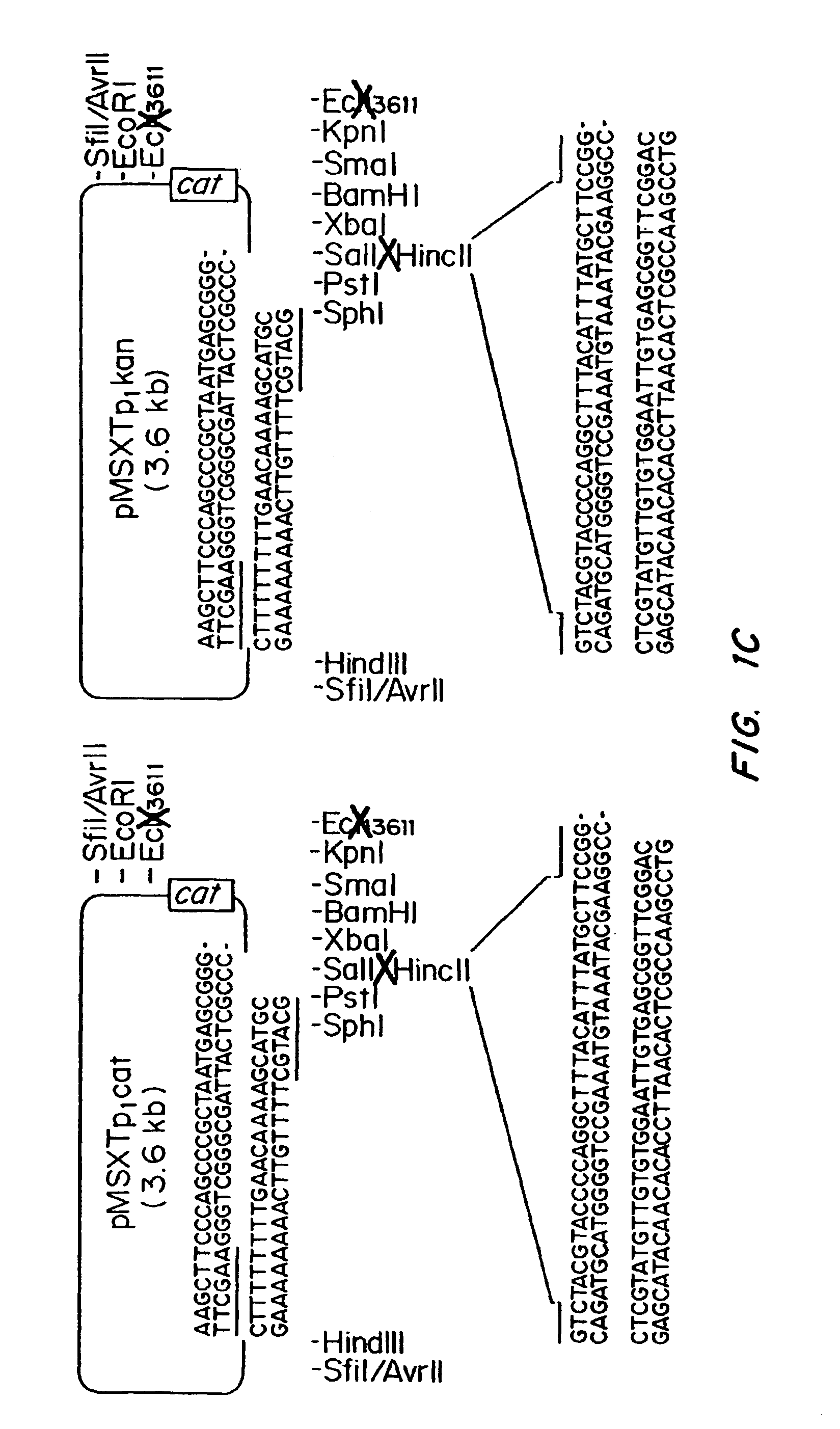
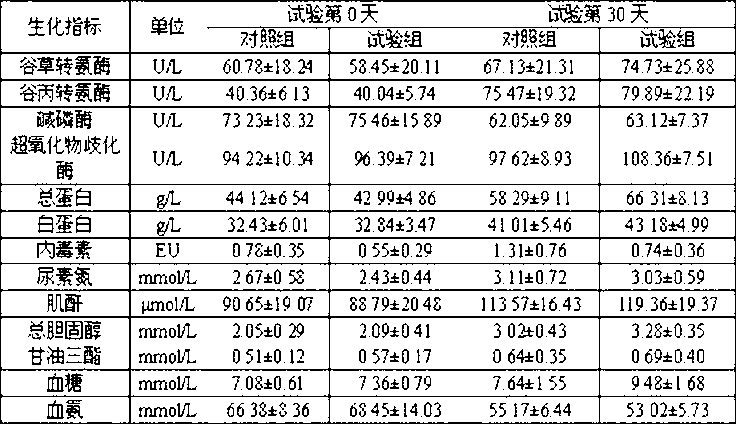
Patents
Literature
519 results about "Antimicrobial resistant organism" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
The WHO defines antimicrobial resistance as a microorganism's resistance to an antimicrobial drug that was once able to treat an infection by that microorganism. A person cannot become resistant to antibiotics. Resistance is a property of the microbe, not a person or other organism infected by a microbe.
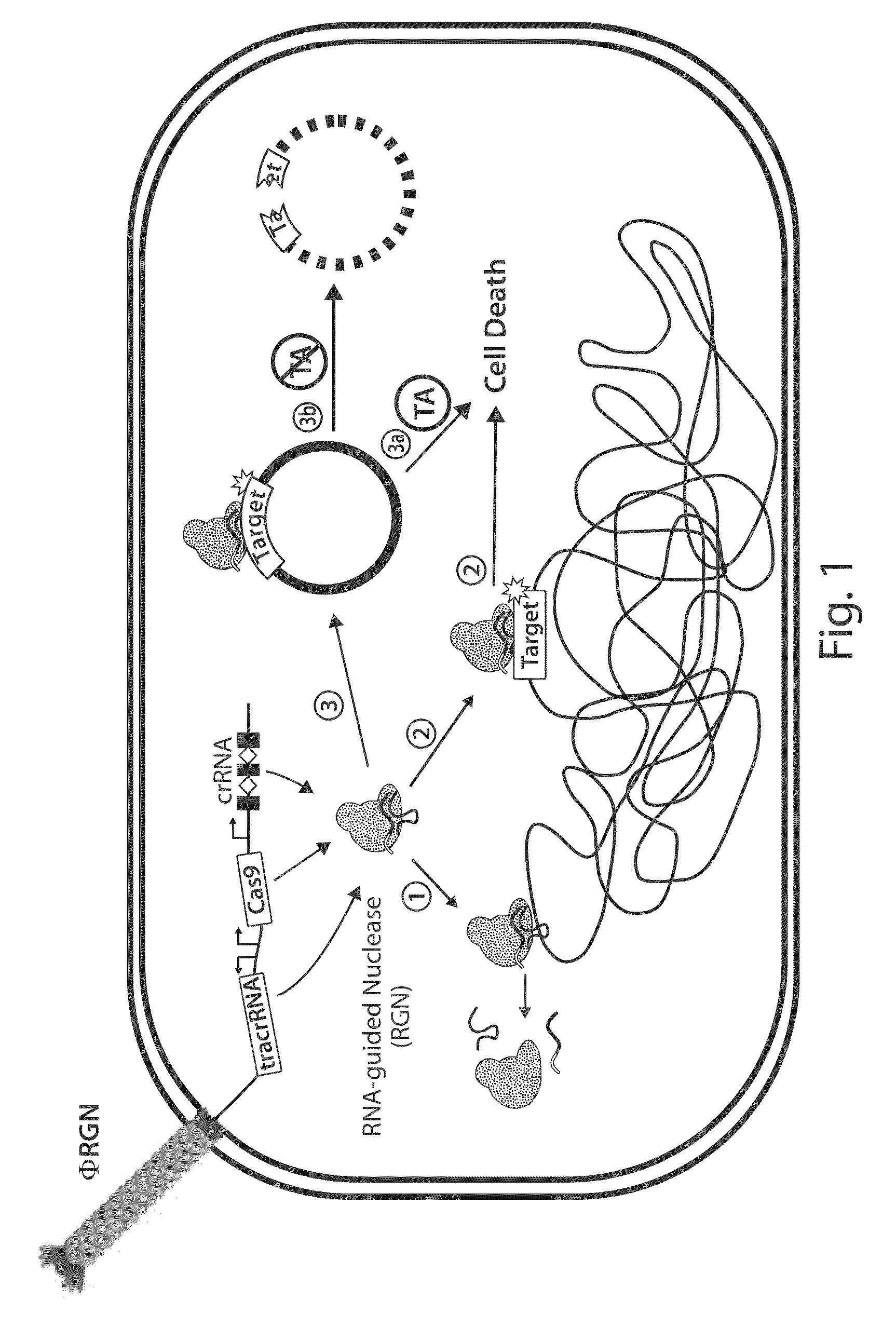
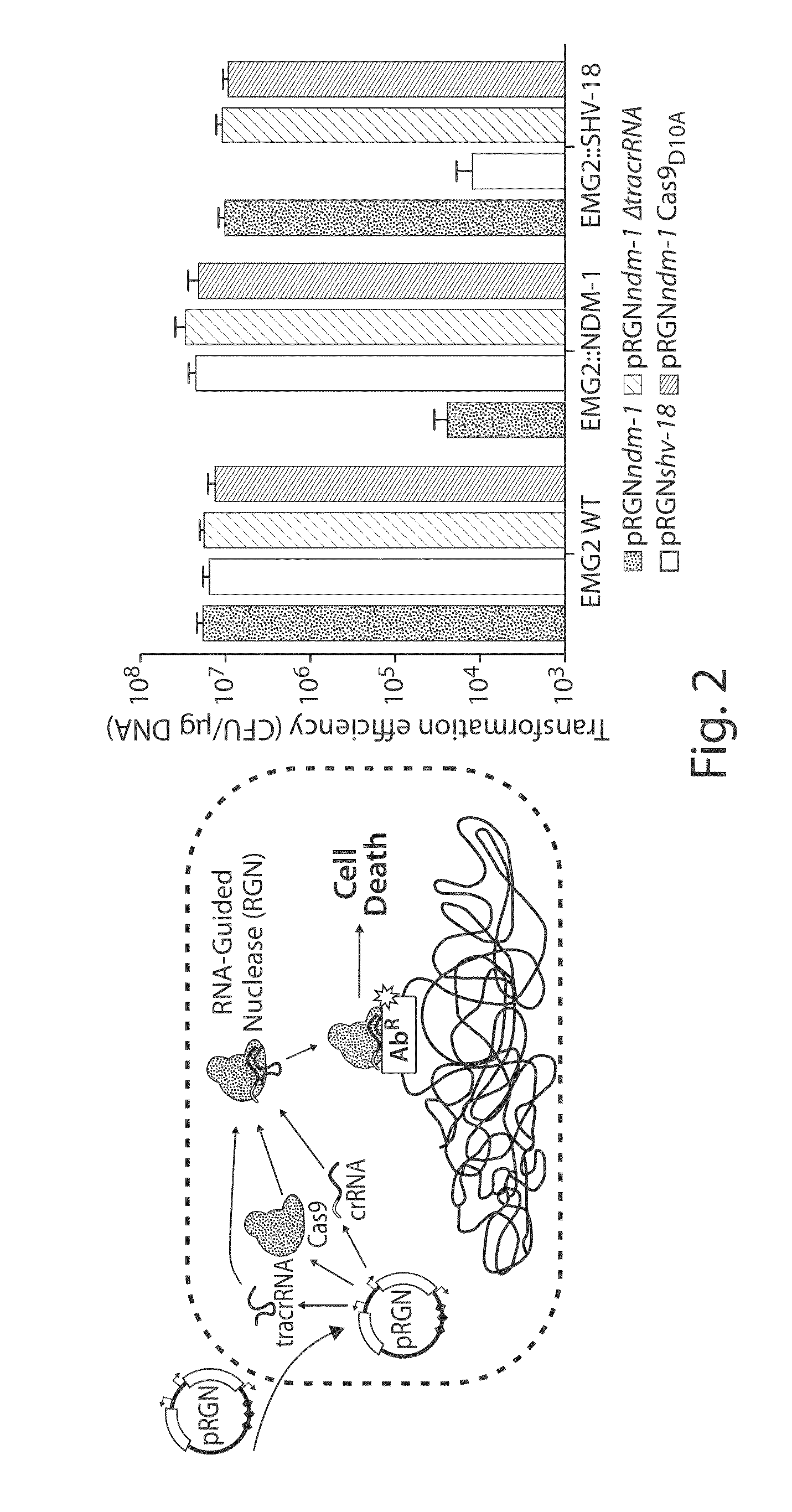
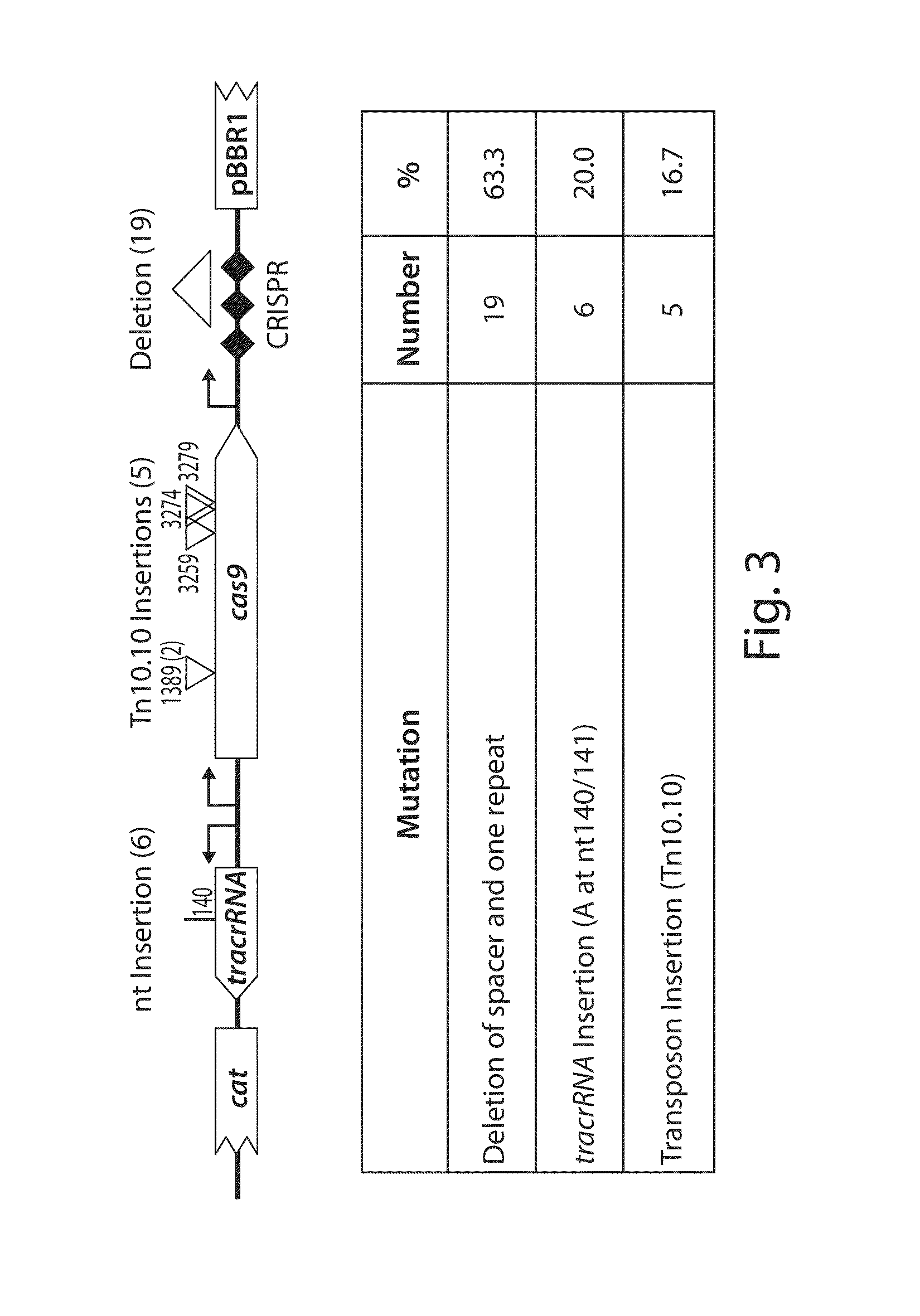
Tuning microbial populations with programmable nucleases
ActiveUS20150064138A1Efficiently and rapidly encoded into synthetic constructBroad deliveryBiocideSugar derivativesMicroorganismVirulent characteristics
Various aspects and embodiments of the invention are directed to methods and compositions for reversing antibiotic resistance or virulence in and / or destroying pathogenic microbial cells such as, for example, pathogenic bacterial cells. The methods include exposing microbial cells to a delivery vehicle with at least one nucleic acid encoding an engineered autonomously distributed circuit that contains a programmable nuclease targeted to one or multiple genes of interest.
Owner:MASSACHUSETTS INST OF TECH
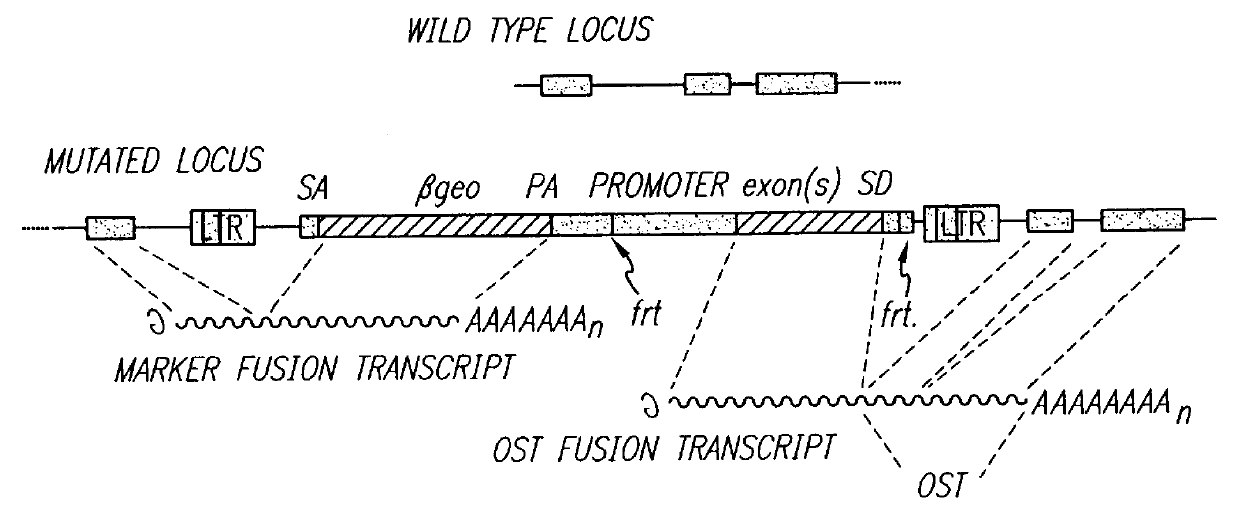
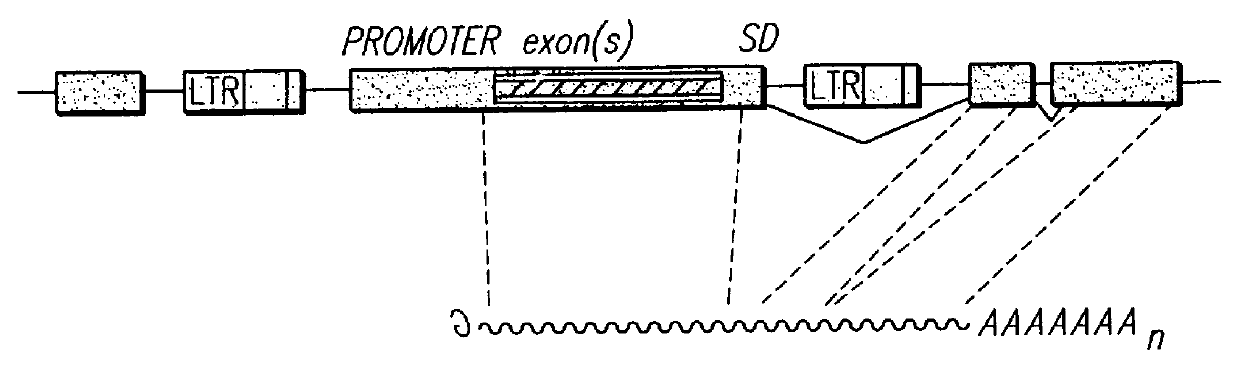
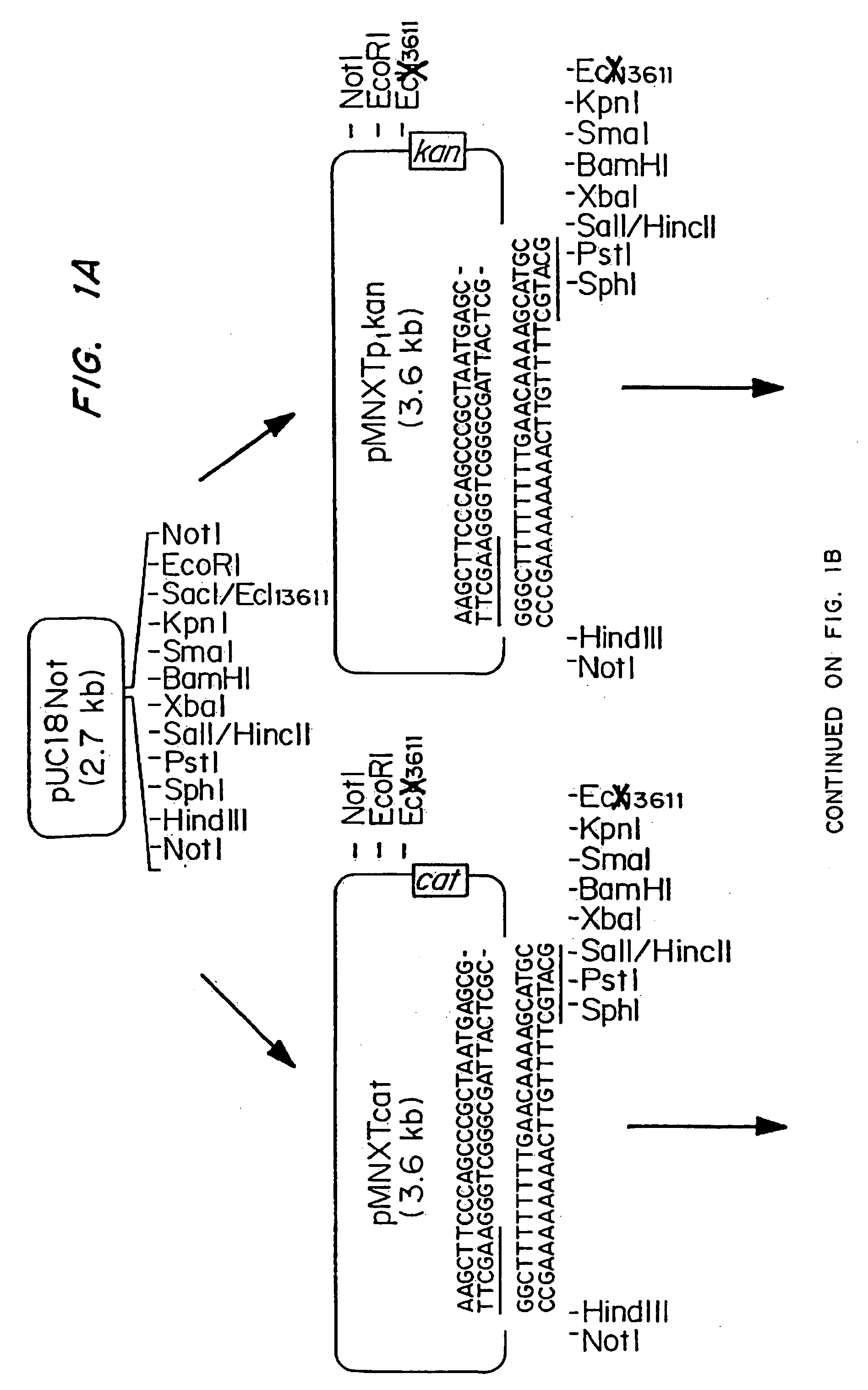
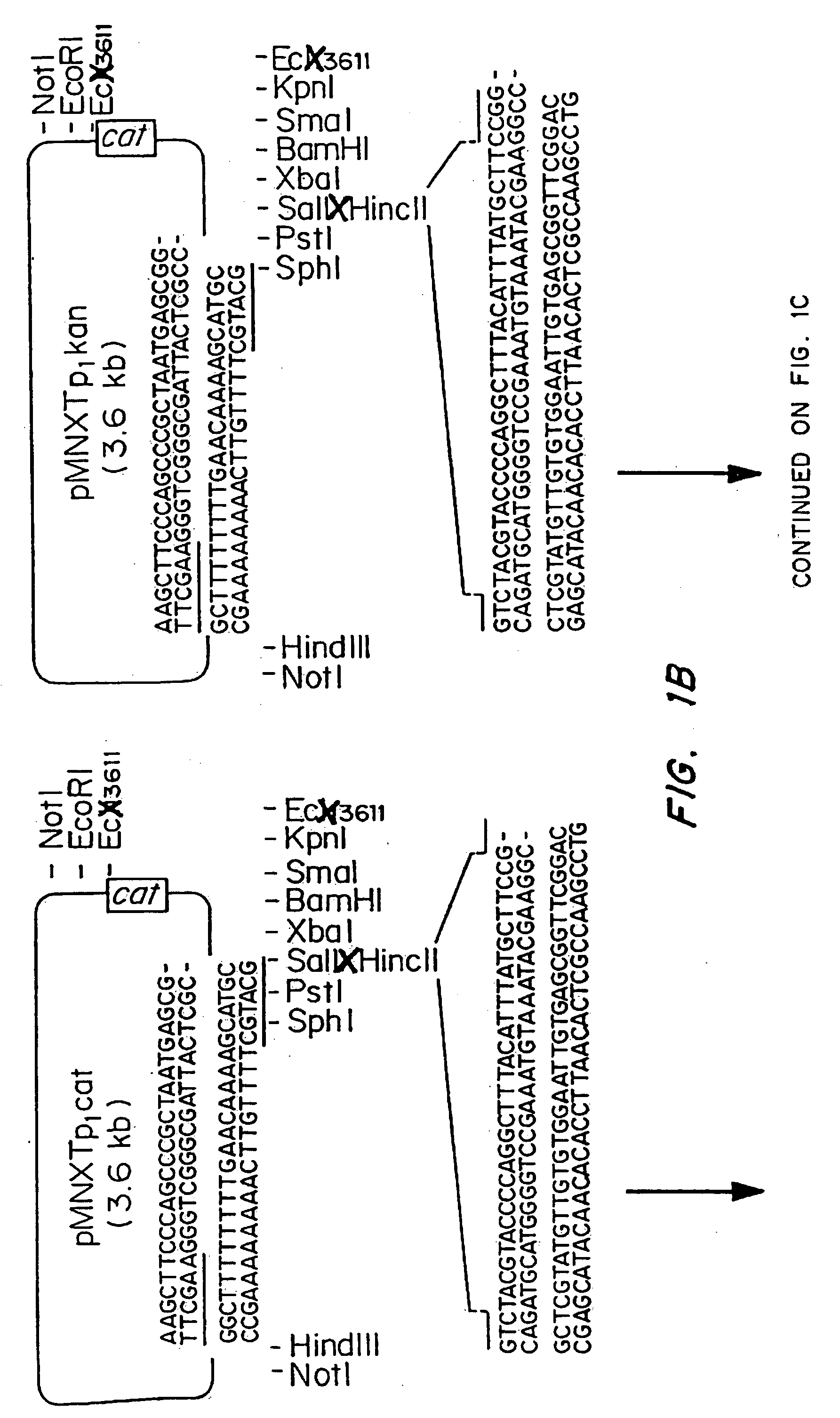
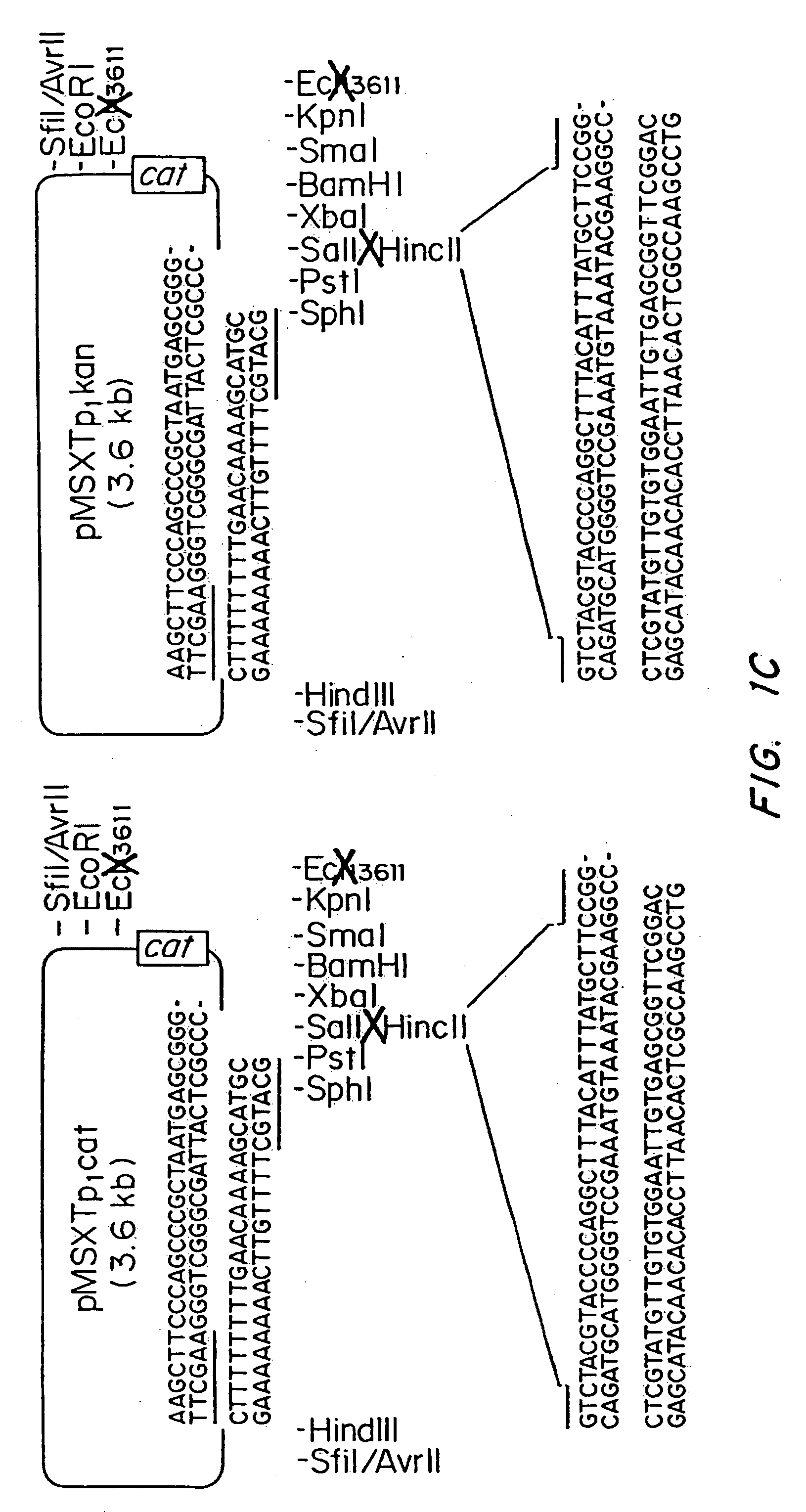
Vectors for gene trapping and gene activation
InactiveUS6080576AImprove efficiencySufficient efficiencyNucleic acid vectorMicroorganism librariesAntibiotic resistanceBiological activation
A novel 3' gene trap cassette is described that does not encode a marker conferring antibiotic resistance and can be used to efficiently trap and identify cellular genes. Vectors incorporating the presently 3' gene trap cassette find particular application in gene discovery, the production of transgenic cells and animals, and gene activation.
Owner:LEXICON PHARM INC
Micro-ecological preparation and application thereof
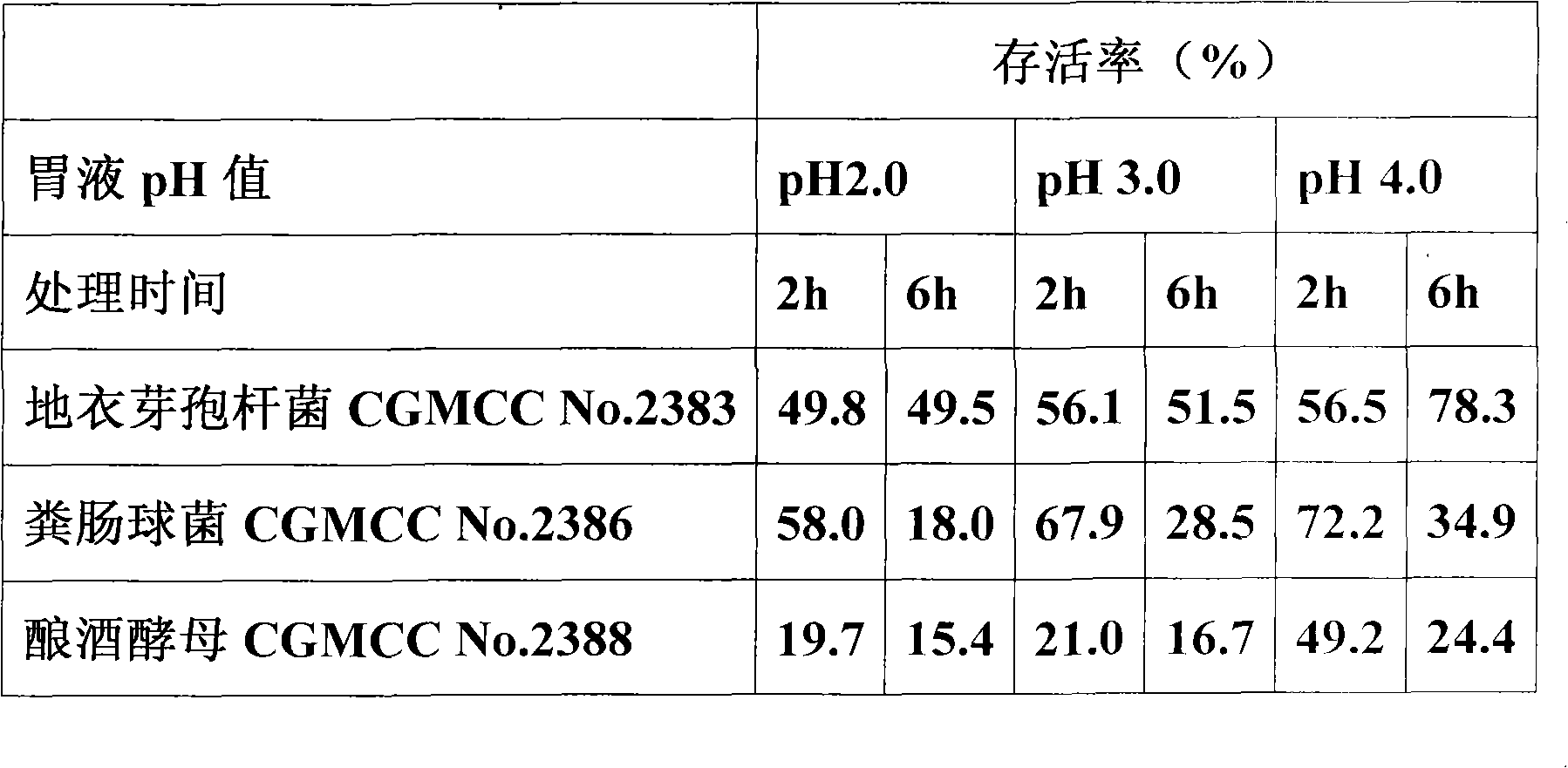
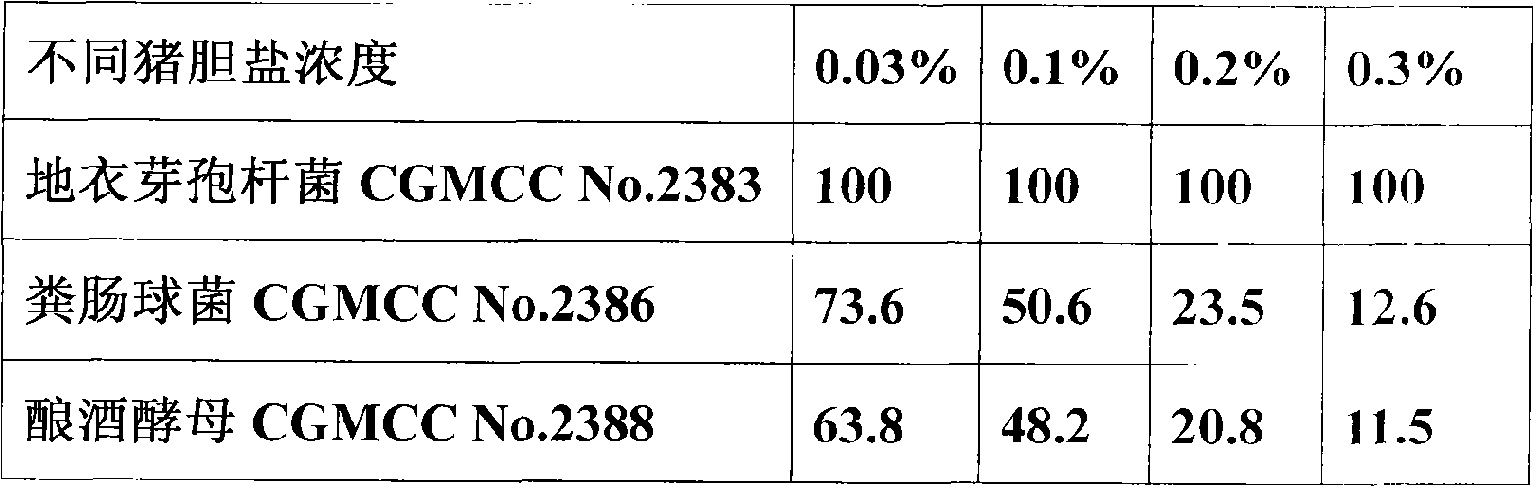
The invention relates to a micro-ecological preparation and the application thereof, in particular to a micro-ecological preparation containing a variety of probiotics and the application thereof. The micro-ecological preparation contains any three or four of the components including CGMCC No. 2383 bacillus licheniformis powder, bacillus subtilis powder, CGMCC No. 2386 enterococcus faecalis powder, lactobacillus acidophilus powder, and CGMCC No. 2388 saccharomyces cerevisiae. The micro-ecological preparation has content of live bacteria, has the adversity resistance such as gastric acid resistance, bile salt resistance, high-temperature resistance, common antibiotic resistance, and the like and the probiotic functions of producing acid and enzyme and resisting pathogenic bacteria. The variety and the proportion of the probiotics and carrier can be determined according to different kinds of animals and different animal growth phase. The micro-ecological preparation can improve the feed utilization efficiency, increase the yield of meat, eggs and milk, promote the growth of the animal, improve the immunity and the disease resistance of the animal, replace the antibiotic and improve the quality of the animal product.
Owner:BEIJING DABEINONG TECH GRP CO LTD +1
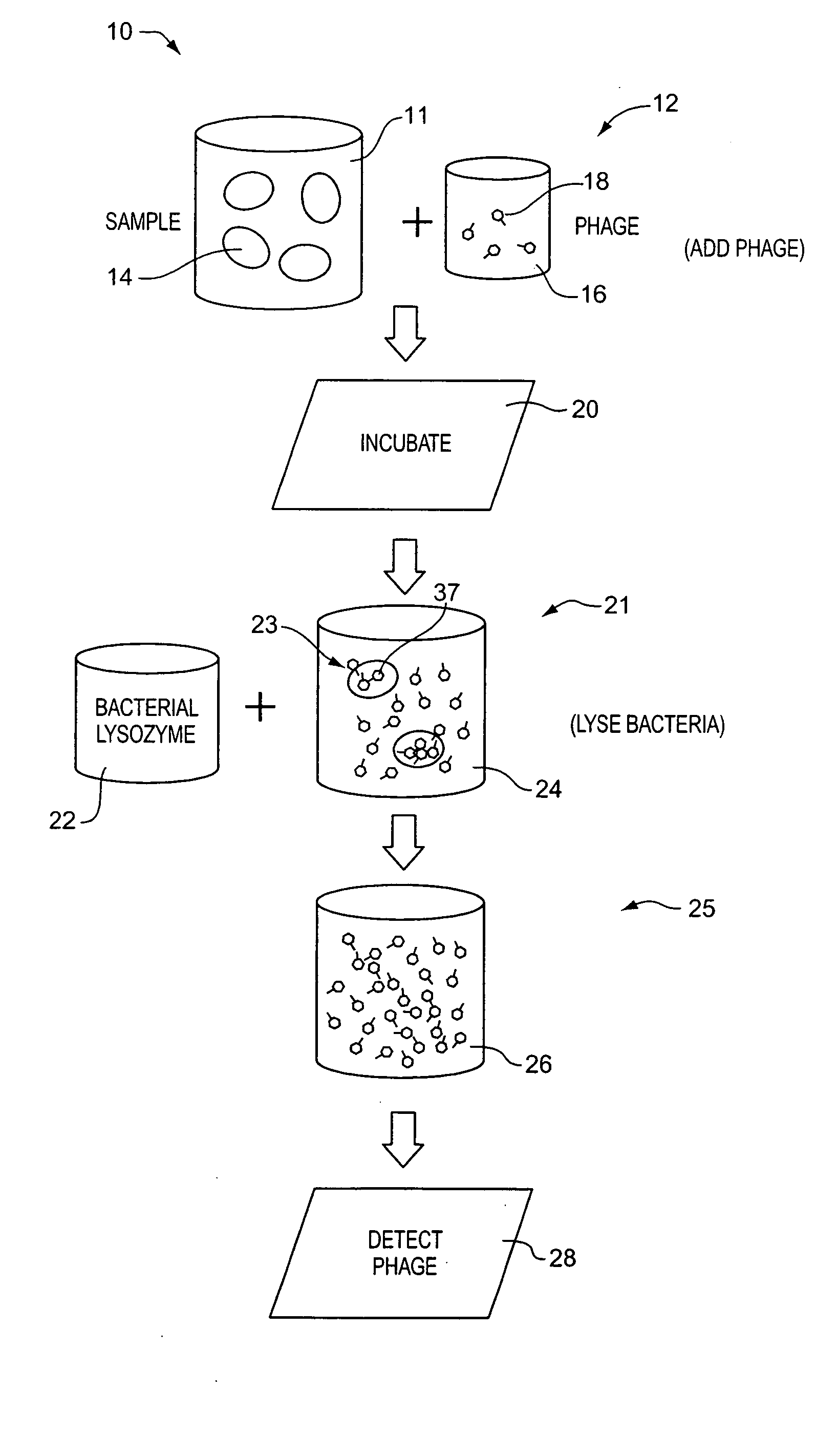
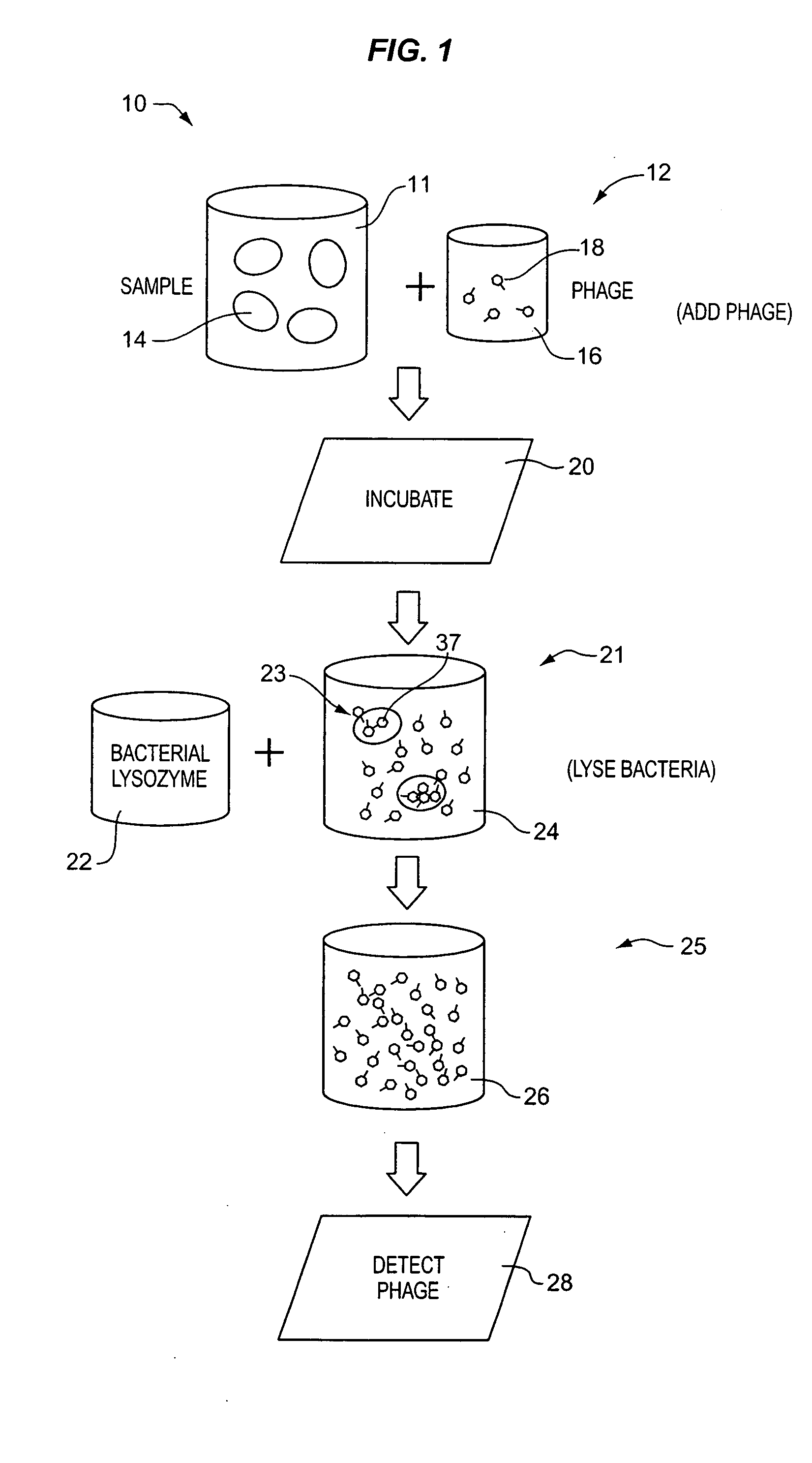
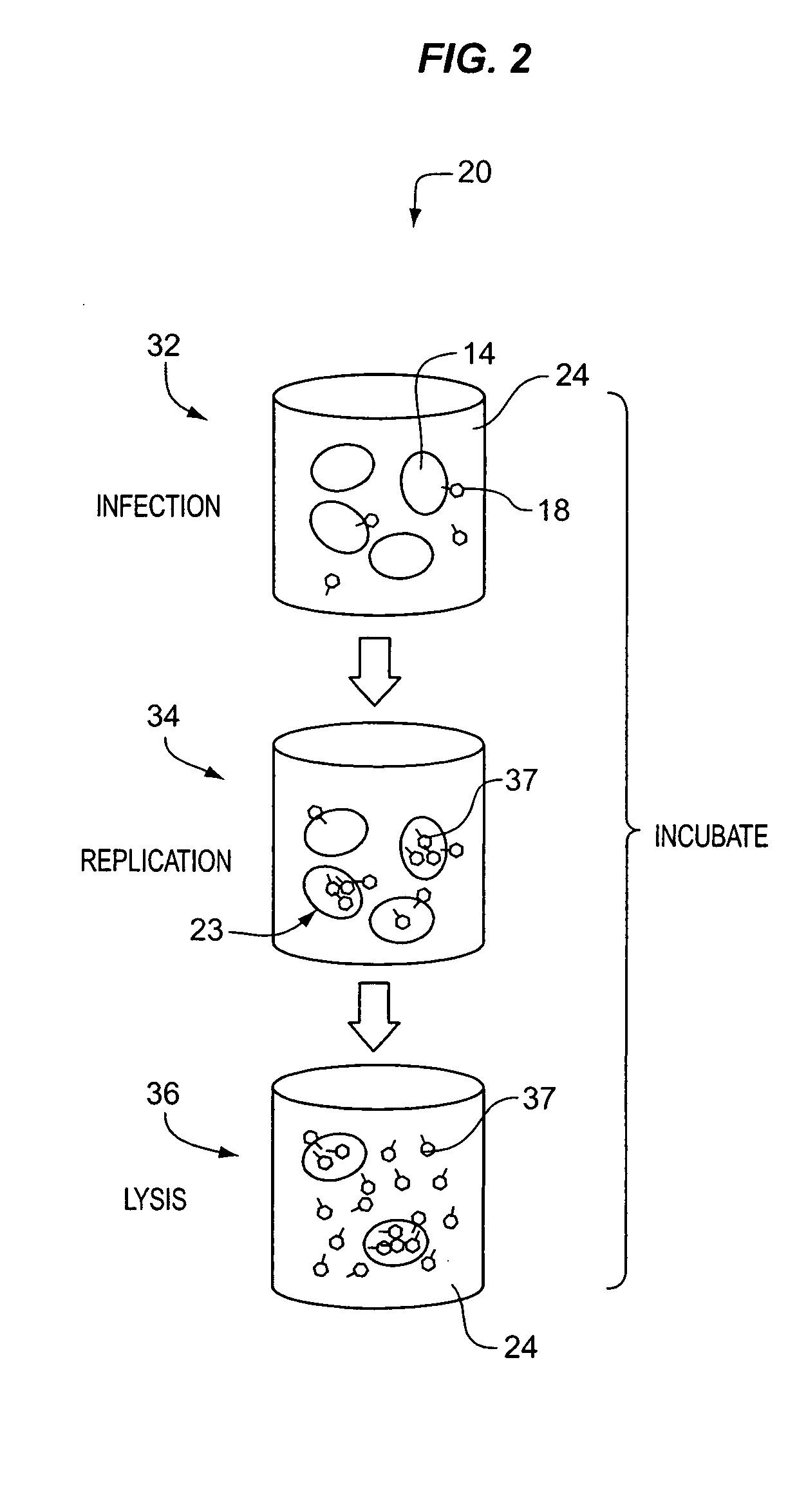
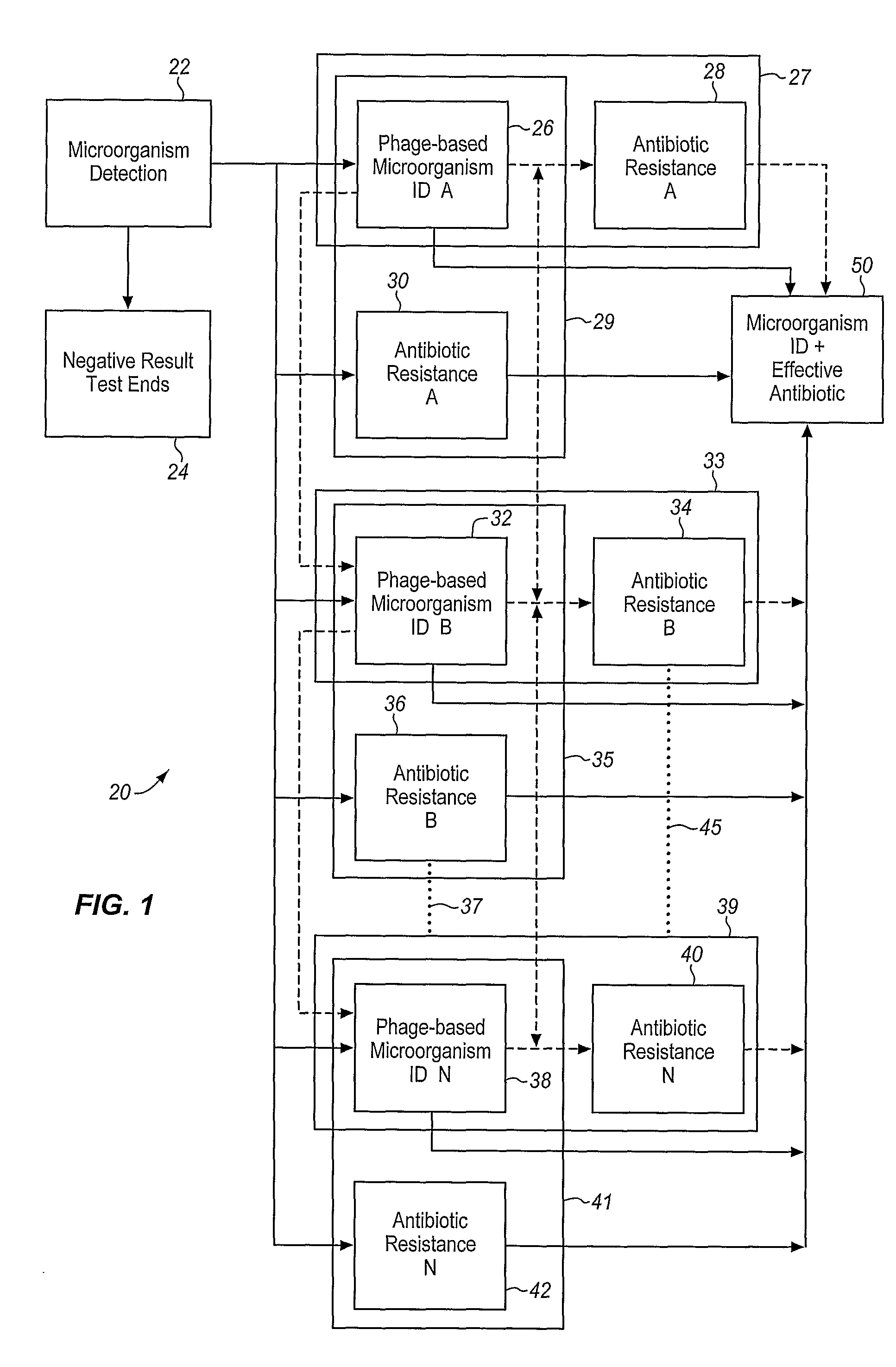
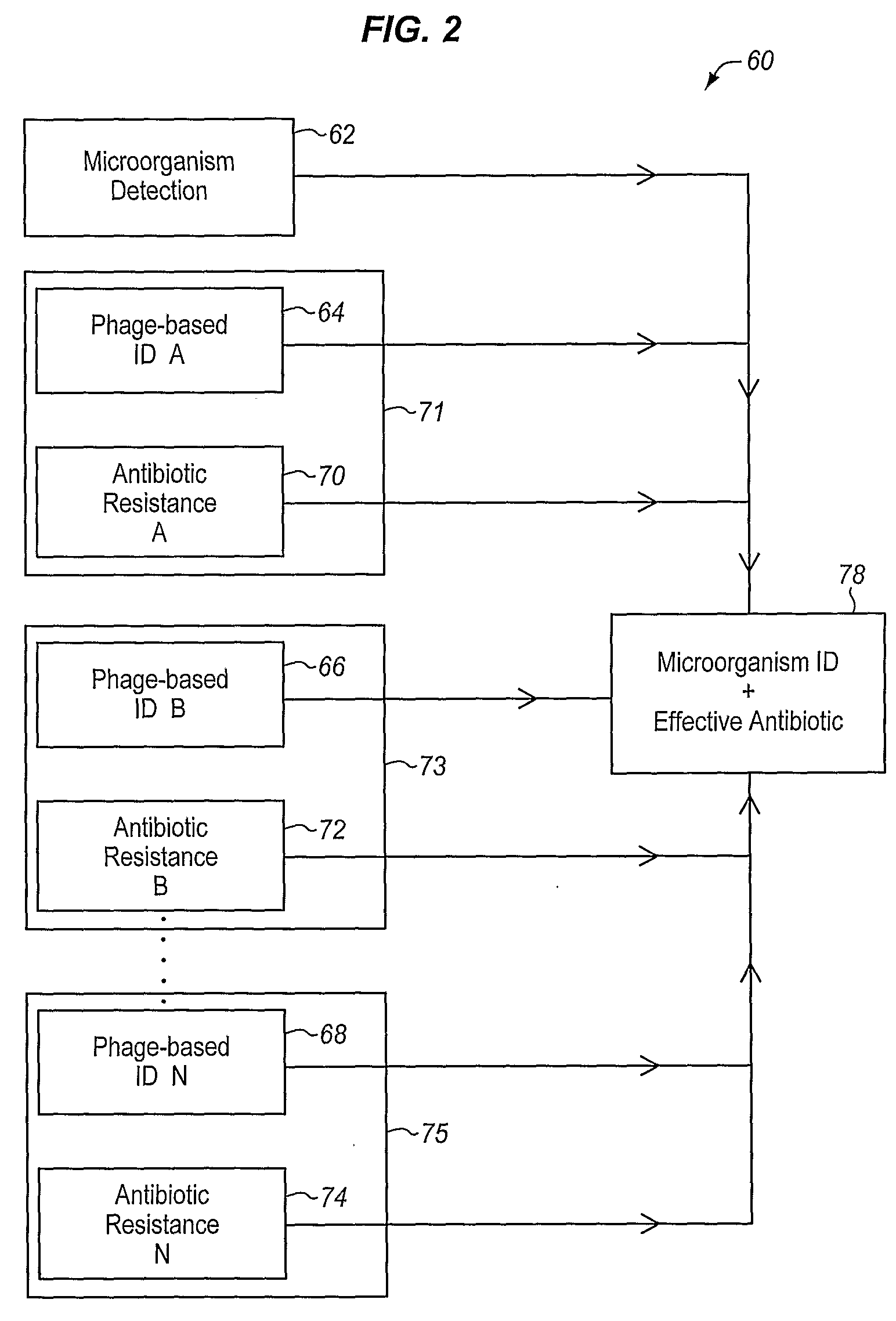
Apparatus and method for detecting microscopic living organisms using bacteriophage
InactiveUS20050003346A1Wide concentration rangeFast resultsMicrobiological testing/measurementMaterial analysisBacteroidesAntibiotic resistance
A method for detecting one or more target bacteria in a raw sample where: 1) bacteriophage(s) specific to each target bacterium are added to the raw sample, 2) the test sample is incubated, and 3) the test sample is tested for the presence of each phage in sufficient numbers to indicate the presence of the associated target bacteria in the raw sample. In one embodiment, each phage is initially added to the raw sample in concentrations below the detection limit of the final phage detection process. In another embodiment, the parent phages are tagged in such a way that they can be separated from the progeny phage prior to the detection process. Preferred phage detection processes are immunoassay methods utilizing antibodies that bind specifically to each phage. Antibodies can be used that bind to the protein capsid of the phage. Alternatively, the phage can by dissociated after the incubation process and the sample tested for the presence of individual capsid proteins or phage nucleic acids. The invention can be used to test target bacteria for antibiotic resistance.
Owner:MICROPHAGE +1
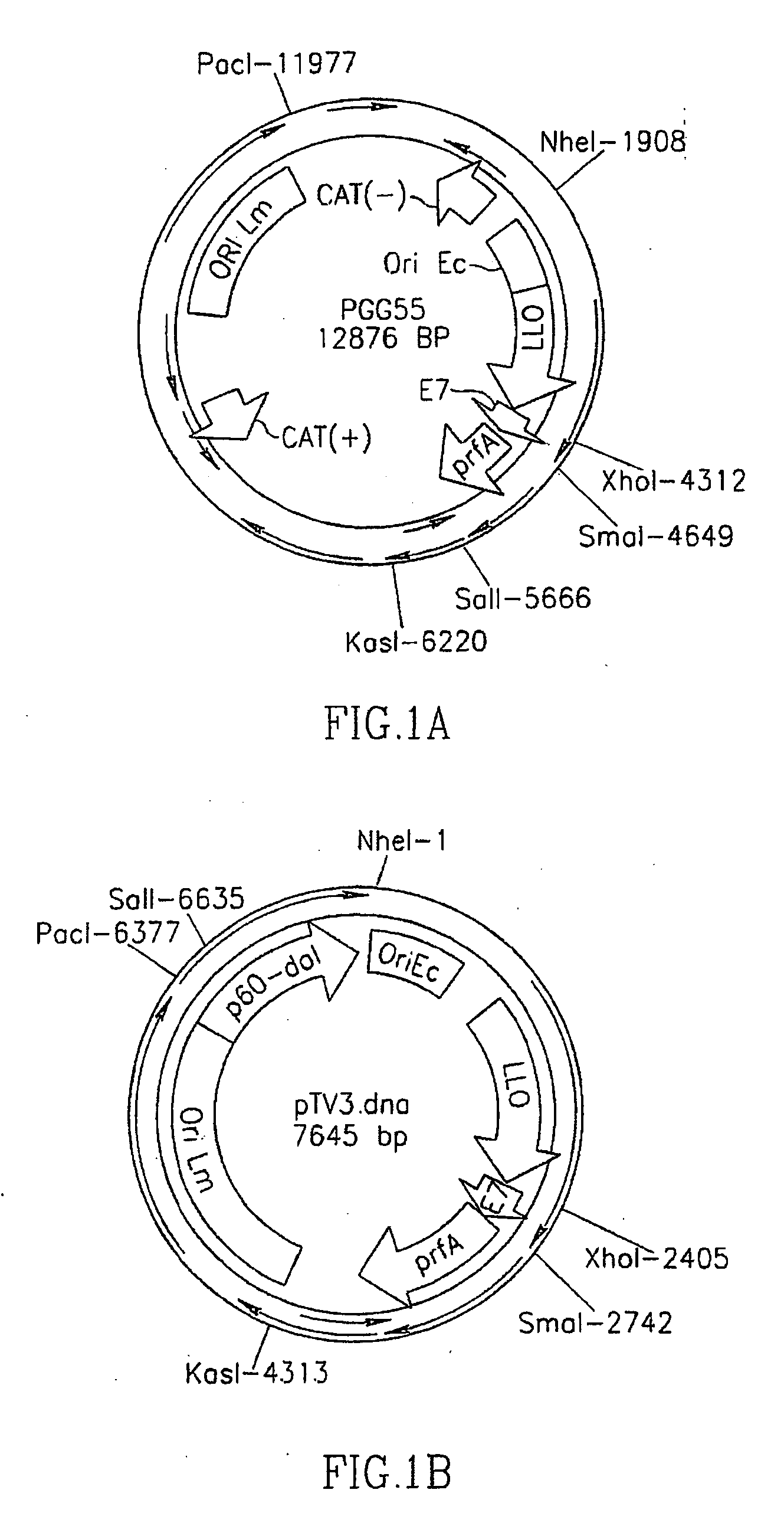
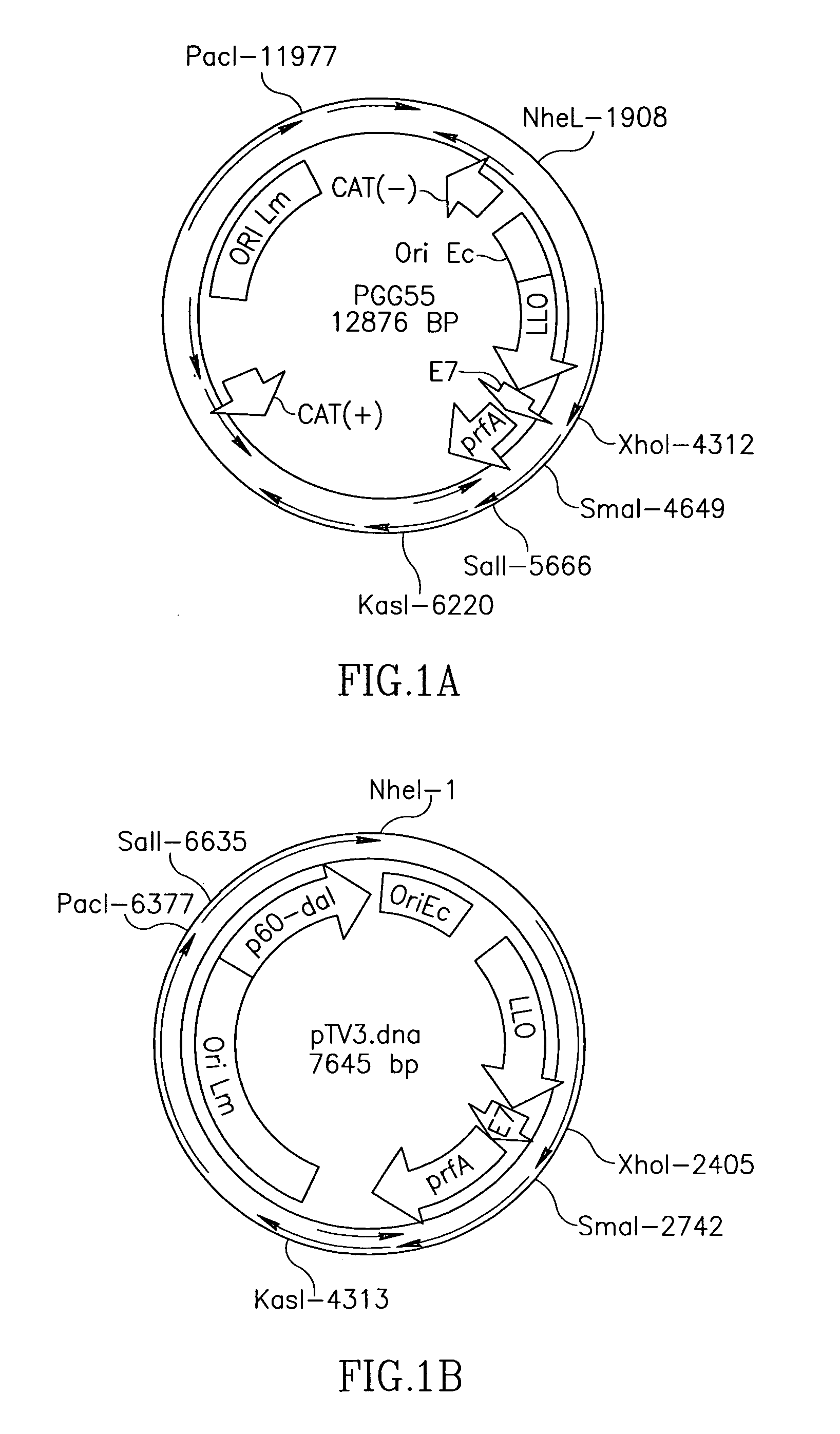
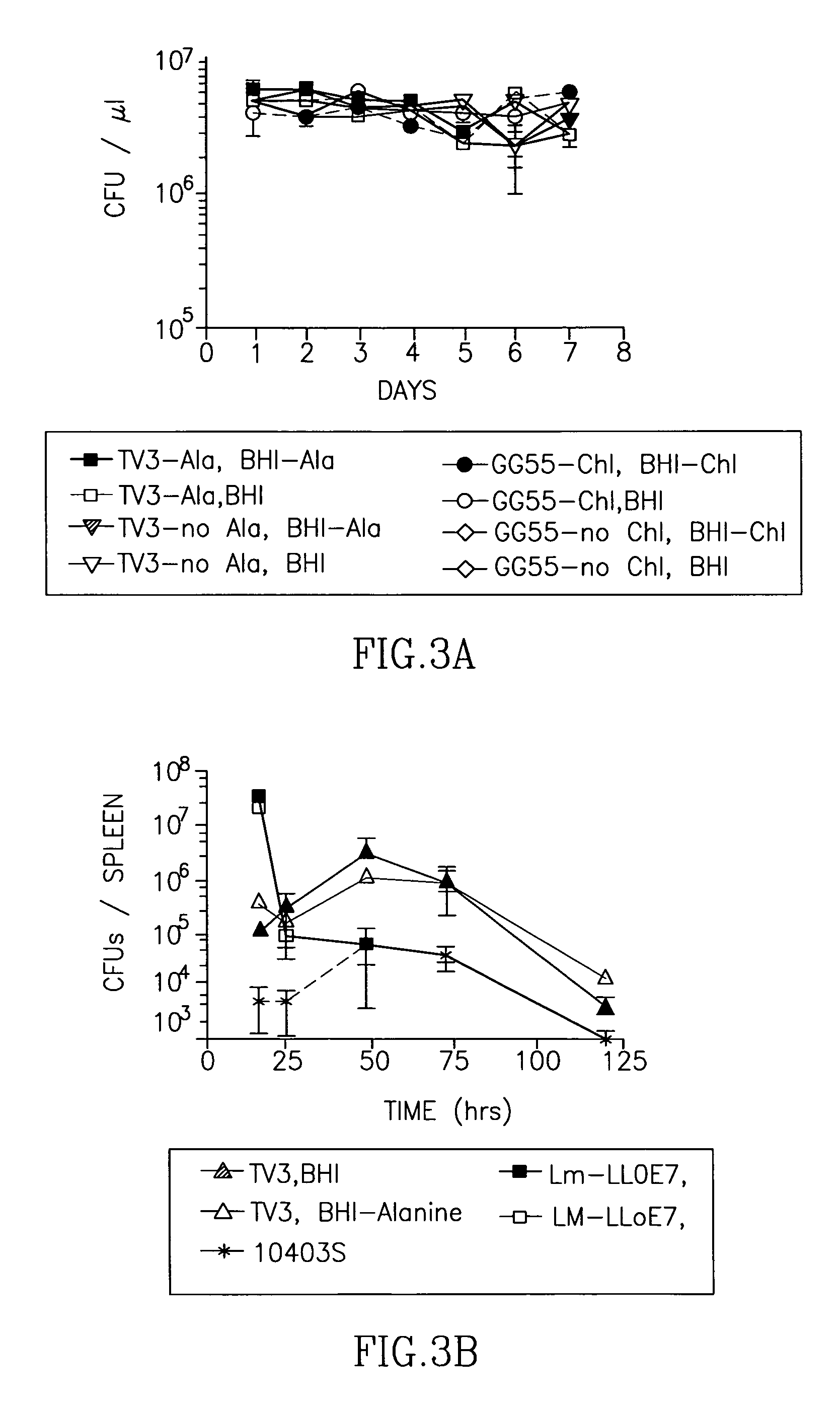
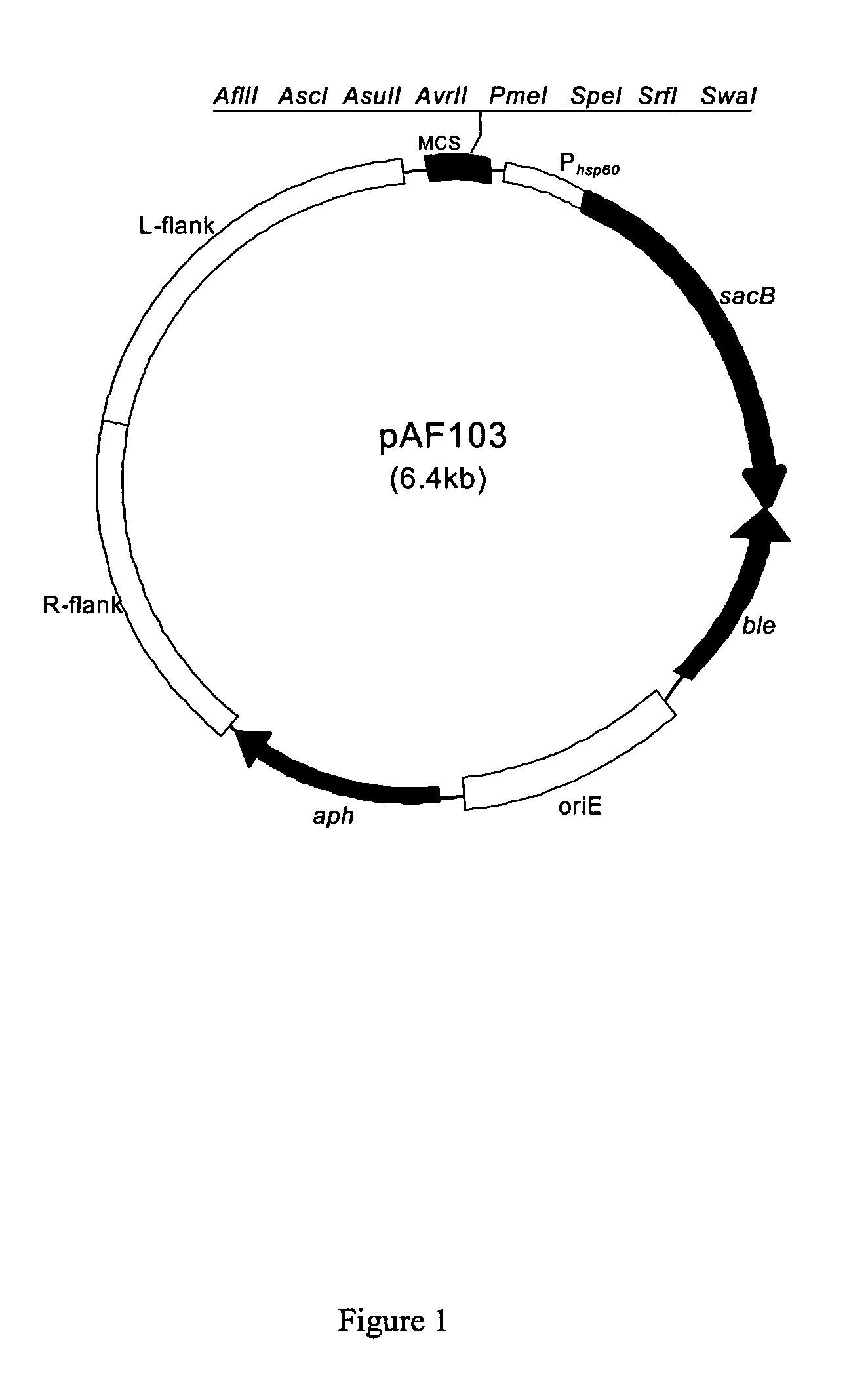
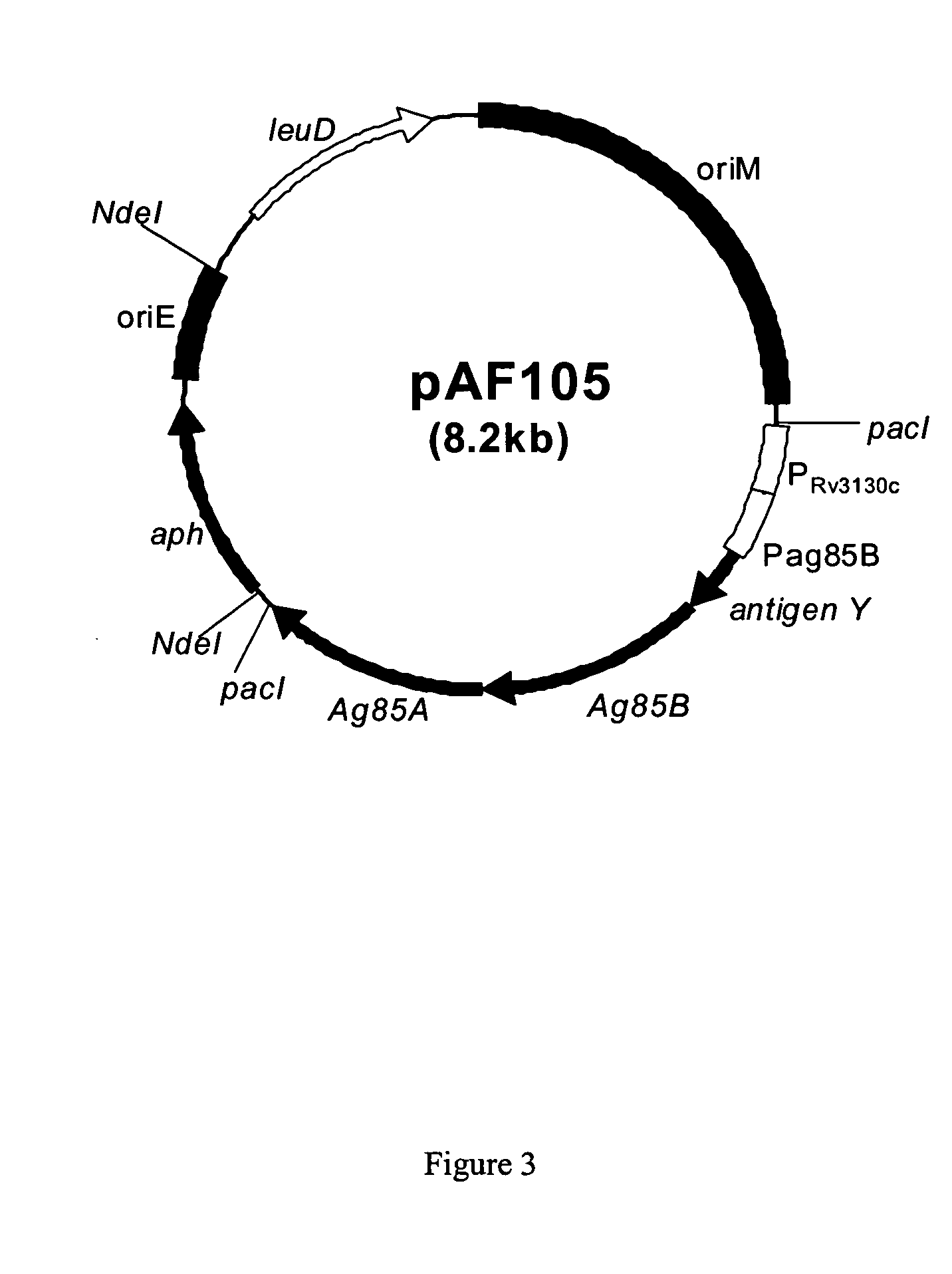
Methods for constructing antibiotic resistance free vaccines
The present invention provides Listeria vaccine strains that express a heterologous antigen and a metabolic enzyme, and methods of generating same.
Owner:THE TRUSTEES OF THE UNIV OF PENNSYLVANIA
Methods and Kits for Detection of Antibiotic Resistance
InactiveUS20120196309A1Promotes bacterial resistanceMicrobiological testing/measurementMass spectrometric analysisResistant bacteriaMass Spectrometry-Mass Spectrometry
The present invention relates to a method of detecting antibiotic resistant bacteria in a sample. The method includes the steps of analyzing a sample derived from bacteria via mass spectrometry to produce a data set, and determining from the data set the presence or absence of a covalently modified antibiotic compound in the sample, wherein the presence of a covalently modified antibiotic compound in the sample is indicative that the bacteria are resistant to the antibiotic. The present invention also relates to a kit for determining the presence or absence of antibiotic resistant bacteria in a sample. The kit includes reagents for preparing and performing the assay, and instructions for the set-up, performance, monitoring, and interpretation of the assay.
Owner:YALE UNIV
Transgenic microbial polyhydroxyalkanoate producers
InactiveUS6913911B2Simple production processInduce expressionSugar derivativesBacteriaBiotechnologyTransposon mutagenesis
Transgenic microbial strains are provided which contain the genes required for PHA formation integrated on the chromosome. The strains are advantageous in PHA production processes, because (1) no plasmids need to be maintained, generally obviating the required use of antibiotics or other stabilizing pressures, and (2) no plasmid loss occurs, thereby stabilizing the number of gene copies per cell throughout the fermentation process, resulting in homogeneous PHA product formation throughout the production process. Genes are integrated using standard techniques, preferably transposon mutagenesis. In a preferred embodiment wherein mutiple genes are incorporated, these are incorporated as an operon. Sequences are used to stabilize mRNA, to induce expression as a function of culture conditions (such as phosphate concentration), temperature, and stress, and to aid in selection, through the incorporation of selection markers such as markers conferring antibiotic resistance.
Owner:CJ CHEILJEDANG CORP
Pig feed additive capable of replacing antibiotics, and preparation method and application thereof
ActiveCN103005210AAcid resistantGuaranteed Oxygen DensityAnimal feeding stuffPhosphoric acidAntibiotic Y
The invention relates to a pig feed additive capable of replacing antibiotics, and a preparation method and an application thereof. Components and weight part ratio thereof in a formula are as follows: 1-5 parts of medicated leaven, 1-5 parts of angelica sinensis, 1-5 parts of radix astragali, 1-5 parts of hawthorn, 1-5 parts of radix codonopsis, 10-20 parts of citric acid, 10-20 parts of lactic acid, 20-30 parts of coated phosphoric acid, 5-10 parts of a micro-ecological preparation, 5-10 parts of yeast cell wall polysaccharide, 1-5 parts of a composite plant essential oil and 5-10 parts of a carrier. The pig feed additive carries out combination collocation of traditional Chinese herbs and functional polysaccharide, the micro-ecological preparation, acidifying agents, plant essential oil and the like, so that synergic effect of the traditional Chinese herbs and other components are maximized; the pig feed additive can replace the antibiotics completely; problems of drug resistance, drug residues, overcommitment and the like of the antibiotics which trouble animal husbandry for a long time is reduced; and at the same time, the pig feed additive plays effects of improving culture environment, reducing feed safety hidden dangers, increasing culture benefits and the like.
Owner:LUOGANG BRANCH GUANGDONG WANGDA GRP
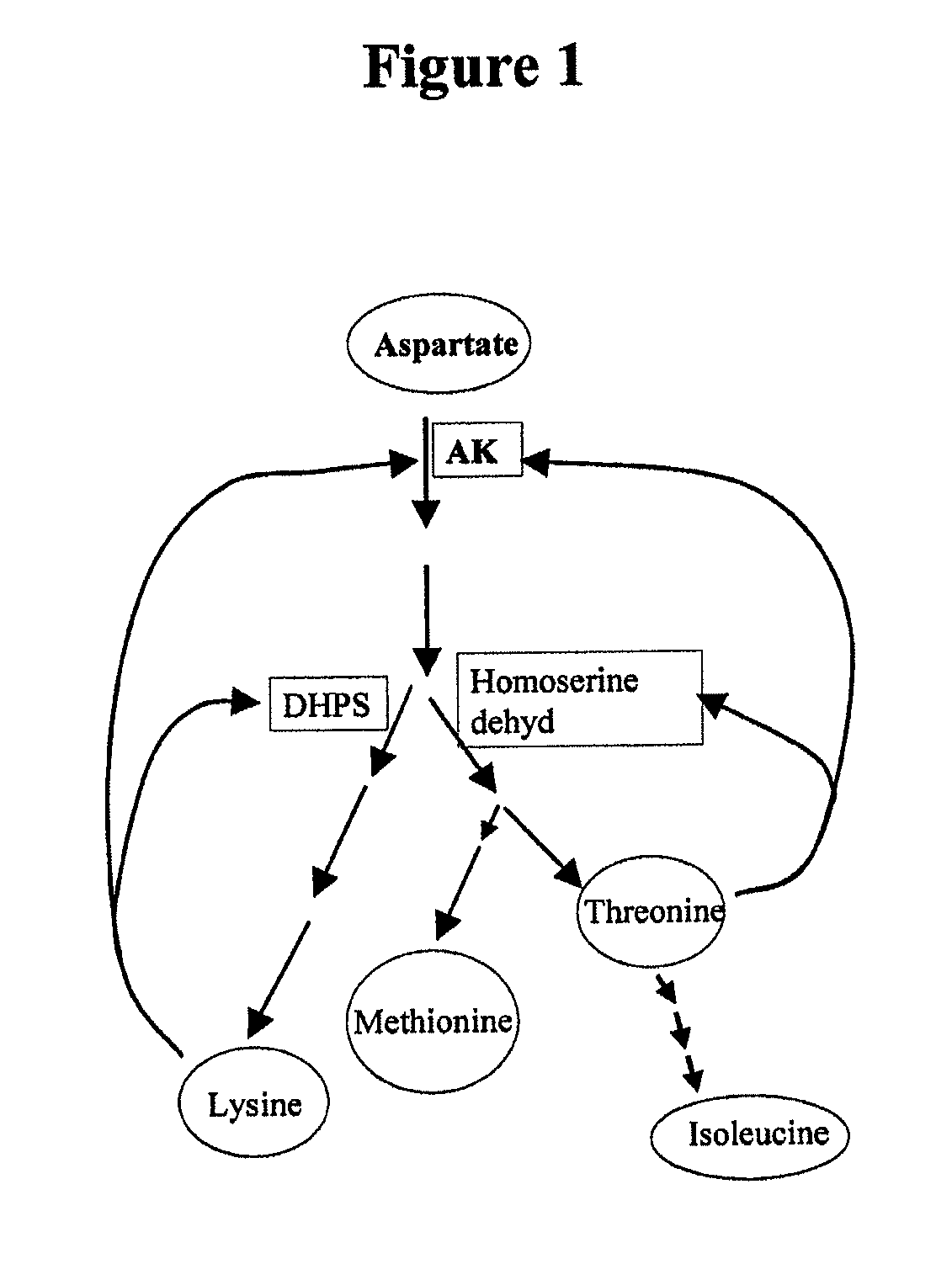
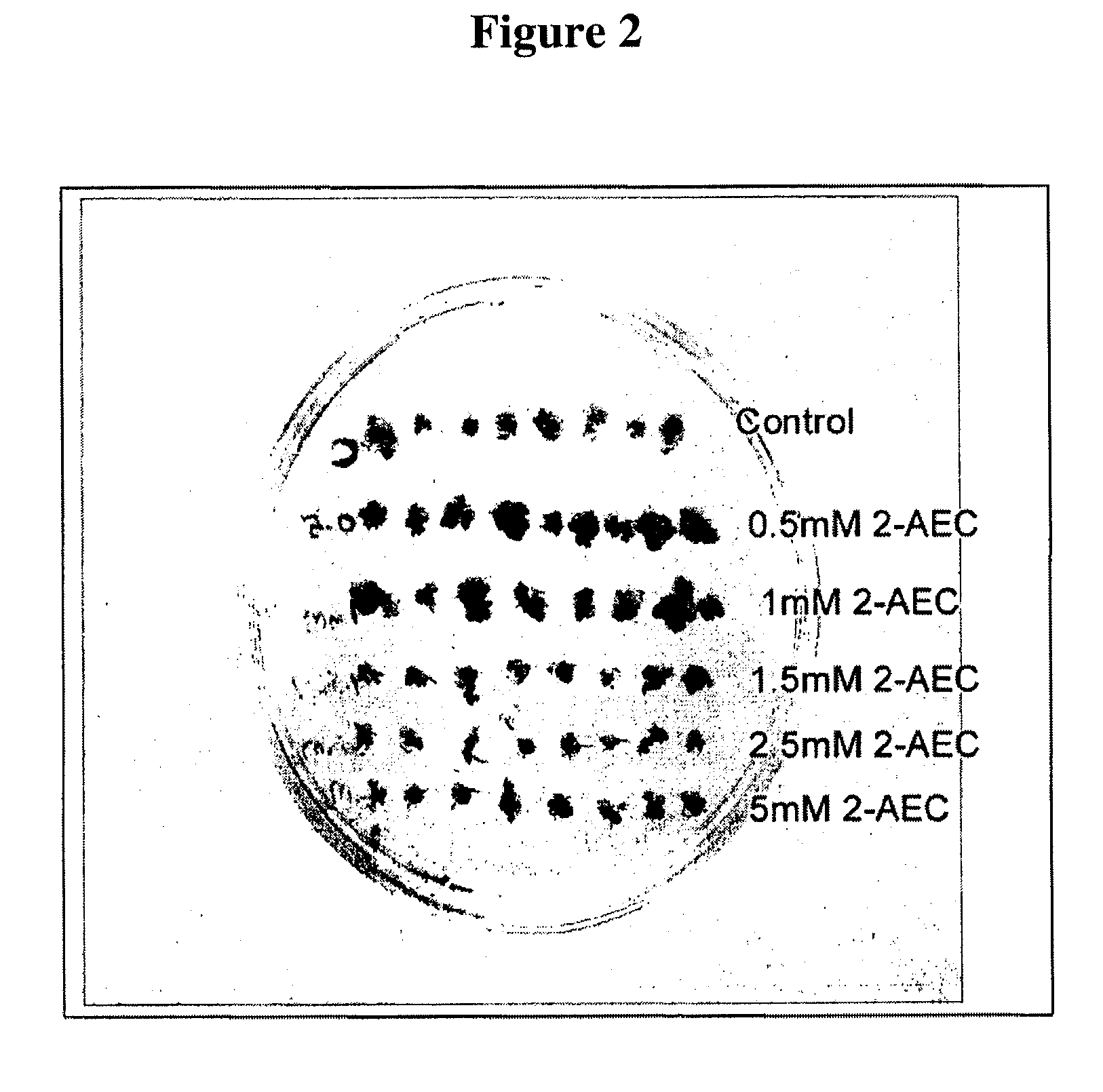
Soybean selection system based on AEC-resistance
InactiveUS7525013B2Improve featuresOther foreign material introduction processesFermentationHeterologousAntibiotic resistance
A method for generating a transgenic soybean plant comprising in its genome a heterologous nucleic acid sequence of interest, comprises introducing into a soybean somatic embryo a polynucleotide encoding a functional dihydrodipicolinate synthase (DHPS) polypeptide, and a polynucleotide encoding a heterologous polypeptide of interest, operably linked to expression control sequences, wherein DHPS expressed from the introduced DHPS-encoding polynucleotide is effective to render the embryo resistant to S-2-aminoethylcysteine (2-AEC), and contacting the embryo with 2-AEC, under conditions effective for an embryo which expresses the DHPS to grow selectably and mature into a soybean plant that expresses a desired trait, and preferably includes no antibiotic resistance marker sequence.
Owner:UNITED SOYBEAN BOARD
Lead and cadmium tolerant bacteria and method for plants renovation of soil pollution by heavy metal
InactiveCN101153272AHas nitrogen fixationWith characteristicsBacteriaContaminated soil reclamationPhosphateAntibiotic resistance
The invention relates to a heavy metal lead and cadmium antibiotic-resistant bacterium and a method for strengthening lead and cadmium heavy metals in the vegetable extraction soil, and belongs to the agricultural and environmental pollution treatment technical field. The J62 culture stain is appraised as the Burkholderia sp, has antibiotic resistance to a plurality of heavy metals, in particular to lead and cadmium, respectively reaching Pb<2+>1000mg / L and Cd<2+>2000mg / L. The invention has the characteristics of nitrogen fixation and phosphate solubilizing, and can promote the growth of the plant and improve the stress resistance of the plant. The effective living microbial numbers of the liquid preparation are above a billion per millimeter, and the effective living microbial numbers of the solid preparation are 200 millions per gram. When the seeds of the plant are seminated in humid soil containing heavy metals, 10 to 20mL of the bacteria liquid of 10<8> bacteria J62 / mLs is inoculated in each grams of soil once or in two times. The invention promotes the growth of the plant, strengthens the enrichment of heavy metal lead and cadmium of the plant, and improves the extraction and recovery efficiency of the plant.
Owner:NANJING AGRICULTURAL UNIVERSITY
Compositions and methods to reduce mutagenesis
InactiveUS20060111302A1Low mutation rateReduce probabilityAntibacterial agentsOrganic active ingredientsAntibiotic resistanceDrug resistance
The present invention provides methods and compositions for said inhibition of drug resistance. In one embodiment, said invention provides methods and compositions for said inhibition of antibiotic resistance. The invention generally involves said administration of achaogens, agents that inhibit said mutational process, to inhibit said evolution of drug resistance. Also, described herein are compositions that are suitable for use as achaogens.
Owner:THE SCRIPPS RES INST +1
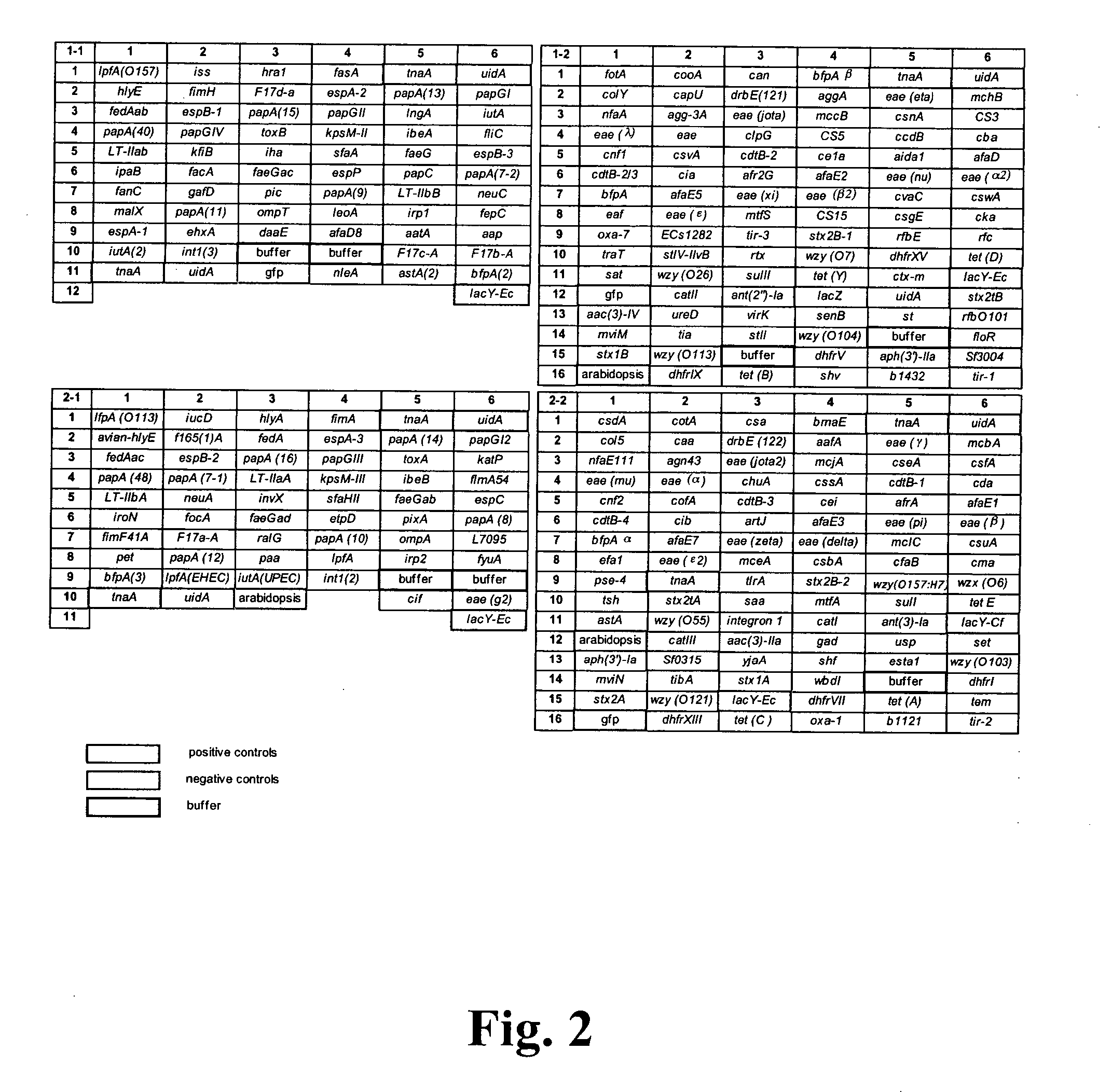
Virulence and antibiotic resistance array and uses thereof
InactiveUS20060094034A1Low costImprove reliabilityBioreactor/fermenter combinationsBiological substance pretreatmentsMicroorganismVirulent characteristics
An array of nucleic acid probes is described for simultaneously identifying or characterizing a pathotype of a microorganism and detecting antibiotic resistance of said microorganism. Methods are also described for detecting the presence of a microorganism in a sample, as well as determining its pathotype and its antibiotic resistance, using the array.
Owner:BROUSU ROLAND +4
Transgenic microbial polyhydroxyalkanoate producers
Transgenic microbial strains are provided which contain the genes required for PHA formation integrated on the chromosome. The strains are advantageous in PHA production processes, because (1) no plasmids need to be maintained, generally obviating the required use of antibiotics or other stabilizing pressures, and (2) no plasmid loss occurs, thereby stabilizing the number of gene copies per cell throughout the fermentation process, resulting in homogeneous PHA product formation throughout the production process. Genes are integrated using standard techniques, preferably transposon mutagenesis. In a preferred embodiment wherein mutiple genes are incorporated, these are incorporated as an operon. Sequences are used to stabilize mRNA, to induce expression as a function of culture conditions (such as phosphate concentration), temperature, and stress, and to aid in selection, through the incorporation of selection markers such as markers conferring antibiotic resistance.
Owner:METABOLIX
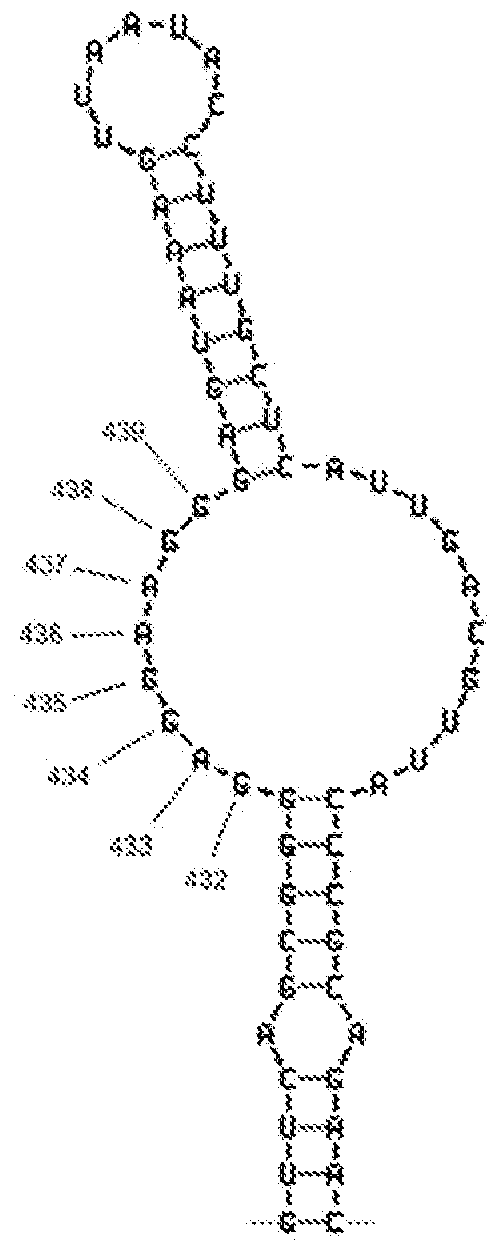
Probes and methods for detection of pathogens and antibiotic resistance
ActiveUS20080199863A1Increase assayRapid detection of antimicrobialSugar derivativesMicrobiological testing/measurementAntibiotic resistanceNucleic acid sequencing
Described are probes and methods for detecting pathogens and antibiotic resistance of a specimen. The method comprises contacting the specimen with a growth medium; and lysing the specimen to release nucleic acid molecules from the specimen. The lysate of the specimen is contacted with a capture probe immobilized on a substrate, wherein the capture probe comprises an oligonucleotide that specifically hybridizes with a first target nucleic acid sequence region of ribosomal RNA. The lysate is in contact with a detector probe that comprises a detectably labeled oligonucleotide that specifically hybridizes with a second target nucleic acid sequence region of ribosomal RNA. The presence or absence of labeled oligonucleotide complexed with the substrate is determined. Detection of labeled oligonucleotide complexed with the substrate is indicative of the presence of pathogen. Performing the method in the presence and absence of an antibiotic permits detection of antibiotic resistance.
Owner:THE GOVERNMENT OF THE UNITED STATES OF AMERICA AS REPRESENTED BY THE DEPT OF VETERANS AFFAIRS +1
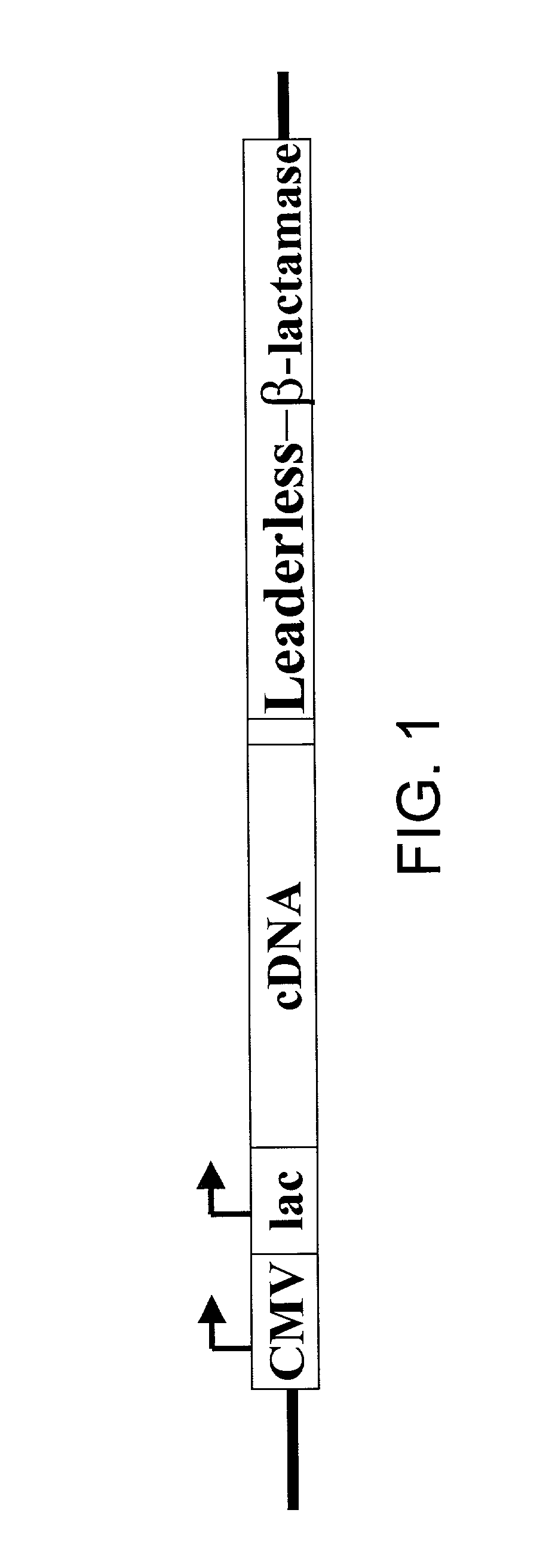
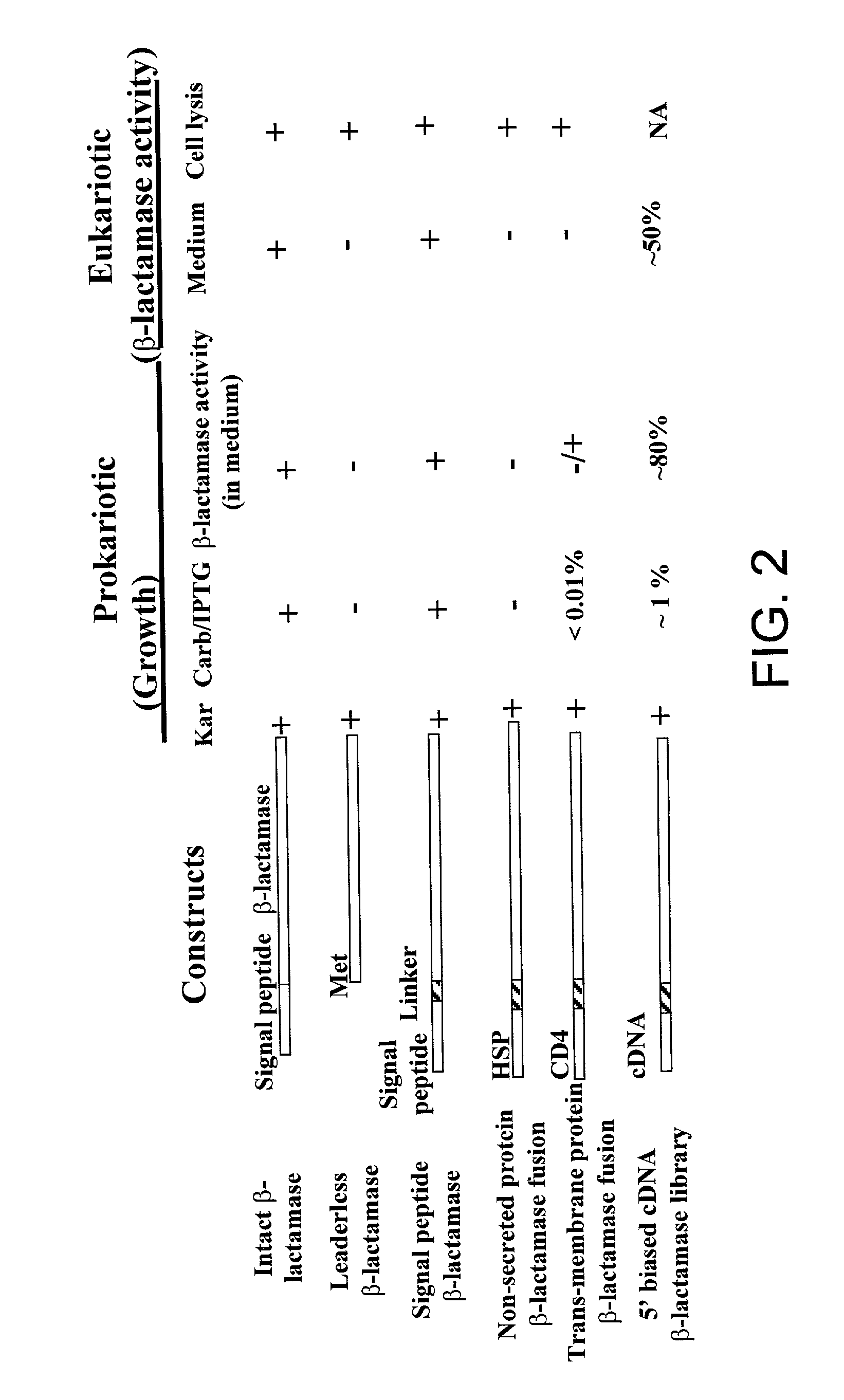
Method for identification of cDNAs encoding signal peptides
InactiveUS20020127557A1Fast wayBacteriaMicrobiological testing/measurementScreening techniquesPresent method
The present invention provides a method in which cDNAs that encode signal sequences for secreted or membrane-associated proteins are isolated using a fusion protein that directs secretion of a molecule that provides antibiotic resistance, e.g., .beta.-lactamase. The present method allows the isolation of signal peptide-associated proteins that may be difficult to isolate with other techniques. Moreover, the present method is amenable to throughput screening techniques and automation, and especially in validating the presence of the signal sequence via expression of the protein in both prokaryotic and eukaryotic cells. This invention provides a powerful and approach to the large scale isolation of novel secreted proteins.
Owner:INCYTE CORP
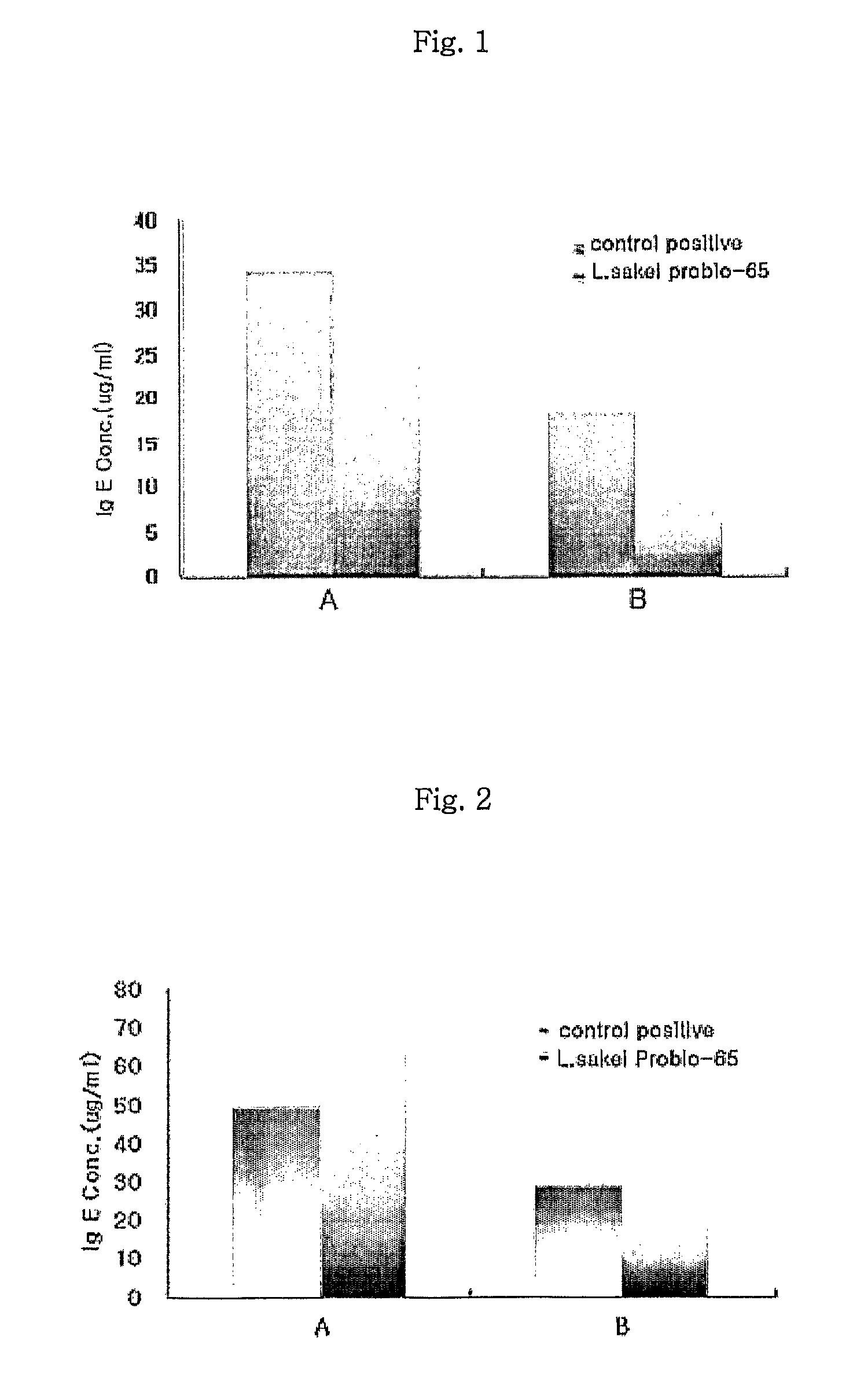
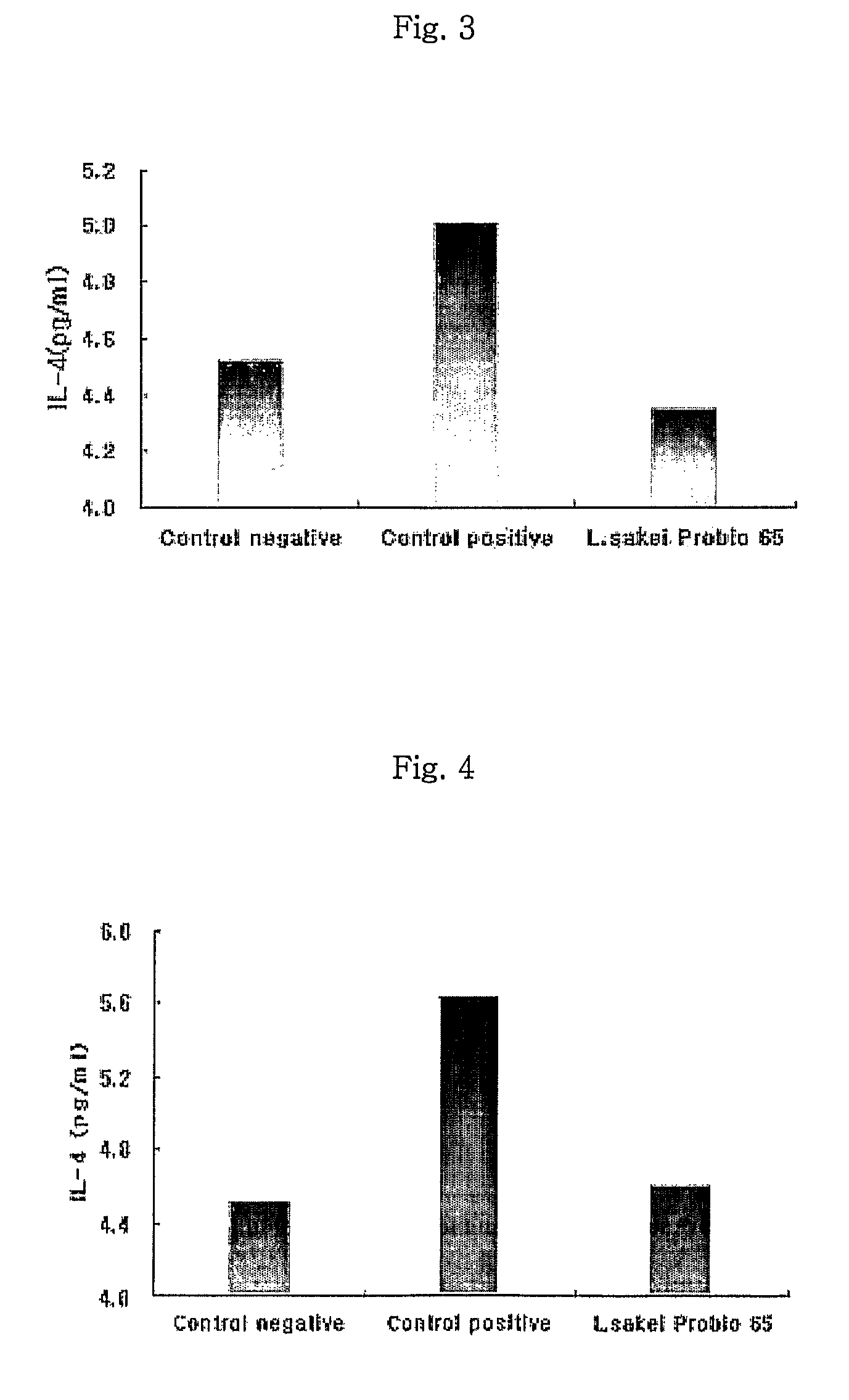
Acid tolerant Lactobacillus sakei probio-65 with the ability of growth suppression of pathogenic microorganisms and the anti-allergic effect
ActiveUS8034606B2Growth inhibitionEffectively inhibit harmful microorganismsBiocideCosmetic preparationsAcid-fastDisease
Disclosed are a novel tactic acid bacterium, Lactobacillus sakei Probio-65, and the use thereof. The L. sakei Probio-65 strain has acid tolerance, bile acid tolerance and antibiotic resistance, inhibits the growth of harmful pathogenic microorganisms in the body and the intestine of animals, and has immunuenhancing activity. In particular, the novel strain inhibits the growth of Staphylocccus aureus, which is known to be a factor aggravating atopic dermatitis. Thus, the novel strain is useful for preventing or treating atopic dermatitis and allergy-related disorders. Also, the novel strain stabilizes intestinal microflors by inhibiting the abnormal proliferation of harmful microorganisms in the intestine. The L. sakei Probio-65 strain or a culture thereof is useful in pharmaceutical, feed, food, and cosmetic compositions.
Owner:PROBIONIC CO LTD
Bifunctional antibiotics for targeting rRNA and resistance-causing enzymes
InactiveUS20050004052A1High activityOvercome resistanceBiocideSugar derivativesAntibiotic resistanceAntibiotic Y
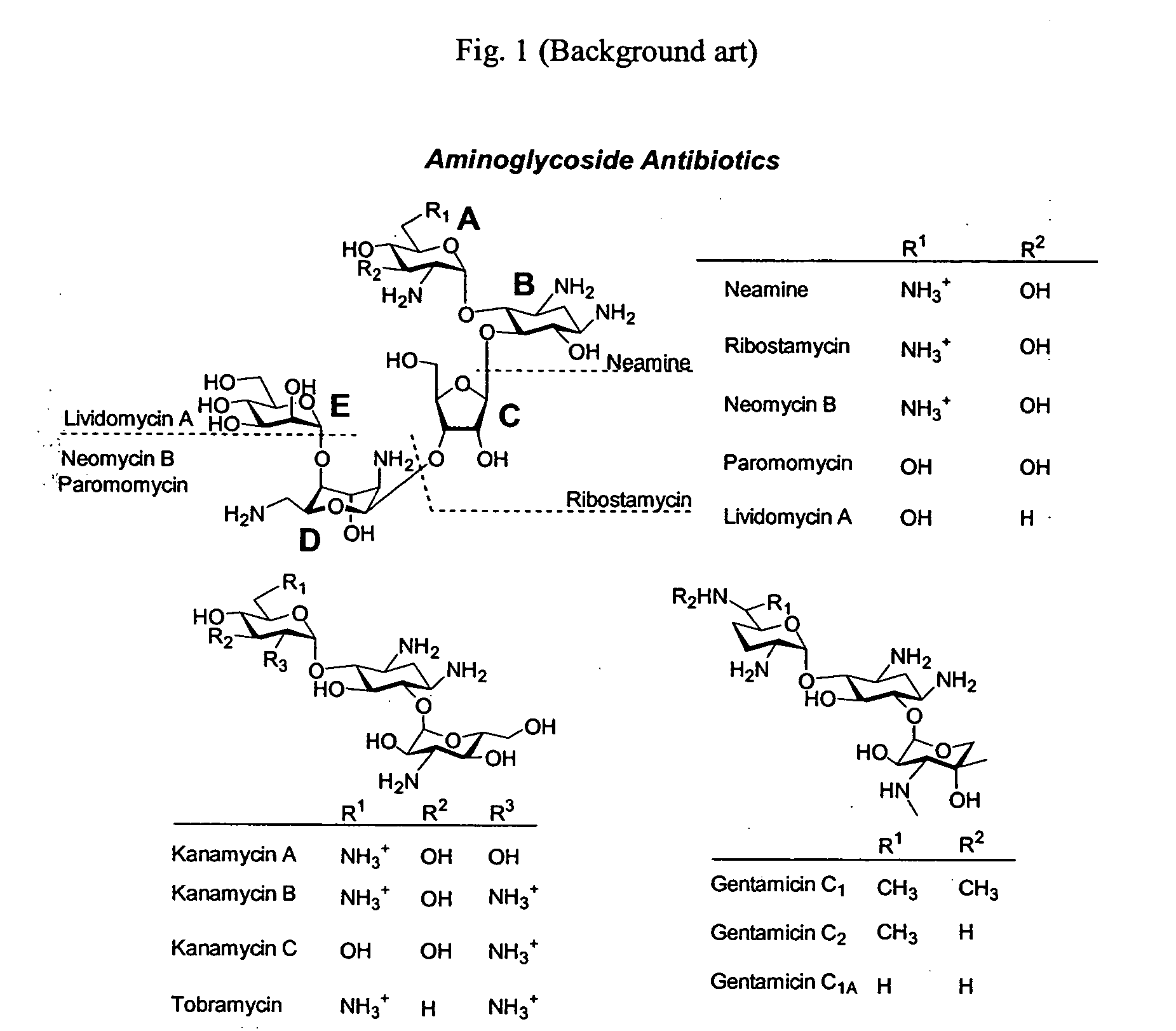
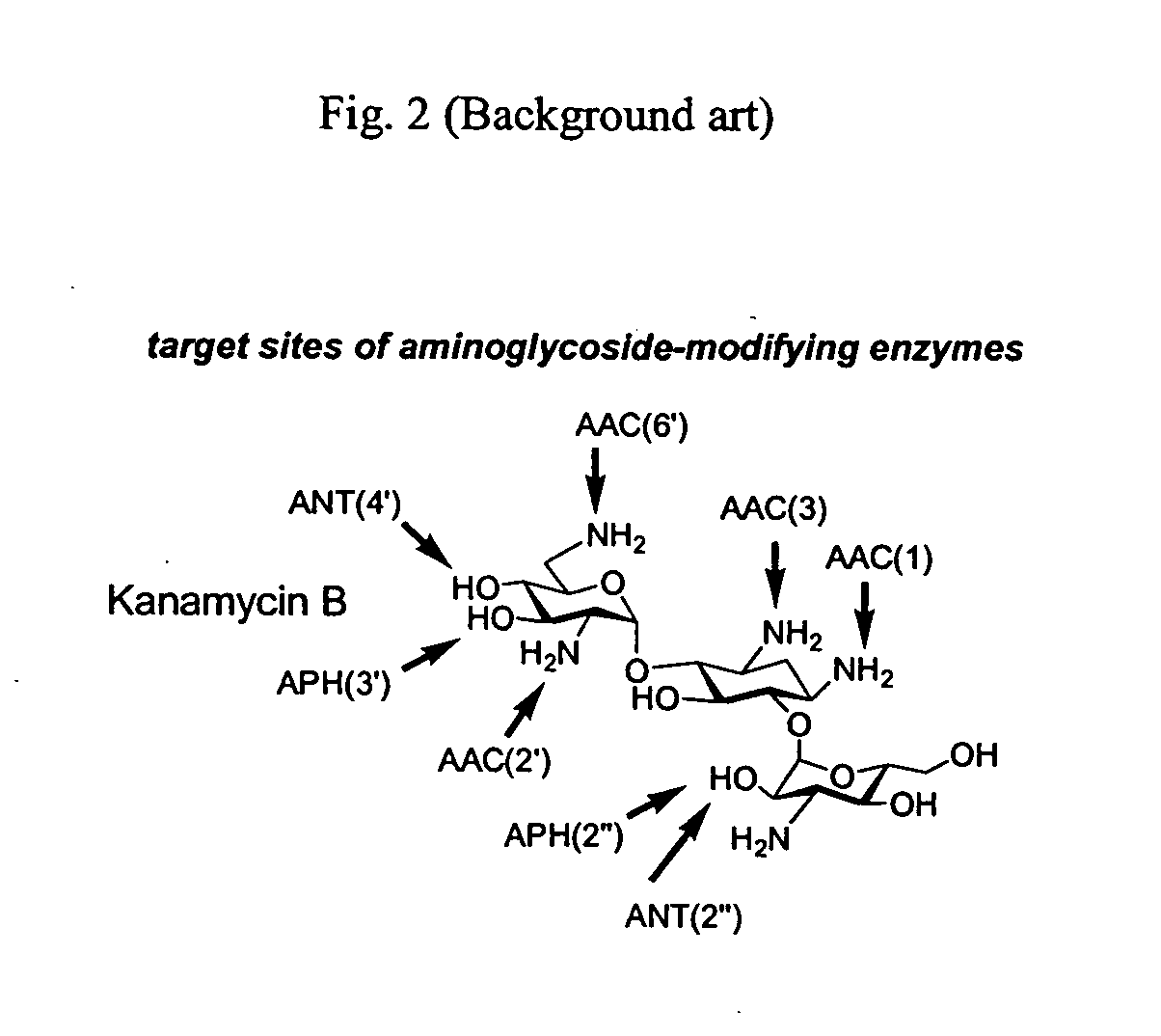
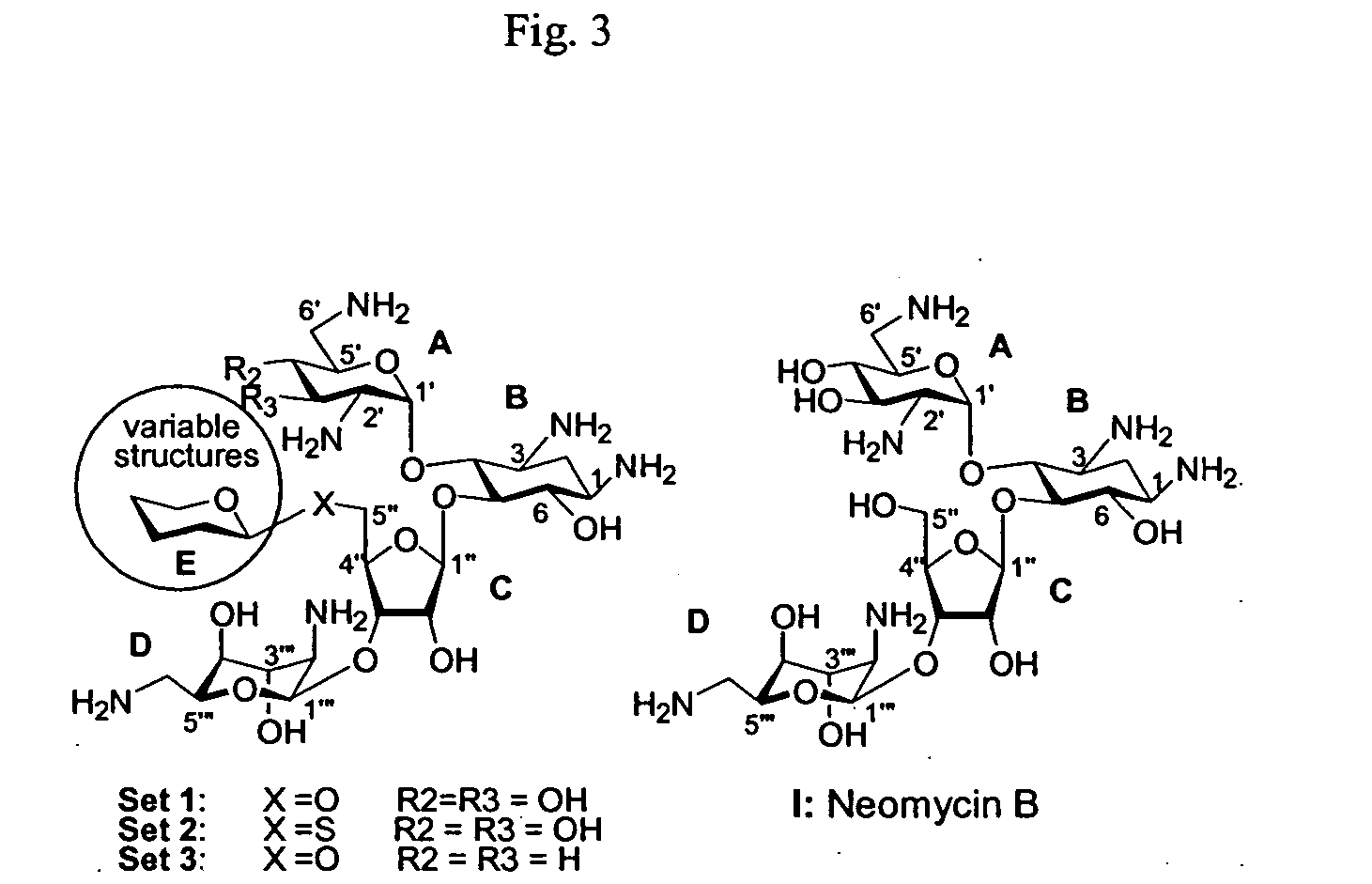
A novel group of aminoglycosides which share some structural features of currently available aminoglycosides with regard to the backbone, while also having significant structural differences. The similarity enables these aminoglycosides to be highly potent and effective antibiotics, while the significant differences enable these aminoglycosides to reduce or even block antibiotic resistance.
Owner:TECHNION RES & DEV FOUND LTD
Double T-DNA carrier and its application in cultivating of non selecting sign transgene rice
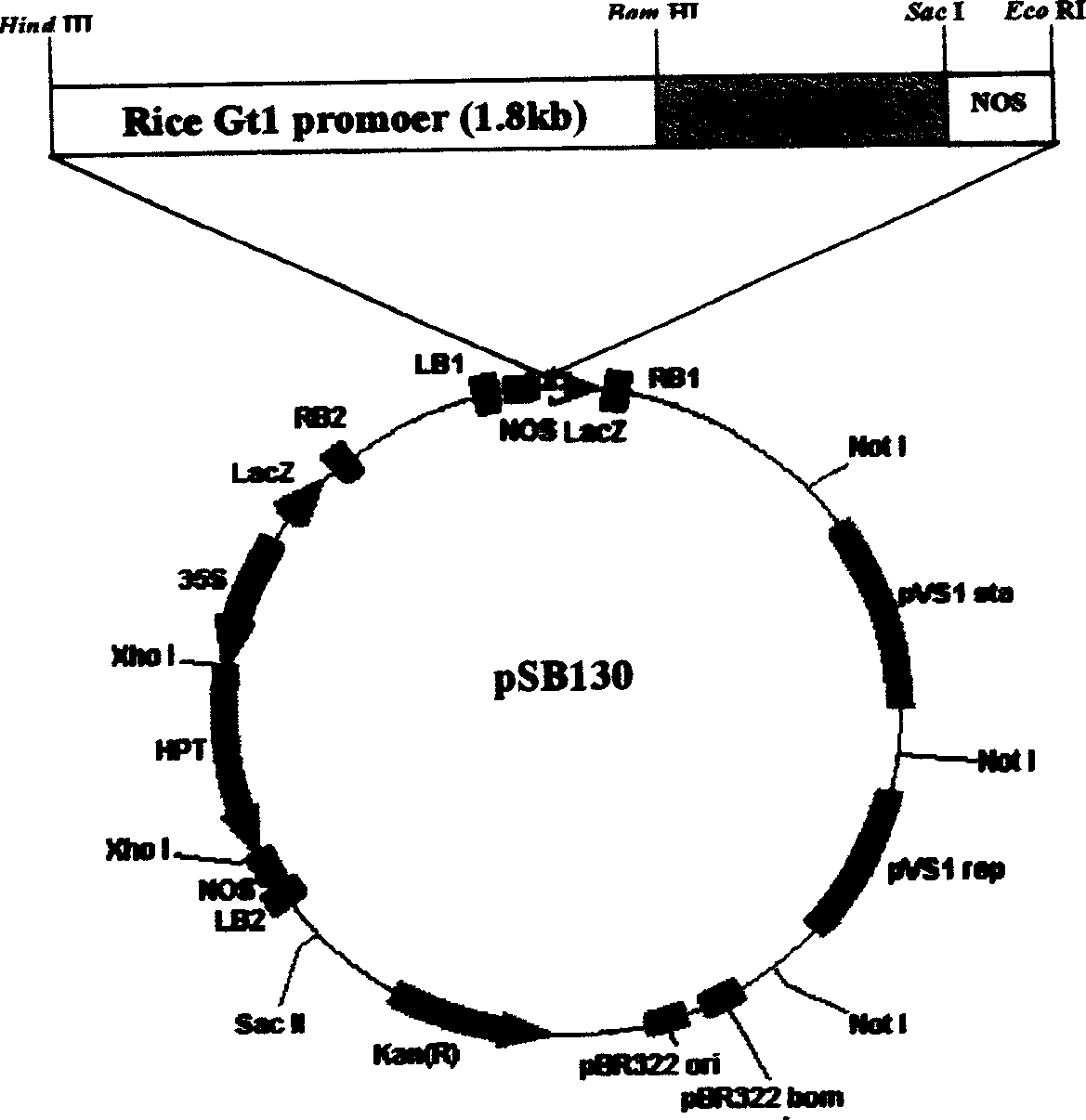
InactiveCN1597969ASmall molecular weightHigh co-transformation efficiencyFermentationVector-based foreign material introductionGenetically modified ricePlant nodule
The invention provides a simple and convenient Agrobacterium Tumefacies dual carrier containing double T-DNA structural regions, and a method of using the carrier system to cultivate transgenic rice without resistance selection label. With the help of the principle of co-conversion mediated by root nodule Agrobacterium Tumefacies, the system contains two separate T-DNA structural sections, where the first T-DNA region contains antibiotic resistance selection label gene and the second one contains a universal polyclonal site able to be arbitrarily inserted with destination gene. The double-T-DNA carrier has small molecular weight, easy to clone and after the destination gene and necessary regulation and control series are cloned in the T-DNA region containing the polyclonal site, it realizes rice co-conversion; by selfing, it selects transgenic individual with destination gene but without selectin label gene from the self-bred progeny, thus eliminating the negative effect on transgenic plant commercialized production, etc, possibly caused by selection label gene.
Owner:YANGZHOU UNIV
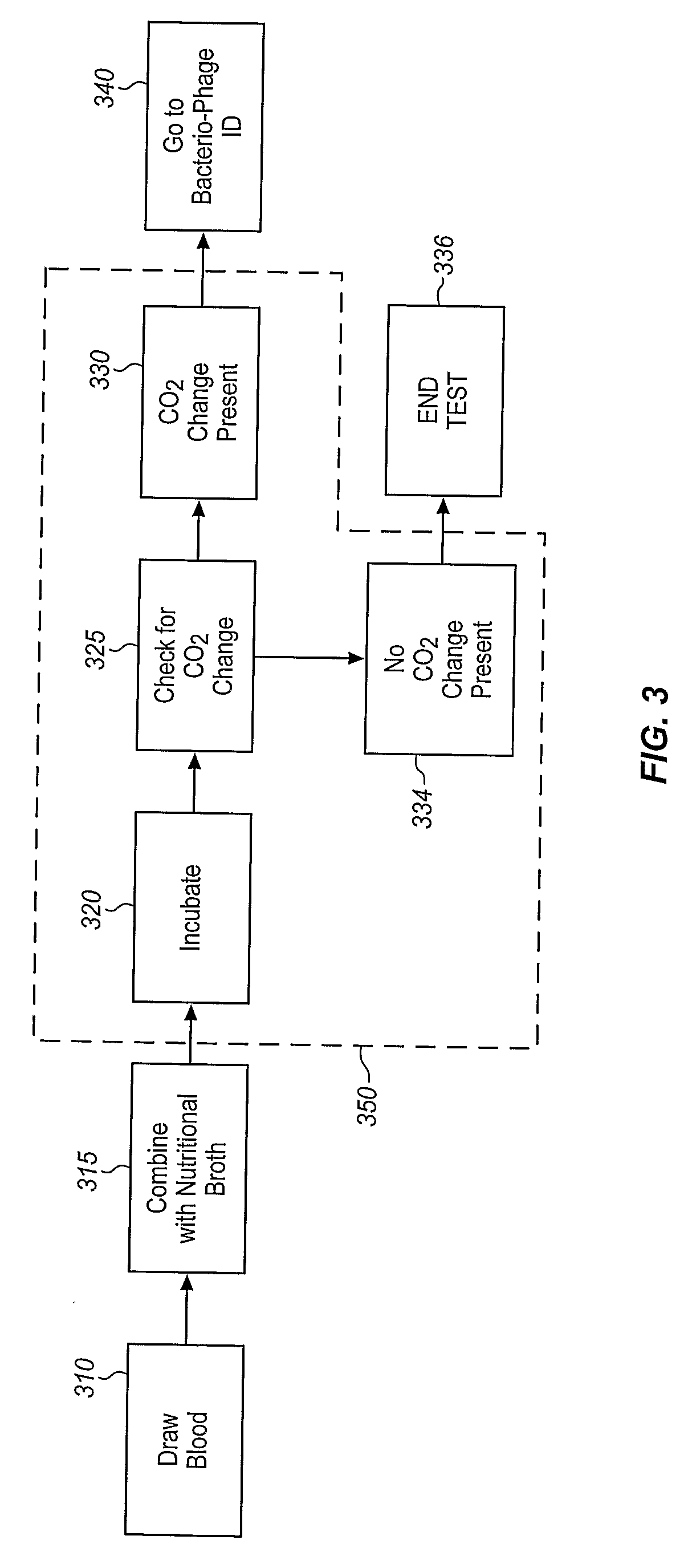
Method and apparatus for bacteriophage-based diagnostic assays
InactiveUS20090286225A1Quick checkQuick identificationBioreactor/fermenter combinationsBiological substance pretreatmentsAssayTest sample
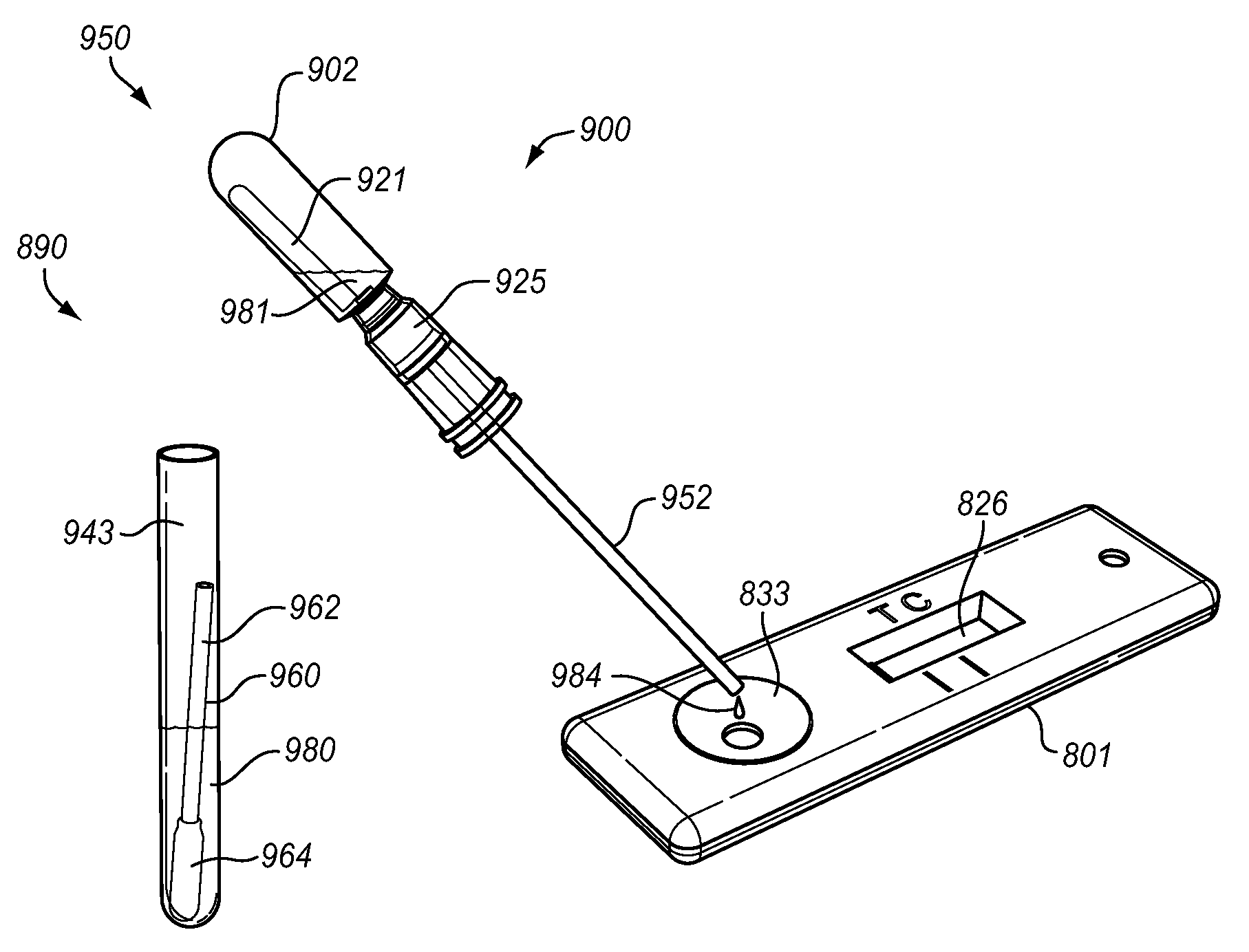
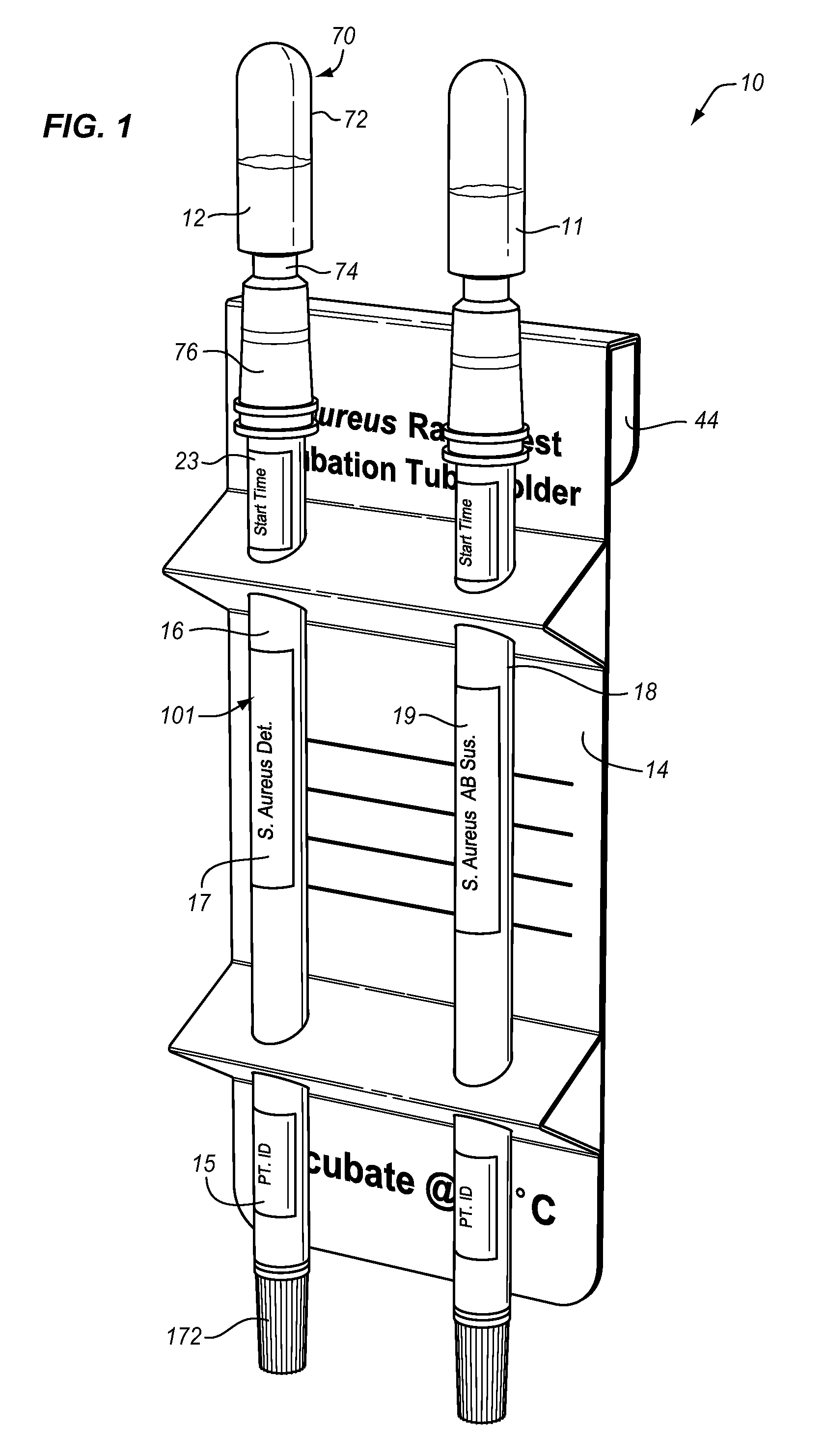
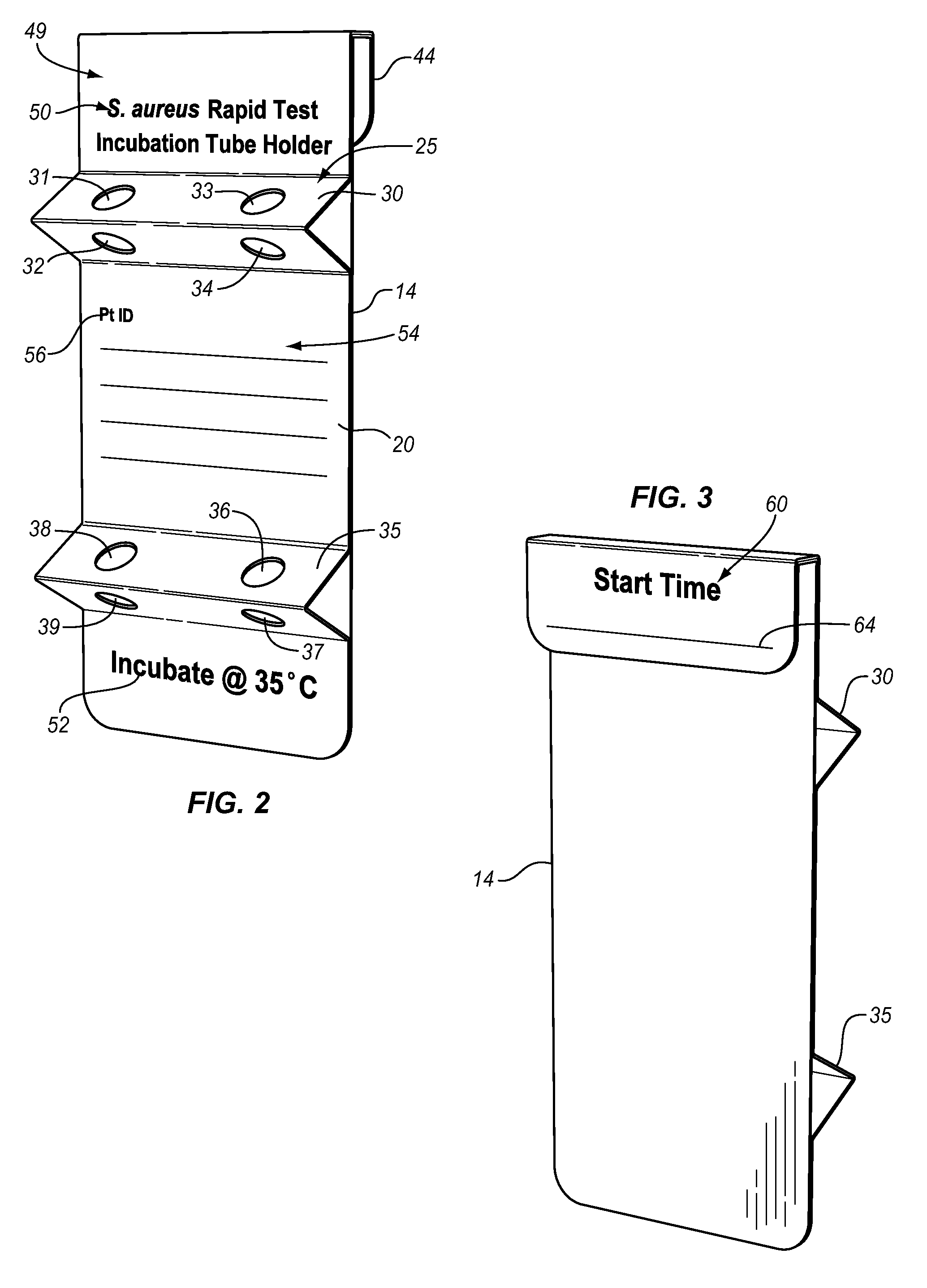
Bacteriophage are combined with a test sample in an incubator, and the bacteriophage-exposed test sample is conjugated and applied to a sample pad in contact with a lateral flow strip to determine the presence or absence of a target bacterium. The conjugation may be performed in the sample pad or prior to application of the bacteriophage-exposed test sample to the pad. The incubator comprises a bacteriophage container and an incubation container separated by a valve. The test sample may be inserted into the incubation chamber using a swab or a rod with a piercing tip and a sample collection eye. The valve comprises a breakable stem. An antibiotic may be added to the test sample to determine the antibiotic resistance or susceptibility of the bacterium.
Owner:MICROPHAGE
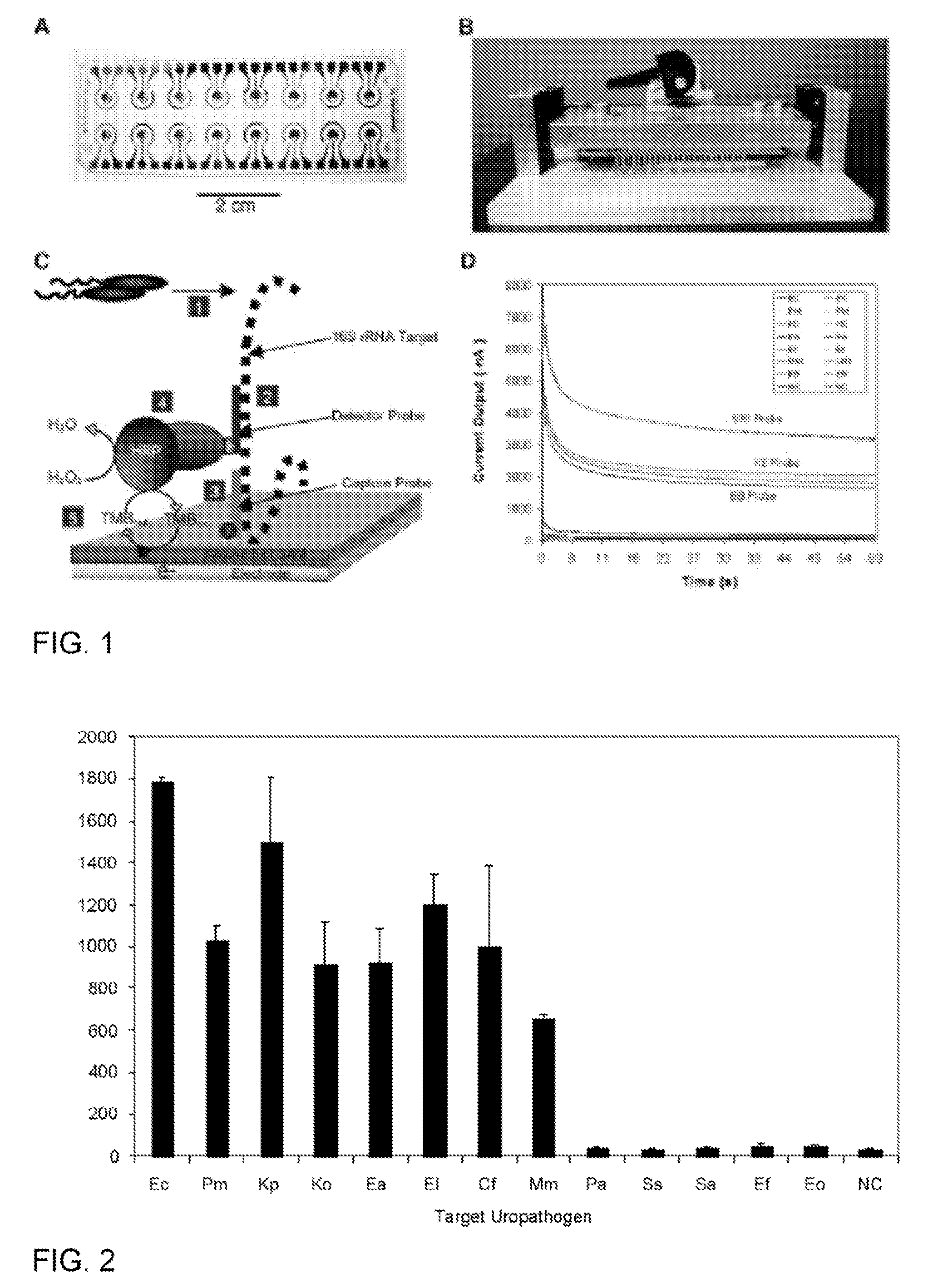
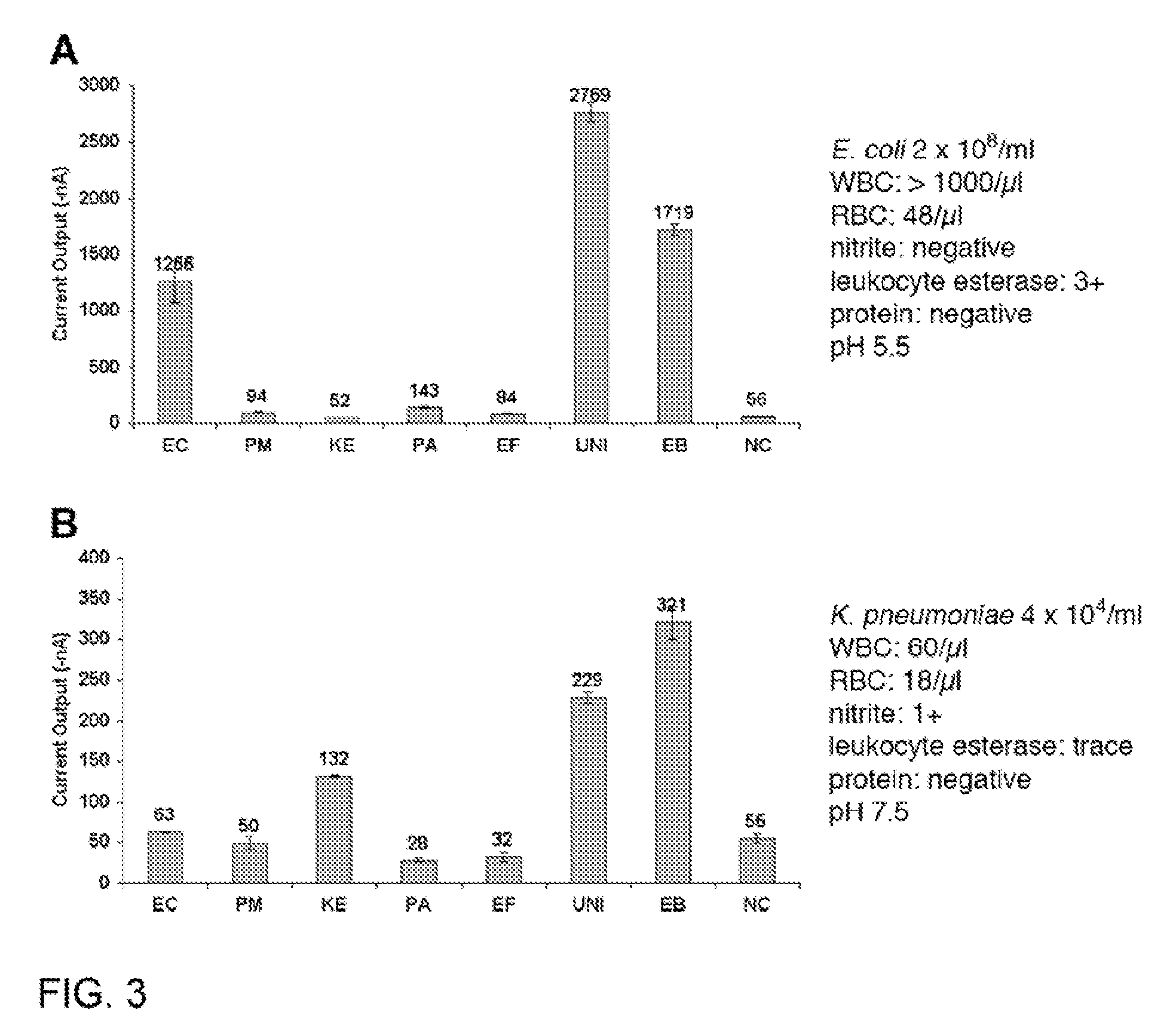
Specific and universal probes and amplification primers to rapidly detect and identify common bacterial pathogens and antibiotic resistance genes from clinical specimens for routine diagnosis in microbiology laboratories
InactiveUS20050042606A9Rapid bacterial identificationShorten the timeMicrobiological testing/measurementDepsipeptidesGenomic SegmentMoraxella catarrhalis
The present invention relates to DNA-based methods for universal bacterial detection, for specific detection of the common bacterial pathogens Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Proteus mirabilis, Streptococcus pneumoniae, Staphylococcus aureus, Staphylococcus epidermidis, Enterococcus faecalis, Staphylococcus saprophyticus, Streptococcus pyogenes, Haemophilus influenzae and Moraxella catarrhalis as well as for specific detection of commonly encountered and clinically relevant bacterial antibiotic resistance genes directly from clinical specimens or, alternatively, from a bacterial colony. The above bacterial species can account for as much as 80% of bacterial pathogens isolated in routine microbiology laboratories. The core of this invention consists primarily of the DNA sequences from all species-specific genomic DNA fragments selected by hybridization from genomic libraries or, alternatively, selected from data banks as well as any oligonucleotide sequences derived from these sequences which can be used as probes or amplification primers for PCR or any other nucleic acid amplification methods. This invention also includes DNA sequences from the selected clinically relevant antibiotic resistance genes. With these methods, bacteria can be detected (universal primers and / or probes) and identified (species-specific primers and / or probes) directly from the clinical specimens or from an isolated bacterial colony. Bacteria are further evaluated for their putative susceptibility to antibiotics by resistance gene detection (antibiotic resistance gene specific primers and / or probes). Diagnostic kits for the detection of the presence, for the bacterial identification of the above-mentioned bacterial species and for the detection of antibiotic resistance genes are also claimed. These kits for the rapid (one hour or less) and accurate diagnosis of bacterial infections and antibiotic resistance will gradually replace conventional methods currently used in clinical microbiology laboratories for routine diagnosis. They should provide tools to clinicians to help prescribe promptly optimal treatments when necessary. Consequently, these tests should contribute to saving human lives, rationalizing treatment, reducing the development of antibiotic resistance and avoid unnecessary hospitalizations.
Owner:GENEOHM SCI CANADA
Method and Apparatus for Identification of Microorganisms Using Bacteriophage
InactiveUS20080286757A1Improve reliabilityImprove signal-to-noise ratioMicrobiological testing/measurementBacteria identificationMicroorganism
A sample is tested for the presence of bacteria, such as in an automatic blood culturing apparatus. If bacteria are determined to be present, a bacteriophage-based bacteria identification process is performed to identify the bacteria present. A plurality of bacteria detection processes, such as a blood culture test and Gram stain test may be carried out prior to the bacteria identification process. A bacteriophage-based antibiotic resistance test or antibiotic susceptibility test is also conducted on the sample.
Owner:MICROPHAGETM
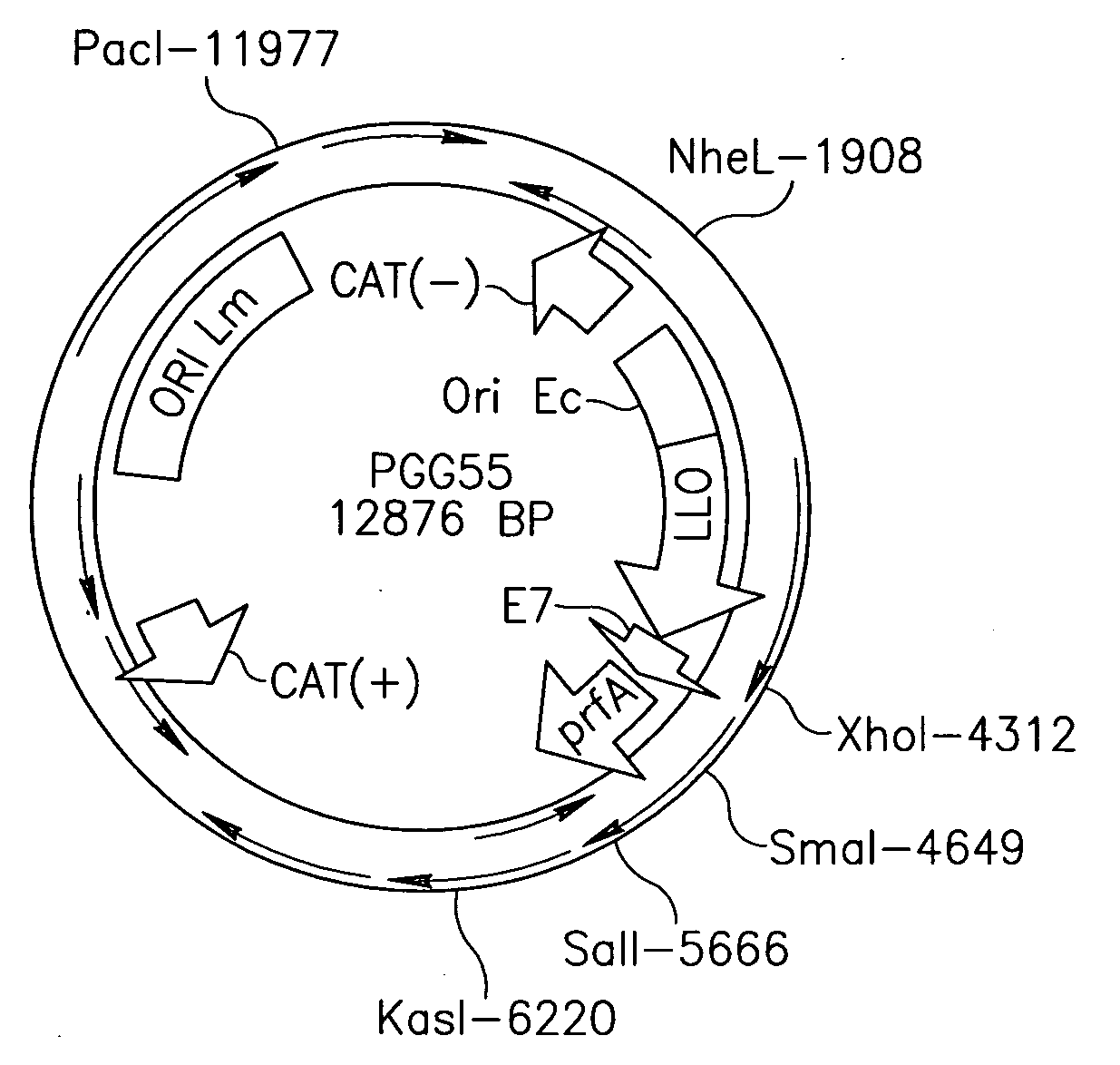
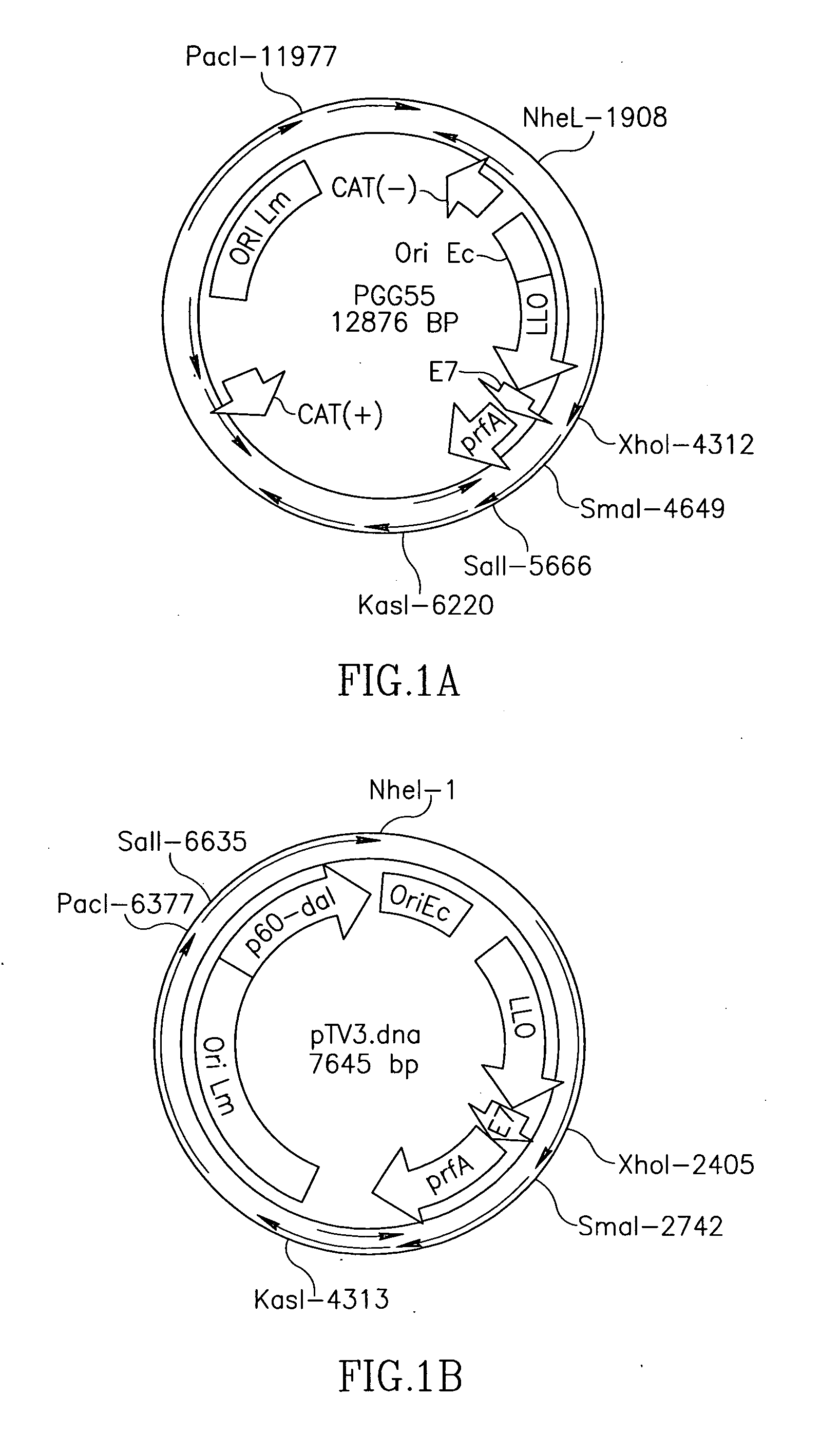
Antibiotic resistance free vaccines and methods for constructing and using same
ActiveUS7855064B2Reduce morbidityBacterial antigen ingredientsBacteriaHeterologousHeterologous Antigens
The present invention provides Listeria strains that express a heterologous antigen and a metabolic enzyme, and methods of generating same.
Owner:THE TRUSTEES OF THE UNIV OF PENNSYLVANIA
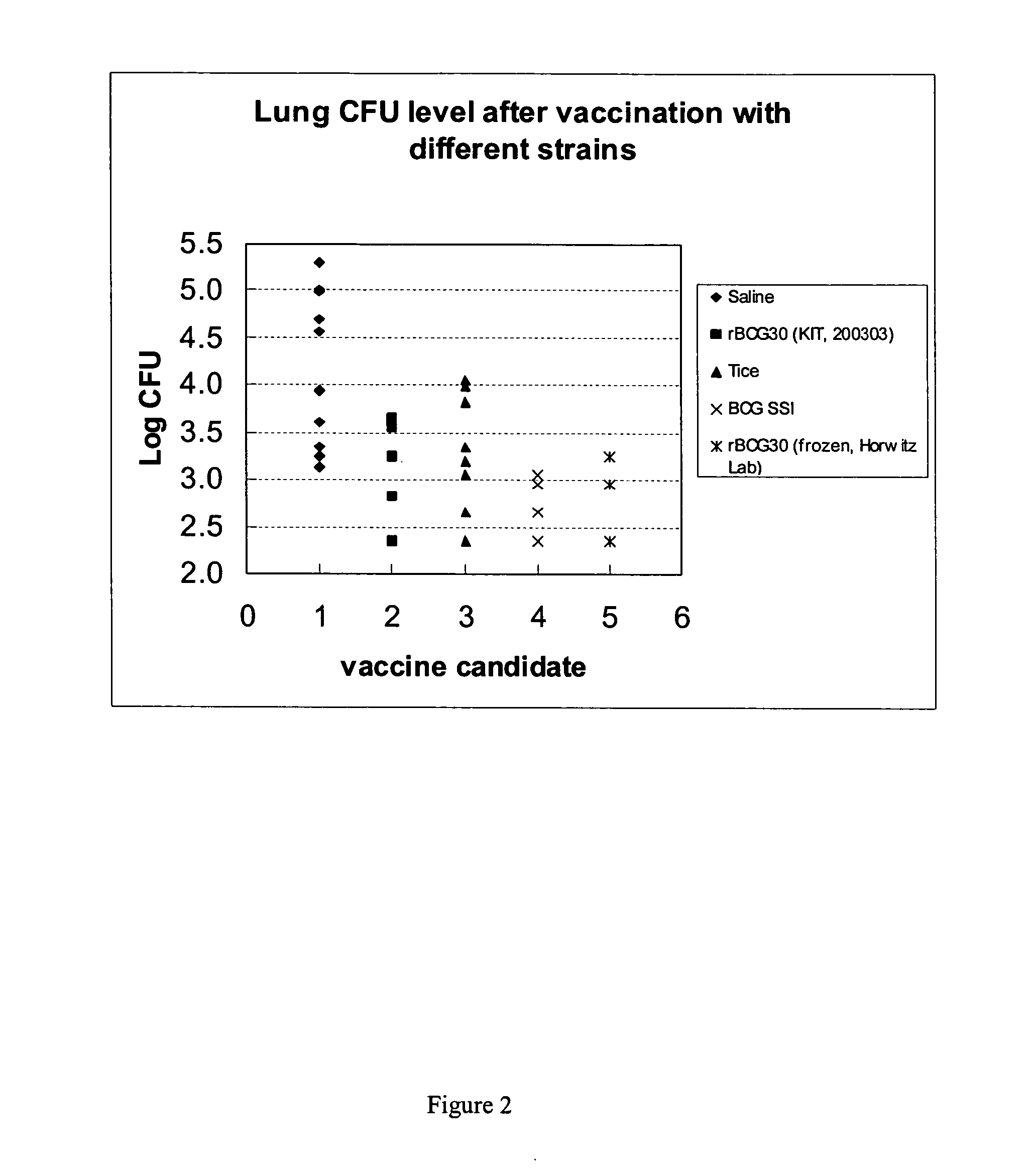
Electroporation of mycobacterium and overexpression of antigens in mycobacteria
ActiveUS20060121054A1Improve effectivenessIncrease strainAntibacterial agentsBiocideAntigenVaccination
Recombinant Mycobacterium strains with improved vaccinal properties for use as vaccinating agents are provided. The parent strains of the recombinant Mycobacterium strains are selected for their potent immunogenicity. The Mycobacterium strains do not display antibiotic resistance, and do not exhibit horizontal transfer to gram-negative bacteria.
Owner:INT AIDS VACCINE INITIATIVE INC
Antibiotic resistance free vaccines and methods for constructing and using same
ActiveUS20080131456A1Reduce morbidityBacterial antigen ingredientsBacteriaHeterologousHeterologous Antigens
The present invention provides Listeria strains that express a heterologous antigen and a metabolic enzyme, and methods of generating same.
Owner:THE TRUSTEES OF THE UNIV OF PENNSYLVANIA
Methods for Antimicrobial Resistance Determination
ActiveUS20100285447A1Rapid diagnosisReduce riskBioreactor/fermenter combinationsBiological substance pretreatmentsMicroorganismAntibiotic resistance
The present invention relates to methods and systems for determining the antibiotic-resistance status of microorganisms. The invention further provides methods for determining the antibiotic-resistance status of microorganisms in situ within a single system.
Owner:BIOMERIEUX INC
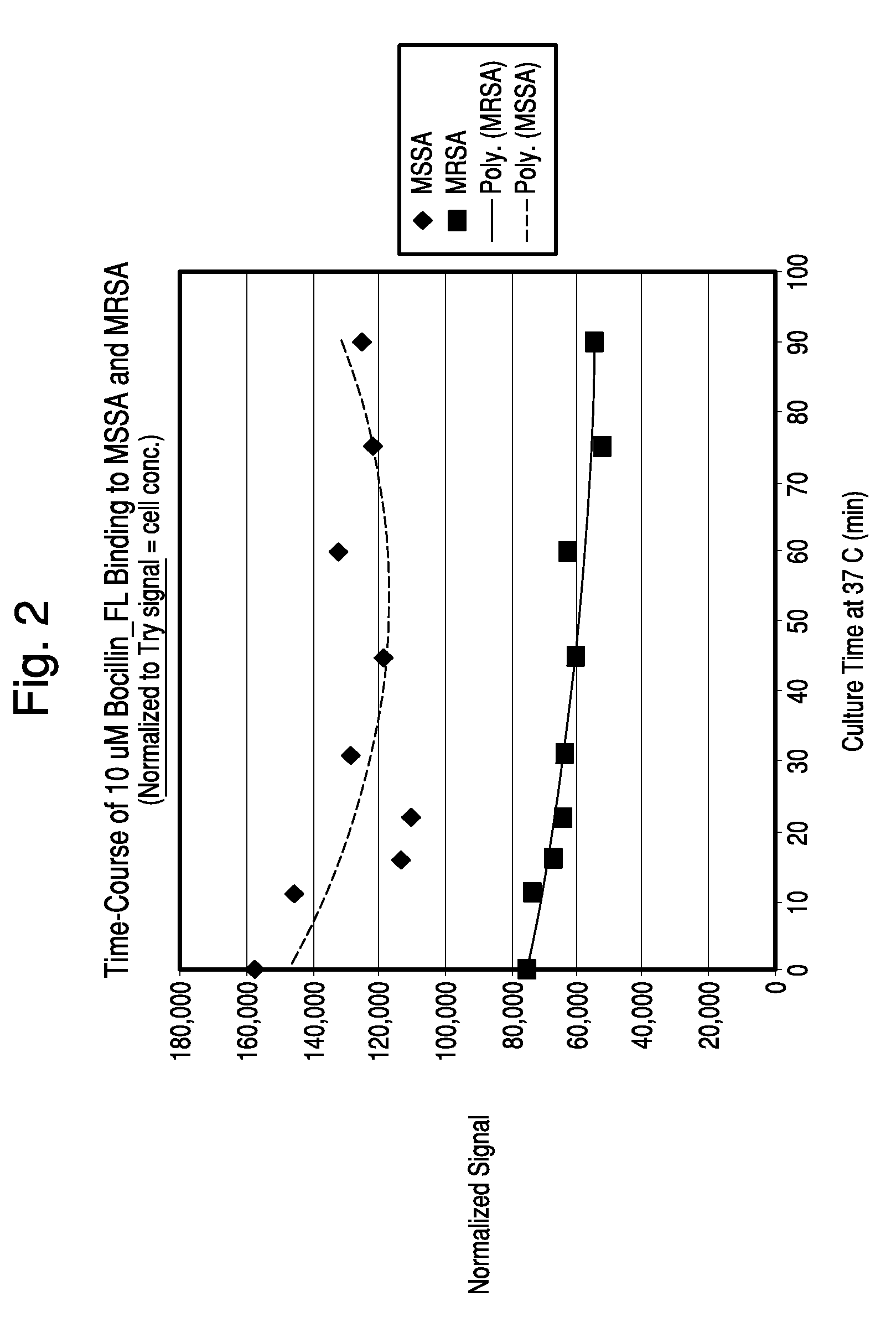
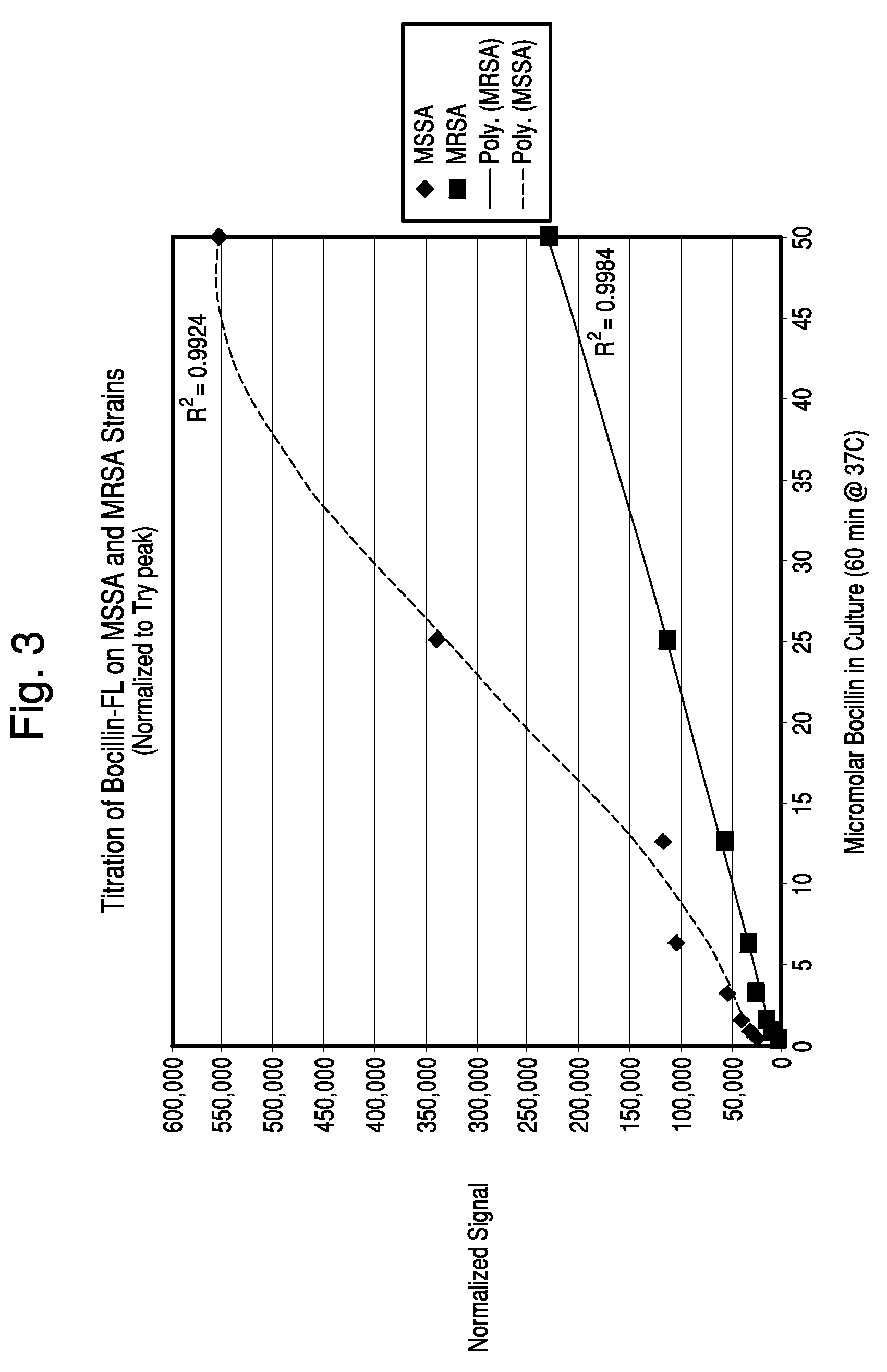
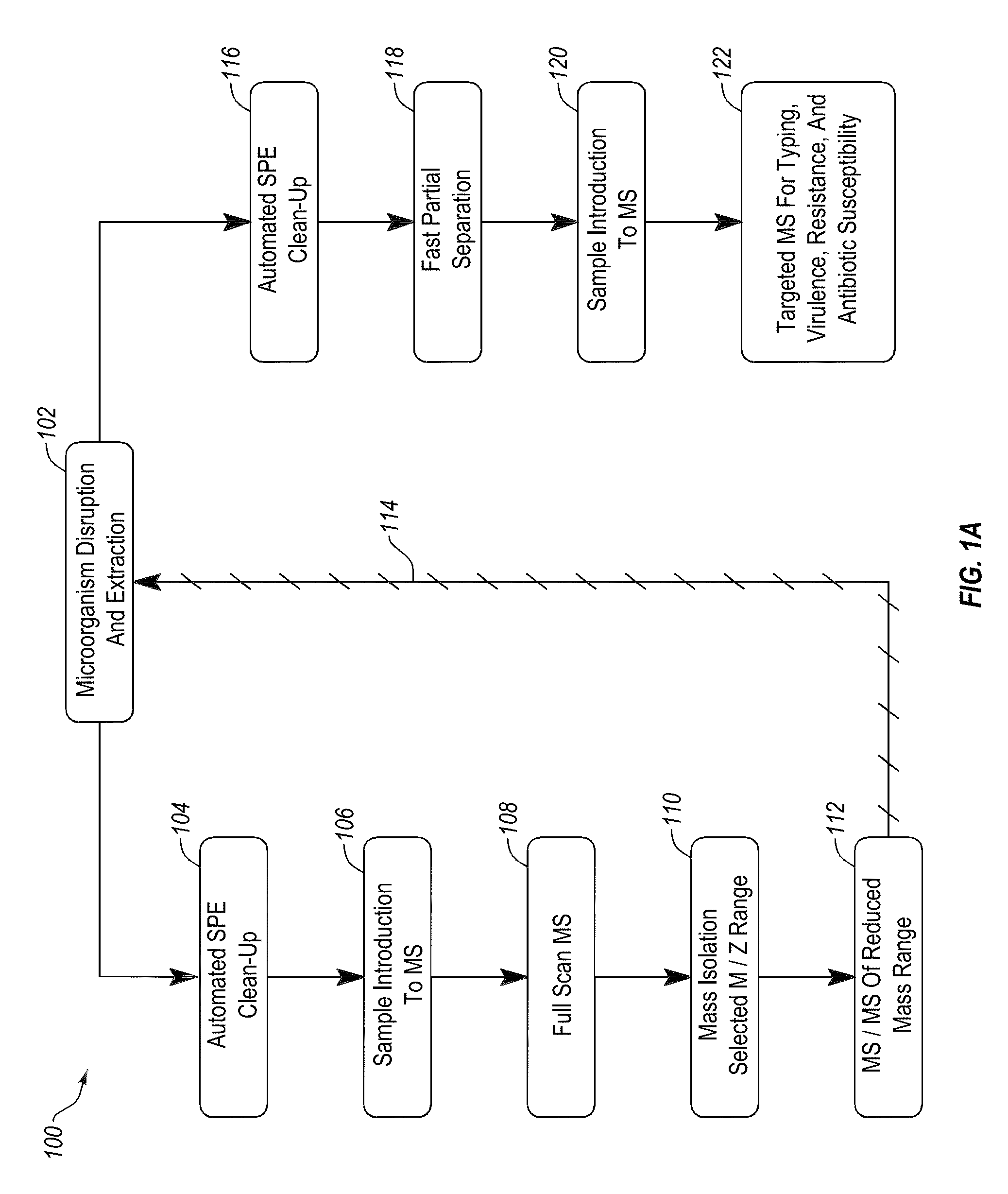
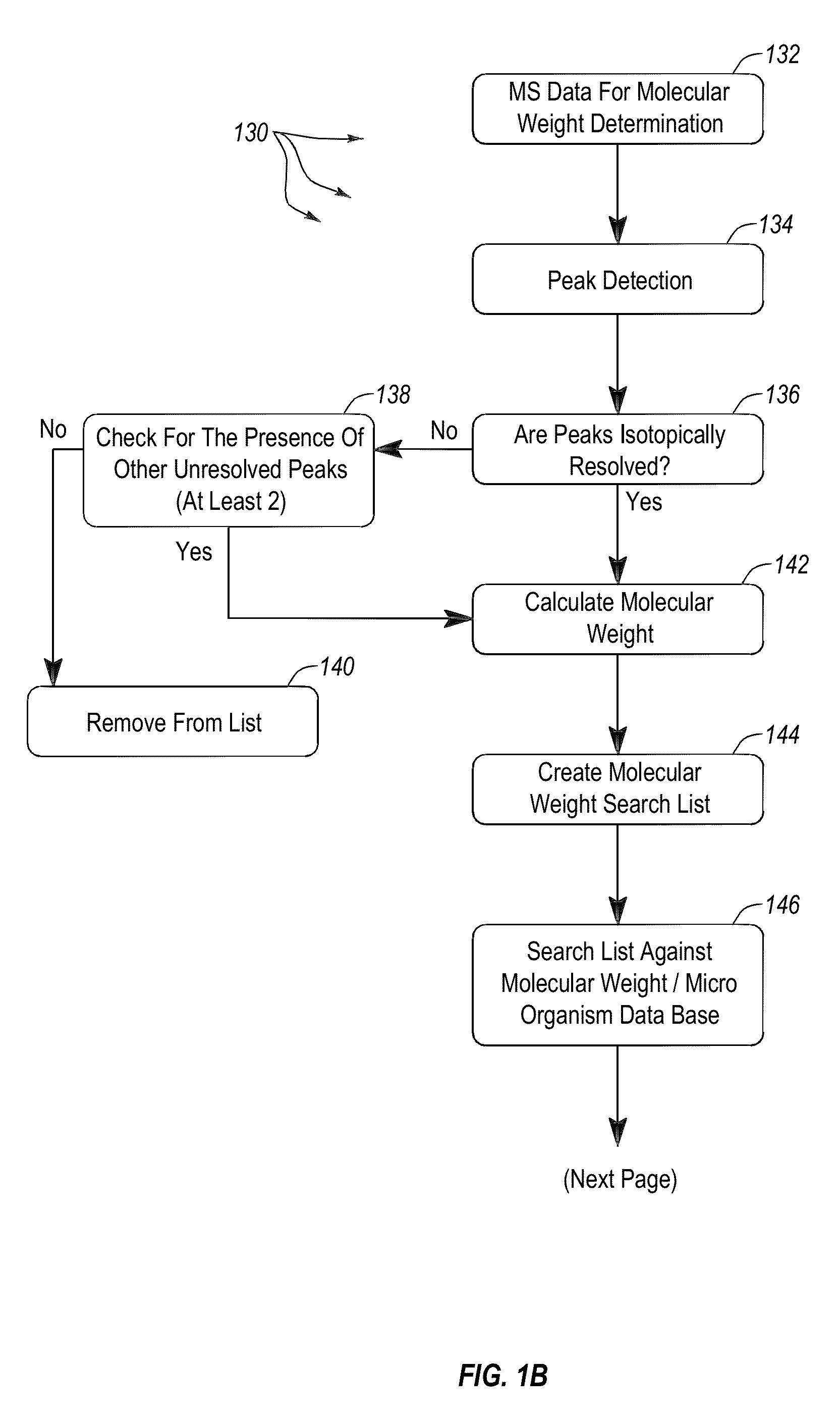
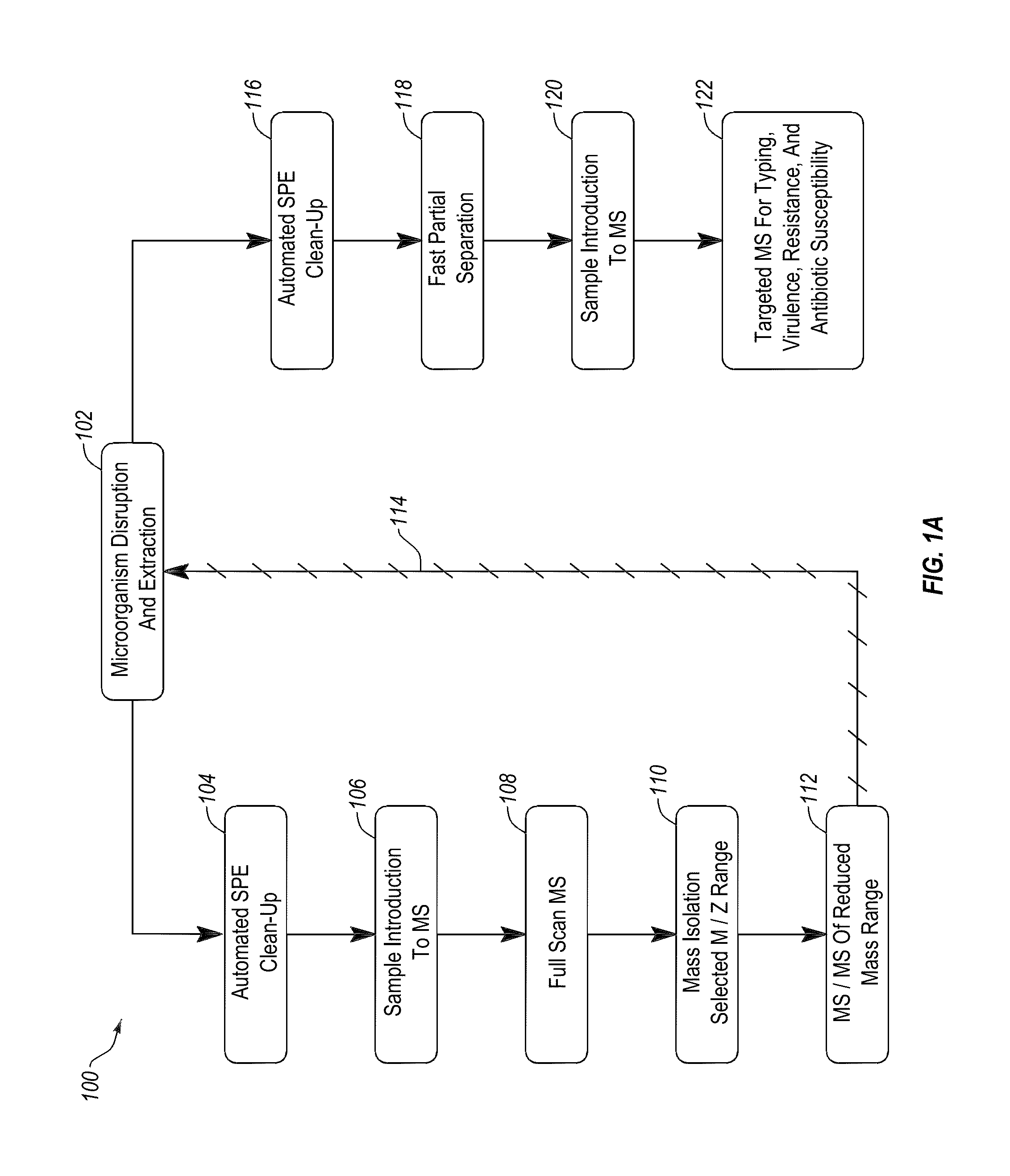
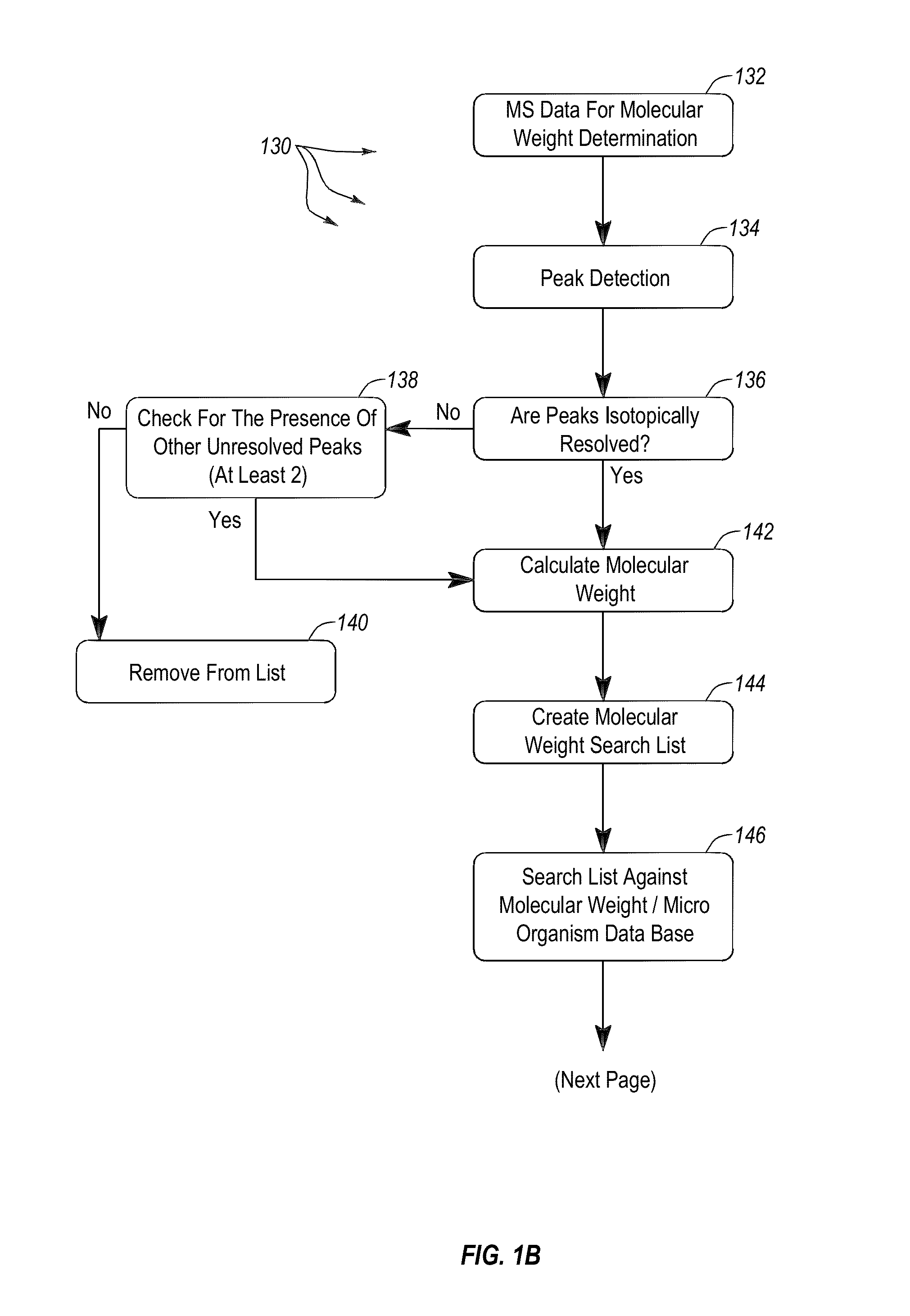
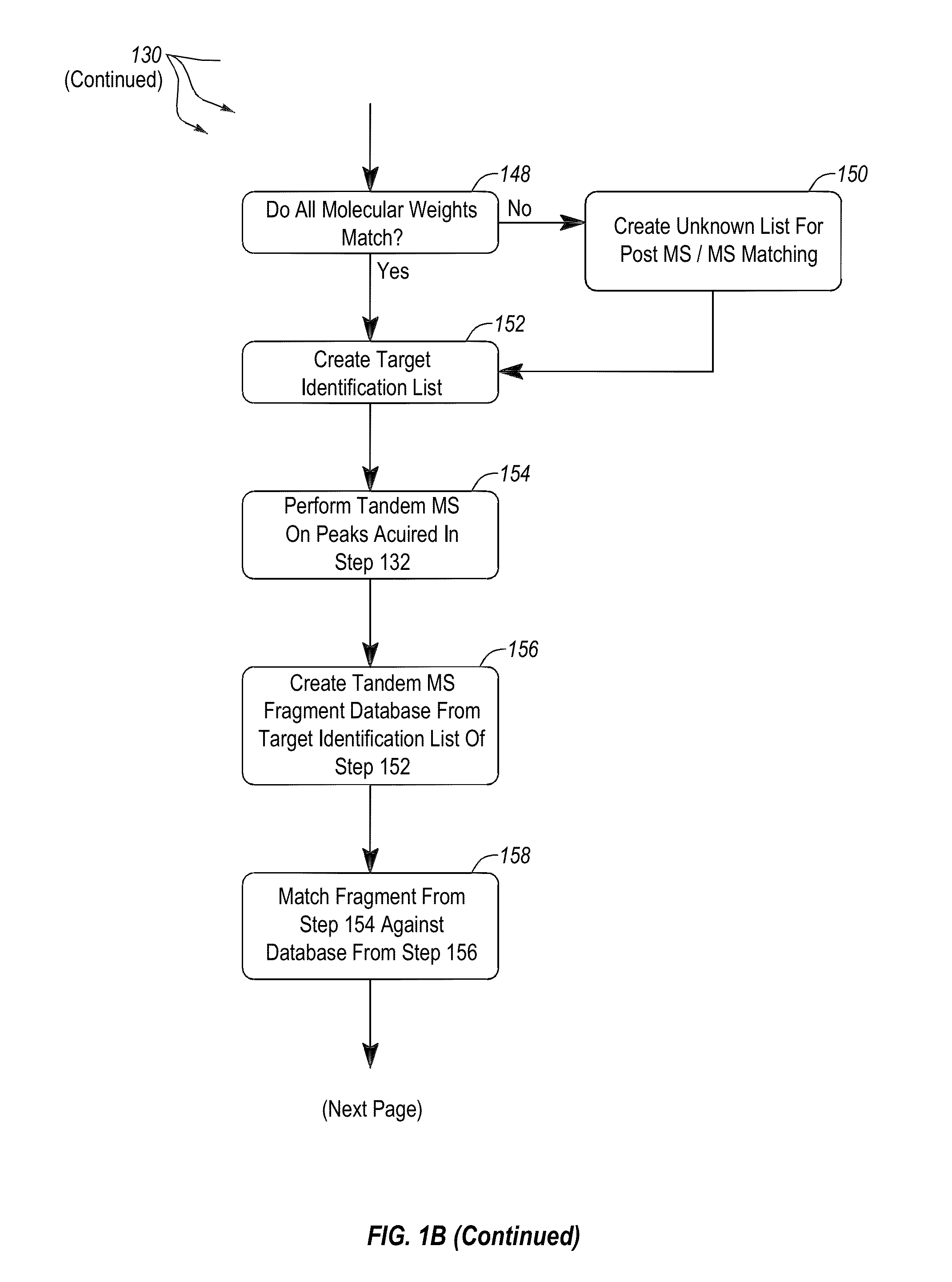
Apparatus and methods for microbial identification by mass spectrometry
ActiveUS9074236B2The process is simple and fastFacilitates epidemiological trackingBioreactor/fermenter combinationsBiological substance pretreatmentsMass spectrometry imagingMass spectrometric
Methods and systems for identification of microorganisms either after isolation from a culture or directly from a sample. The methods and systems are configured to identify microorganisms based on the characterization of proteins of the microorganisms via high-resolution / mass accuracy single-stage (MS) or multi-stage (MSn) mass spectrometry. Included herein are also discussion of targeted detection and evaluation of virulence factors, antibiotic resistance markers, antibiotic susceptibility markers, typing, or other characteristics using a method applicable to substantially all microorganisms and high-resolution / mass accuracy single-stage (MS) or multi-stage (MSn) mass spectrometry.
Owner:OXOID
Specific and universal probes and amplification primers to rapidly detect and identify common bacterial pathogens and antibiotic resistance genes from clinical specimens for routine diagnosis in microbiology laboratories
InactiveUS20070009947A1Rapid and exponential in vitro replicationQuick identificationMicrobiological testing/measurementDepsipeptidesBacteroidesMoraxella catarrhalis
The present invention relates to DNA-based methods for universal bacterial detection, for specific detection of the common bacterial pathogens Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Proteus mirabilis, Streptococcus pneumoniae, Staphylococcus aureus, Staphylococcus epidermidis, Enterococcus faecalis, Staphylococcus saprophyticus, Streptococcus pyogenes, Haemophilus influenzae and Moraxella catarrhalis as well as for specific detection of commonly encountered and clinically relevant bacterial antibiotic resistance genes directly from clinical specimens or, alternatively, from a bacterial colony. The above bacterial species can account for as much as 80% of bacterial pathogens isolated in routine microbiology laboratories. The core of this invention consists primarily of the DNA sequences from all species-specific genomic DNA fragments selected by hybridization from genomic libraries or, alternatively, selected from data banks as well as any oligonucleotide sequences derived from these sequences which can be used as probes or amplification primers for PCR or any other nucleic acid amplification methods. This invention also includes DNA sequences from the selected clinically relevant antibiotic resistance genes. With these methods, bacteria can be detected (universal primers and / or probes) and identified (species-specific primers and / or probes) directly from the clinical specimens or from an isolated bacterial colony. Bacteria are further evaluated for their putative susceptibility to antibiotics by resistance gene detection (antibiotic resistance gene specific primers and / or probes). Diagnostic kits for the detection of the presence, for the bacterial identification of the above-mentioned bacterial species and for the detection of antibiotic resistance genes are also claimed. These kits for the rapid (one hour or less) and accurate diagnosis of bacterial infections and antibiotic resistance will gradually replace conventional methods currently used in clinical microbiology laboratories for routine diagnosis. They should provide tools to clinicians to help prescribe promptly optimal treatments when necessary. Consequently, these tests should contribute to saving human lives, rationalizing treatment, reducing the development of antibiotic resistance and avoid unnecessary hospitalizations.
Owner:GENEOHM SCI CANADA
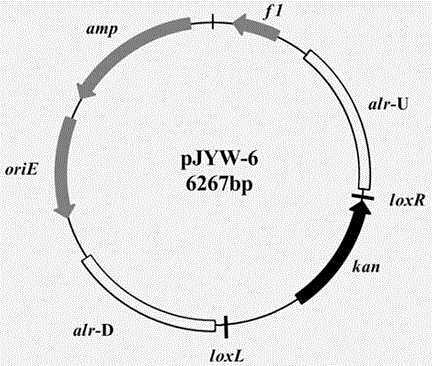
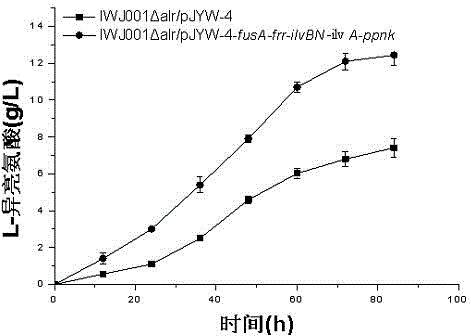
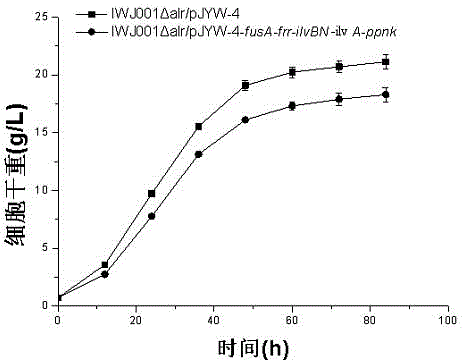
Construction method and application of L-isoleucine producing genetically engineered bacteria
InactiveCN104480057AIncrease productionEase of industrial productionBacteriaMicroorganism based processesBiotechnologyMicroorganism
The invention discloses L-isoleucine producing genetically engineered bacteria and a construction method and application thereof, belonging to the technical field of genetic engineering and microbial fermentation. According to the method, a knockout vector of the gene alr is constructed and electroporated into the host bacteria to obtain a deficient strain IWJ001 delta alr of the alr. The gene segments are connected to an overexpression vector by use of the PCR amplification genes fusA, frr, ilvBN, ilvA and ippnk and then electroporated into corynebacterium glutamicum IWJ001 delta alr, and the target genetically engineered bacteria, namely L-isoleucine producing genetically engineered bacteria IWJ001 delta alr / pJYW-4-fusA-frr-ilvBN-ilvA-ppnk, are obtained through antibiotic resistance screening. The L-isoleucine producing genetically engineered bacteria constructed by the method disclosed by the invention can be used for increasing the yield of L-isoleucine through fermentation in a shake flask or fermentation tank, and the method is conductive to reducing the difficulty in separation and purification and increasing the yield so as to remarkably reduce the production cost of L-isoleucine.
Owner:JIANGNAN UNIV
Apparatus and methods for microbiological analysis
ActiveUS20140051113A1Facilitates epidemiological trackingThe process is simple and fastBioreactor/fermenter combinationsBiological substance pretreatmentsMicroorganismSingle stage
Methods and systems for identification of microorganisms either after isolation from a culture or directly from a sample. The methods and systems are configured to identify microorganisms based on the characterization of proteins of the microorganisms via high-resolution / mass accuracy single-stage (MS) or multi-stage (MSn) mass spectrometry. Included herein are also discussion of targeted detection and evaluation of virulence factors, antibiotic resistance markers, antibiotic susceptibility markers, typing, or other characteristics using a method applicable to substantially all microorganisms and high-resolution / mass accuracy single-stage (MS) or multi-stage (MSn) mass spectrometry.
Owner:OXOID
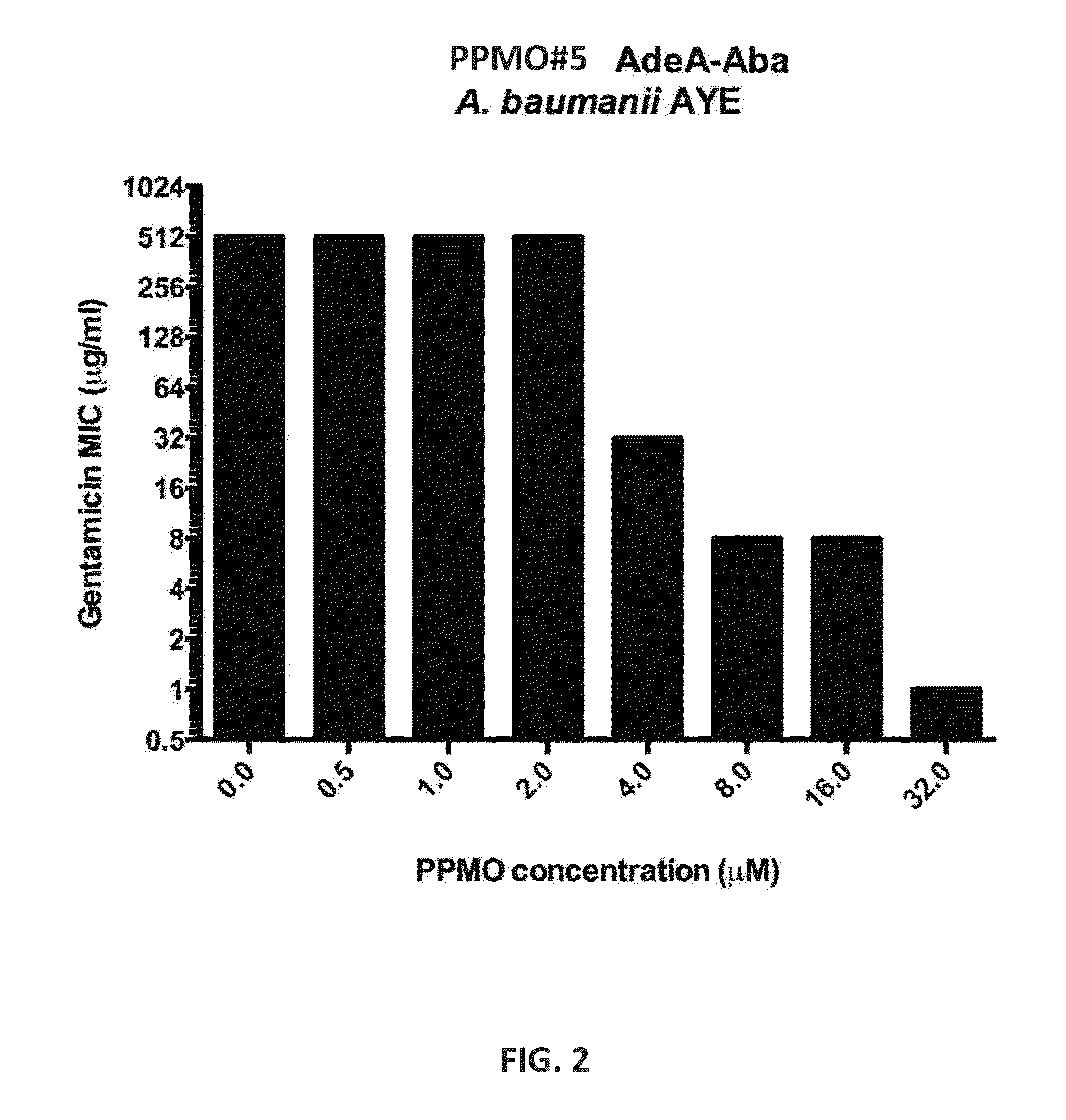
Antisense antibacterial compounds and methods
ActiveUS20150361425A1Increase antibiotic susceptibilityReduce capacityAntibacterial agentsSugar derivativesAntibiotic resistanceMorpholine
Provided are antisense morpholino oligomers targeted against bacterial virulence factors such as genes that contribute to antibiotic resistance or biofilm formation, or genes associated with fatty acid biosynthesis, and related compositions and methods of using the oligomers and compositions, for instance, in the treatment of an infected mammalian subject.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST +1
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com