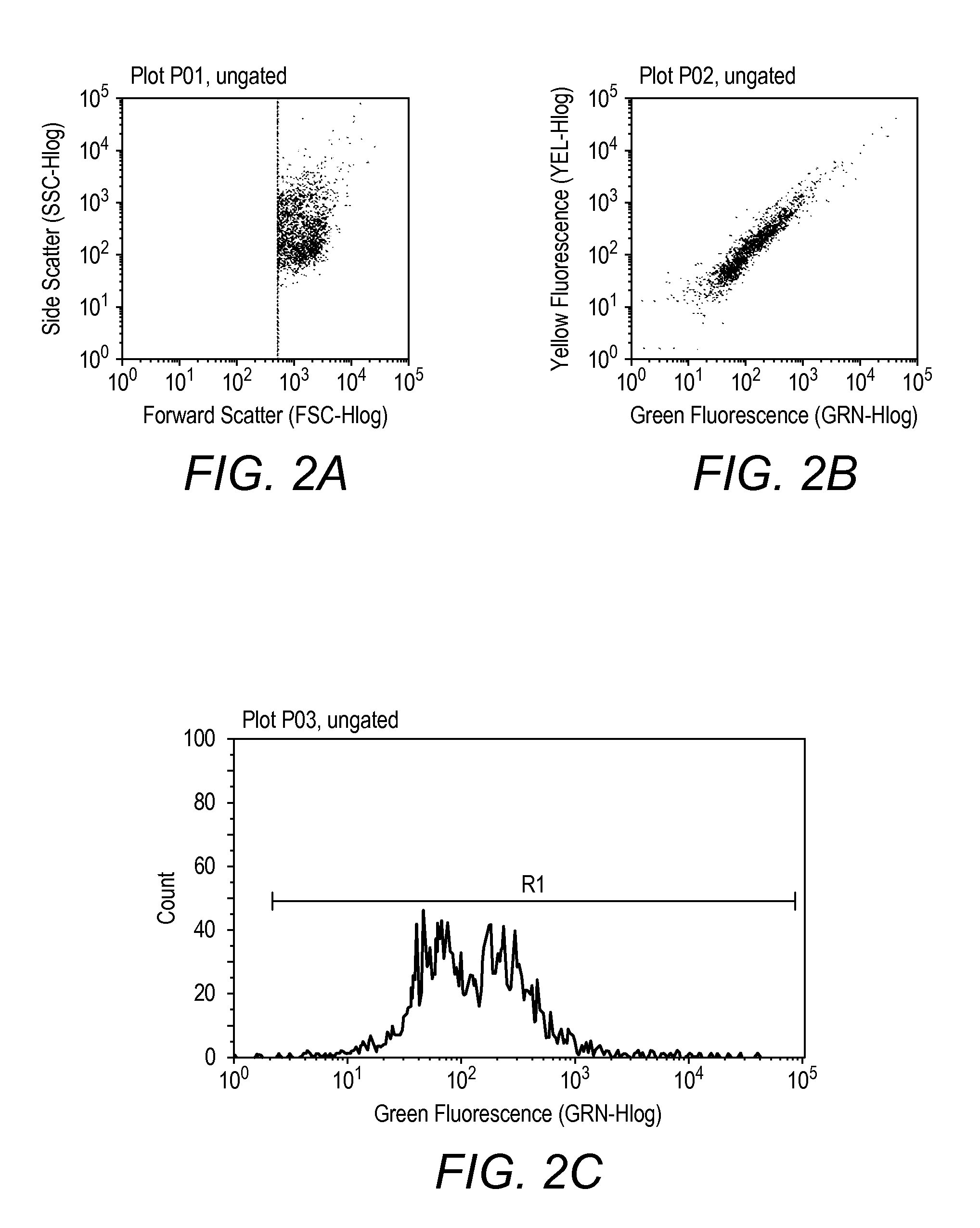
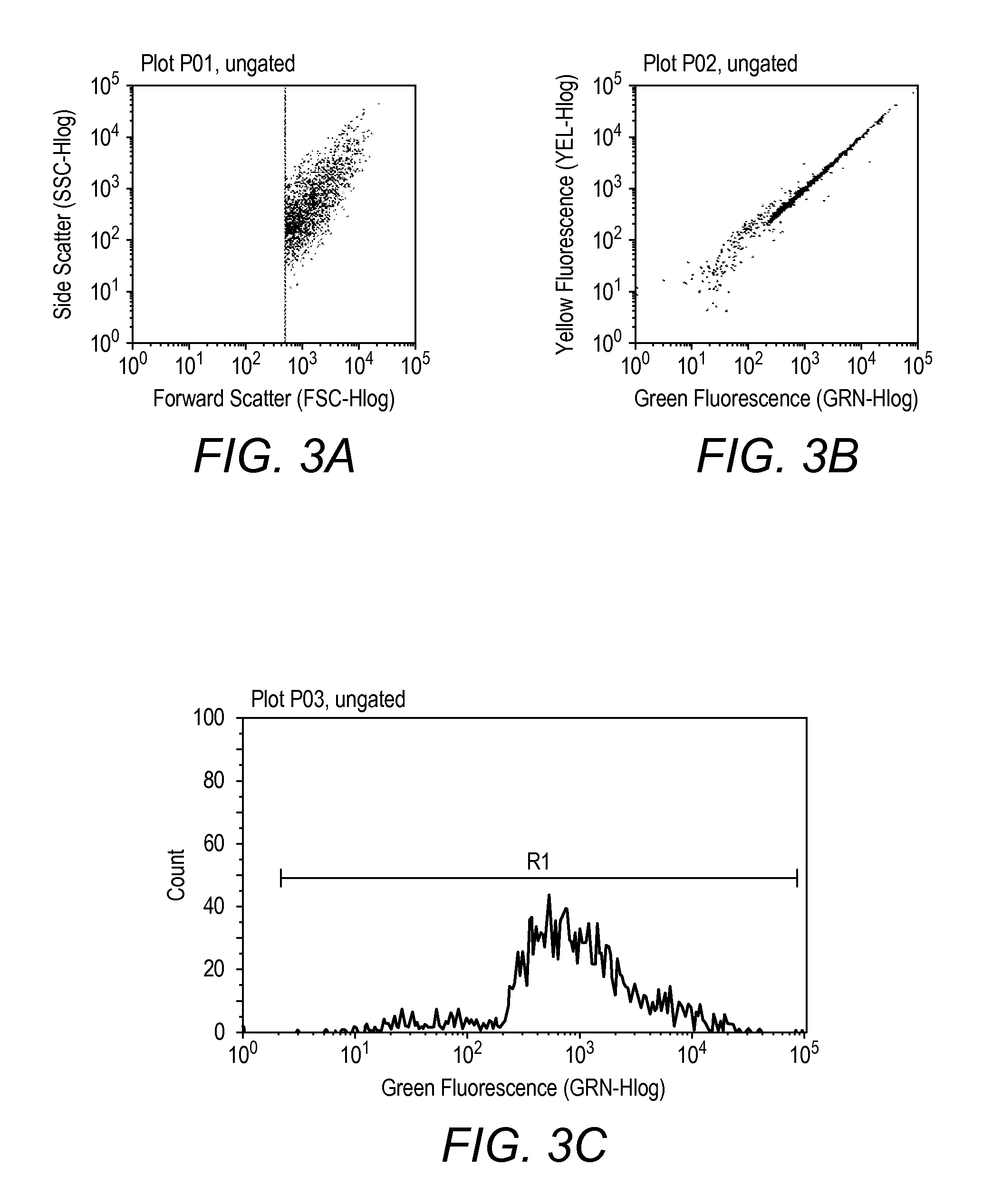
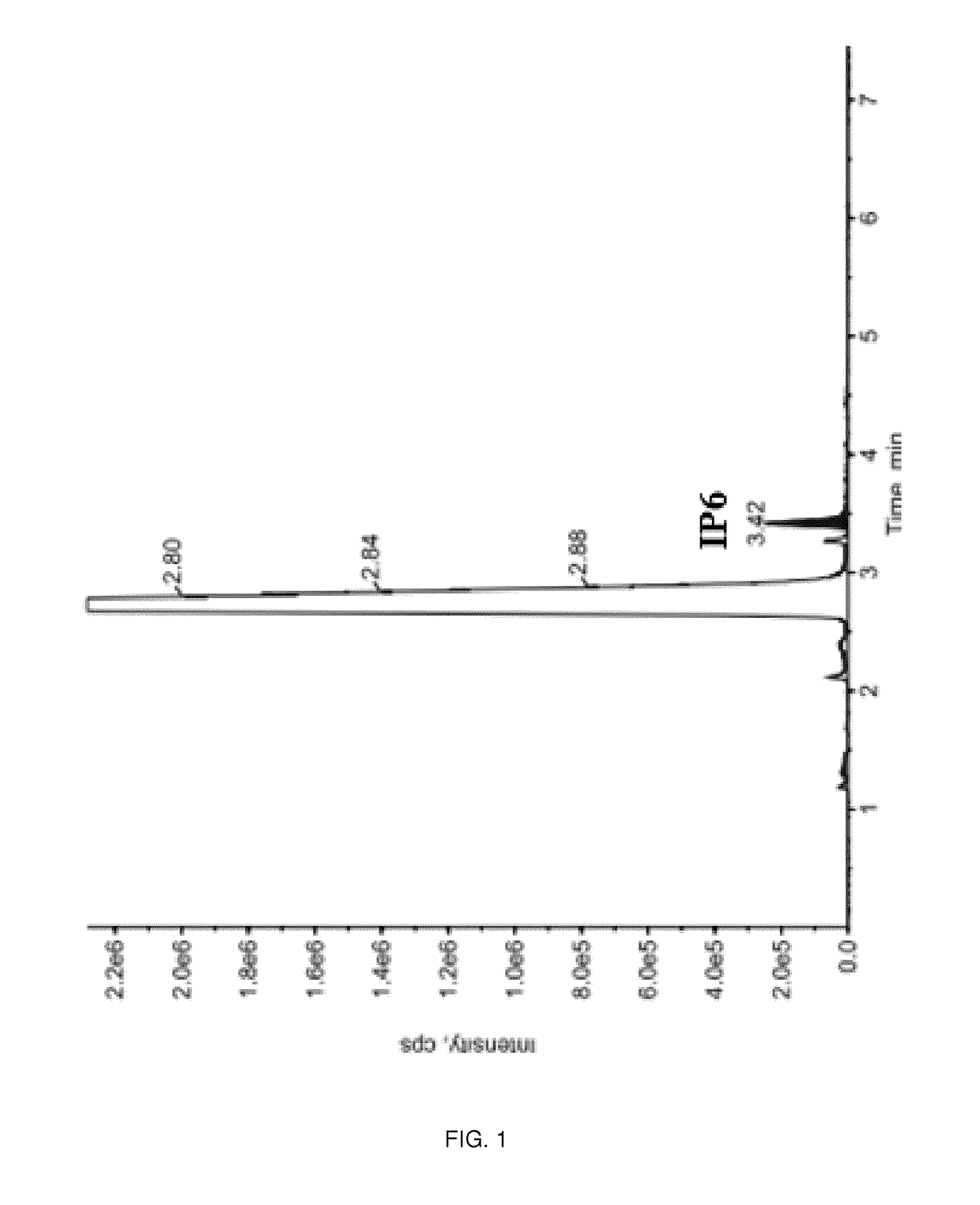
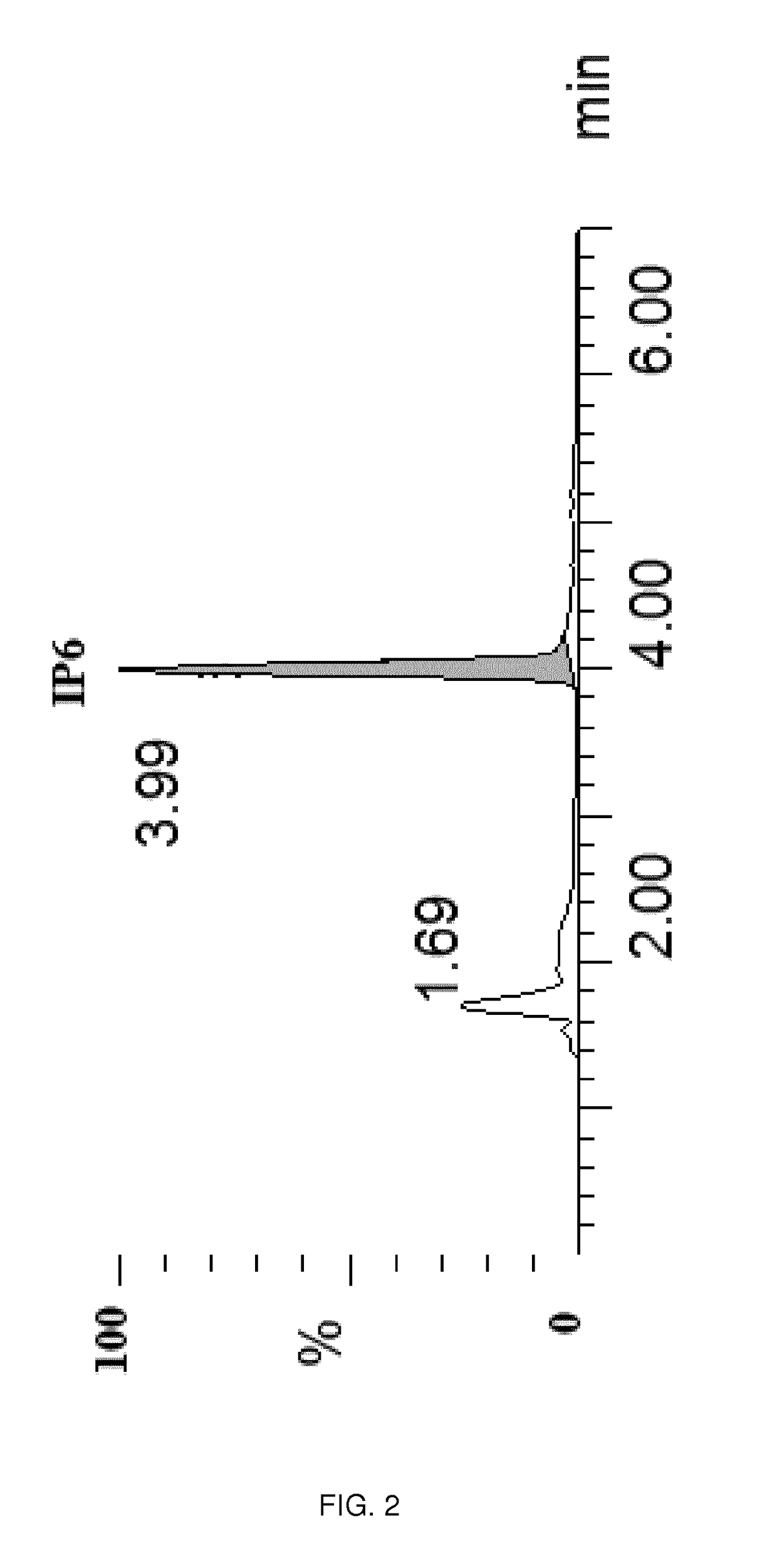
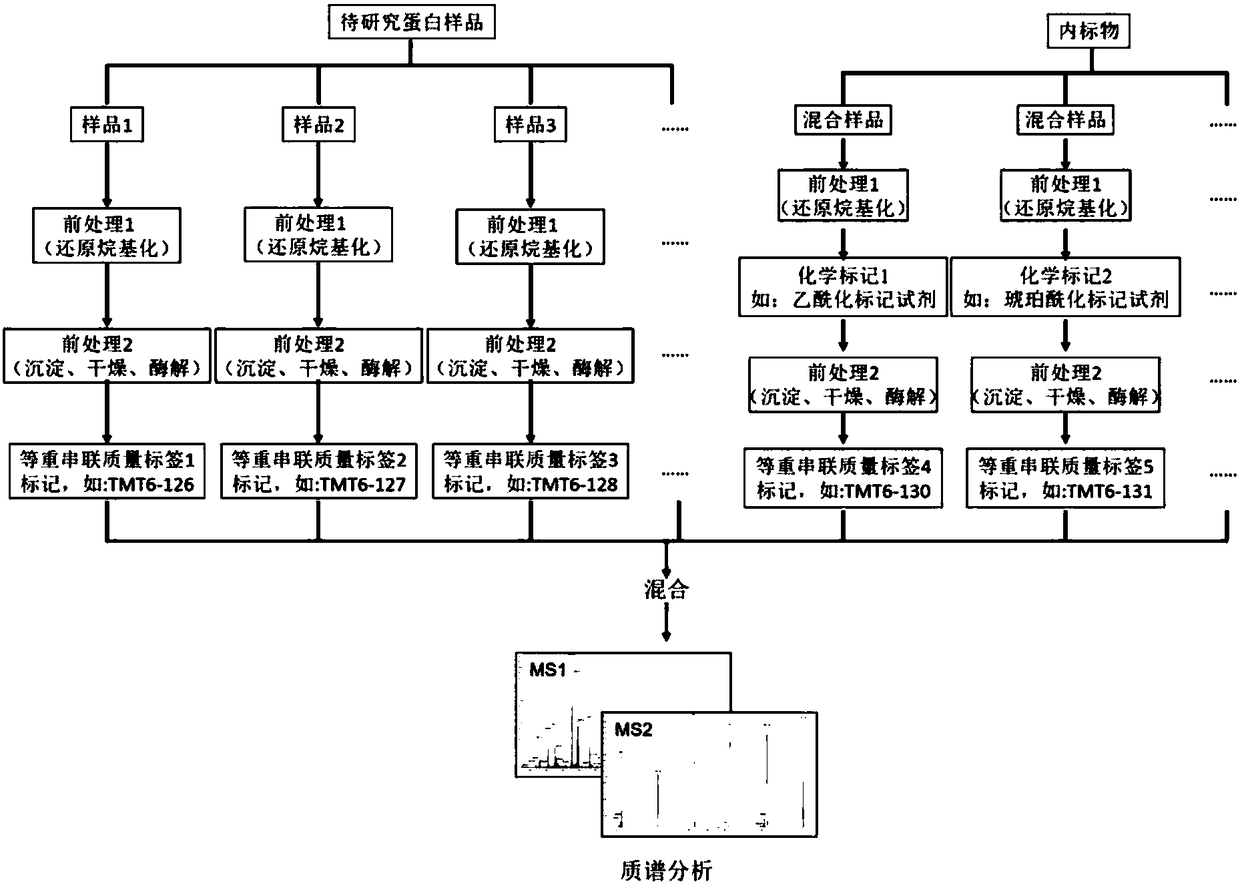
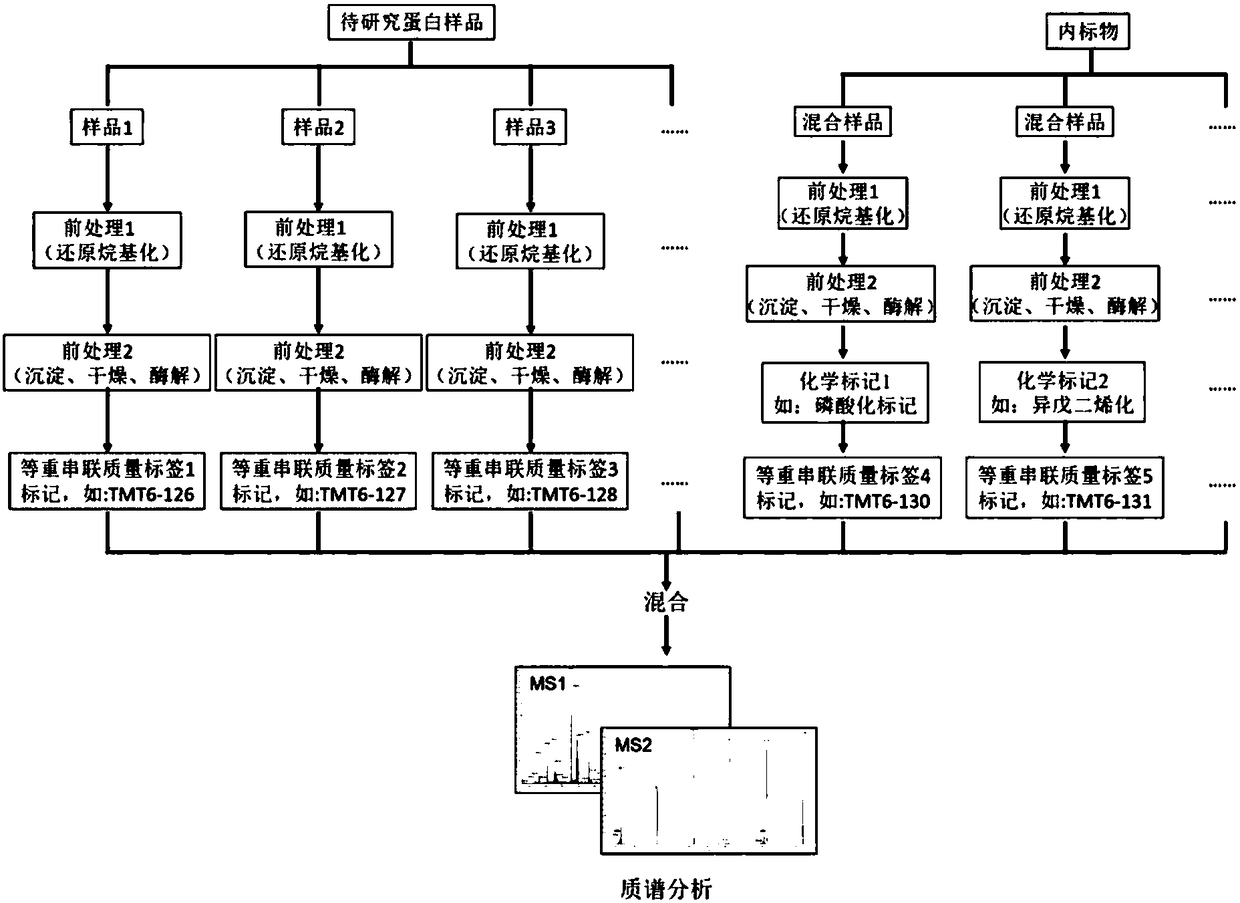
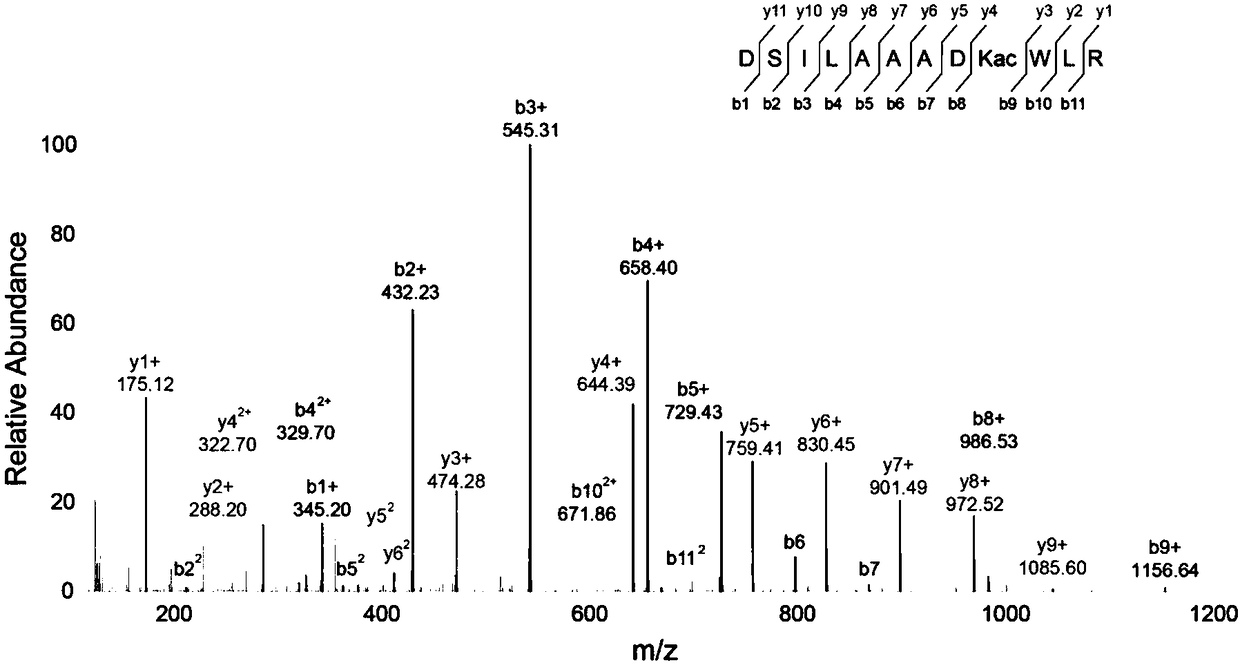
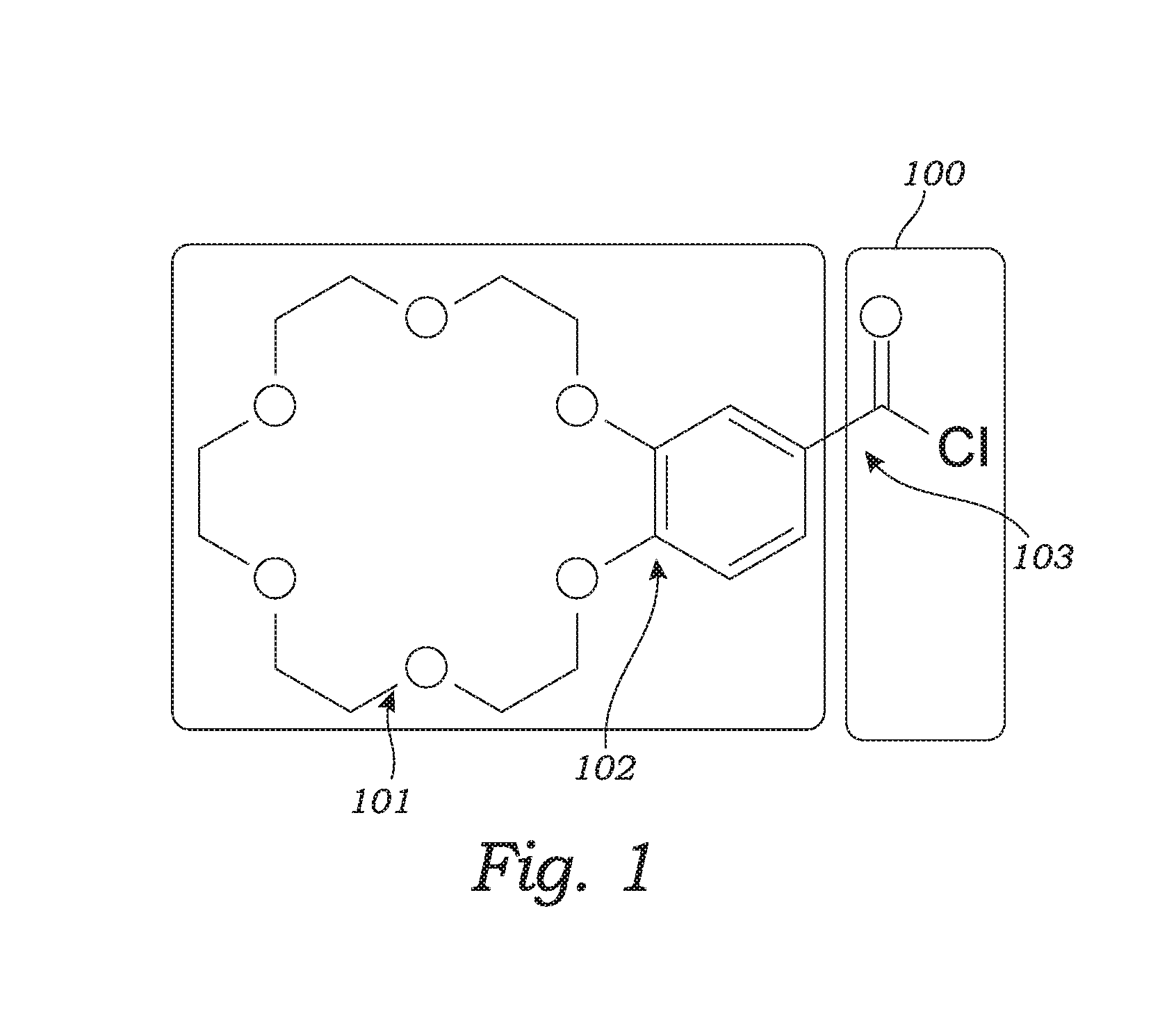
Patents
Literature
644results about "Mass spectrometric analysis" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
High sensitivity quantitation of peptides by mass spectrometry
InactiveUS20040072251A1Loss of substantial specific binding capacityReduce complexitySamplingComponent separationChemical structureProtein target
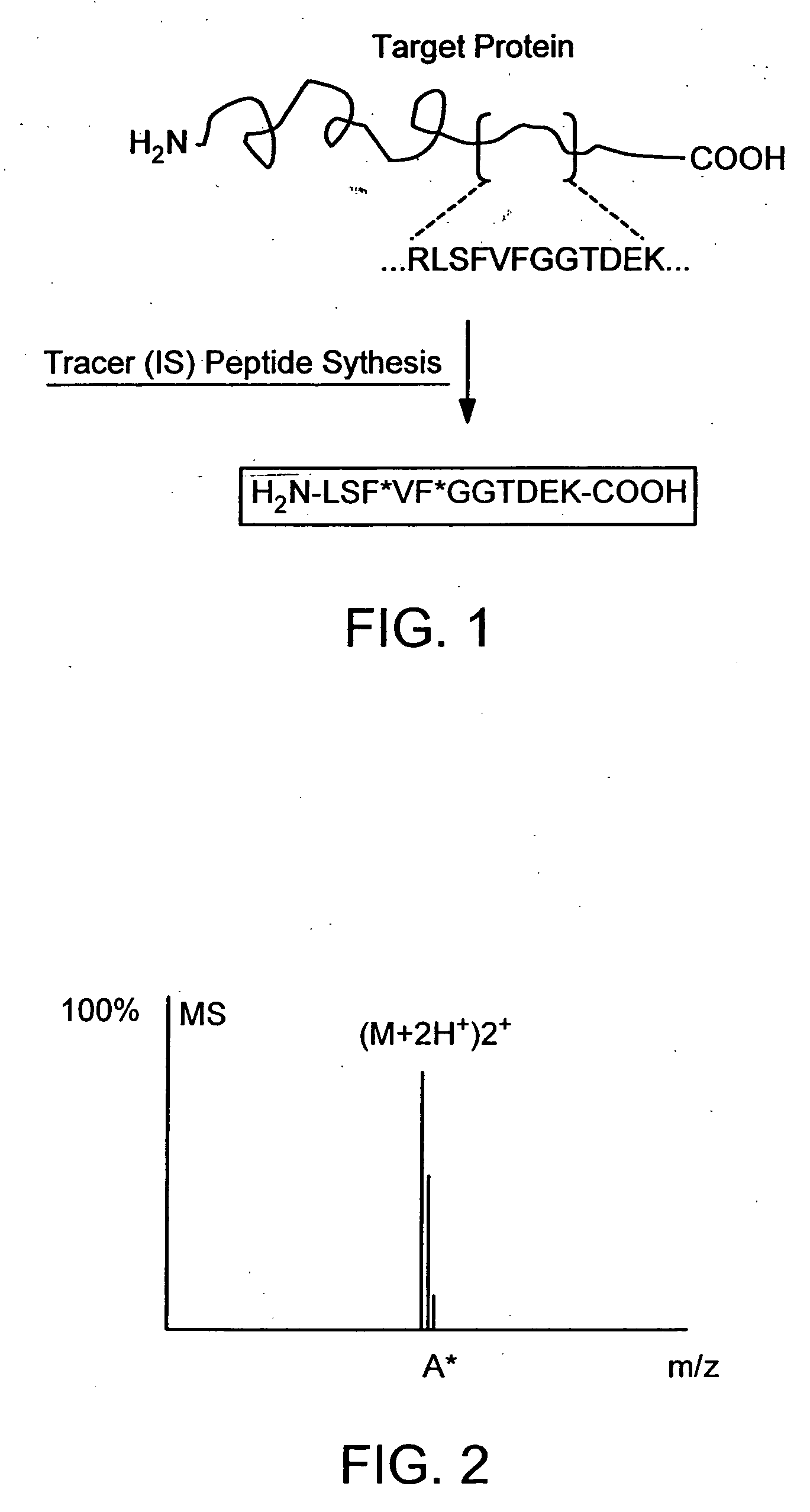
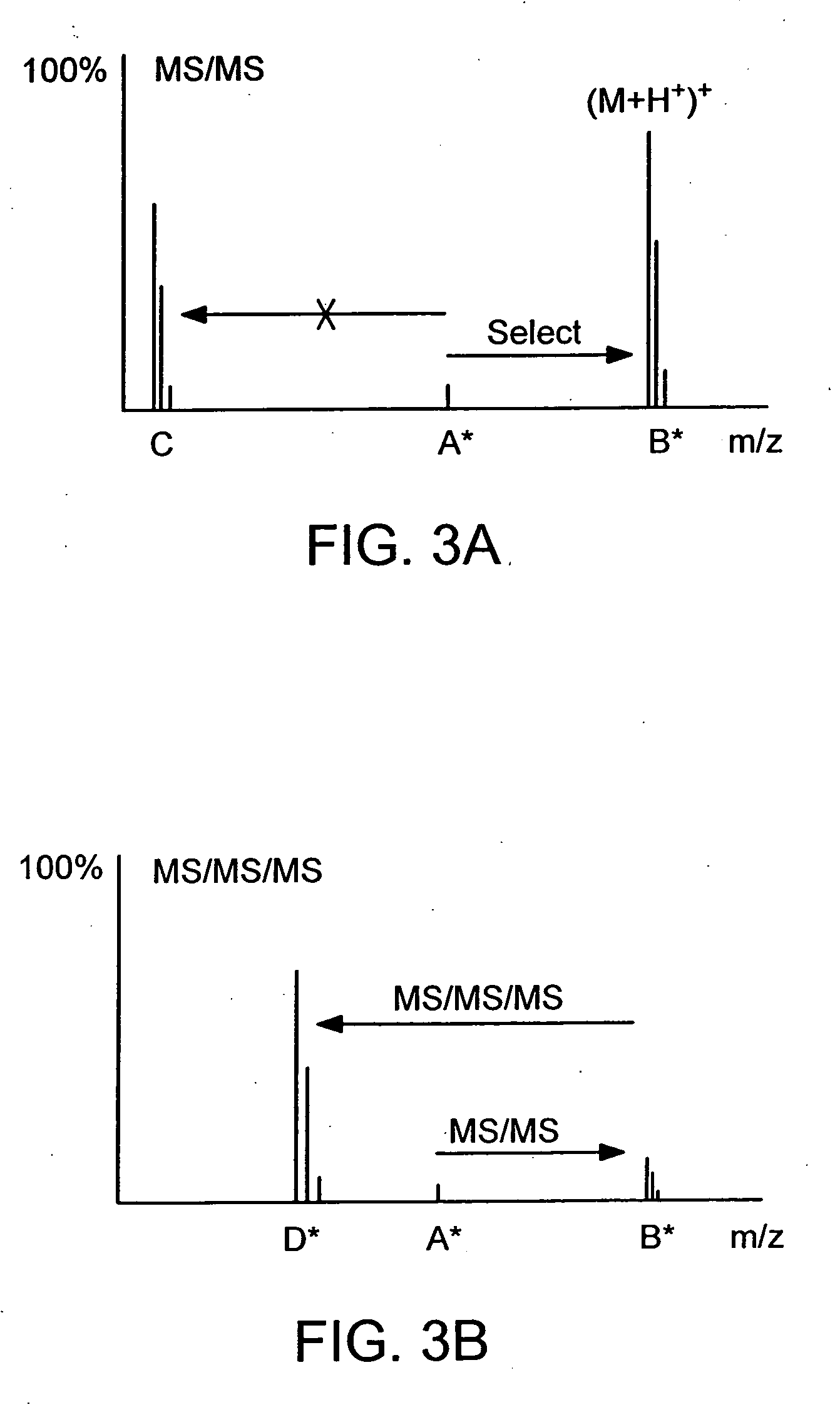
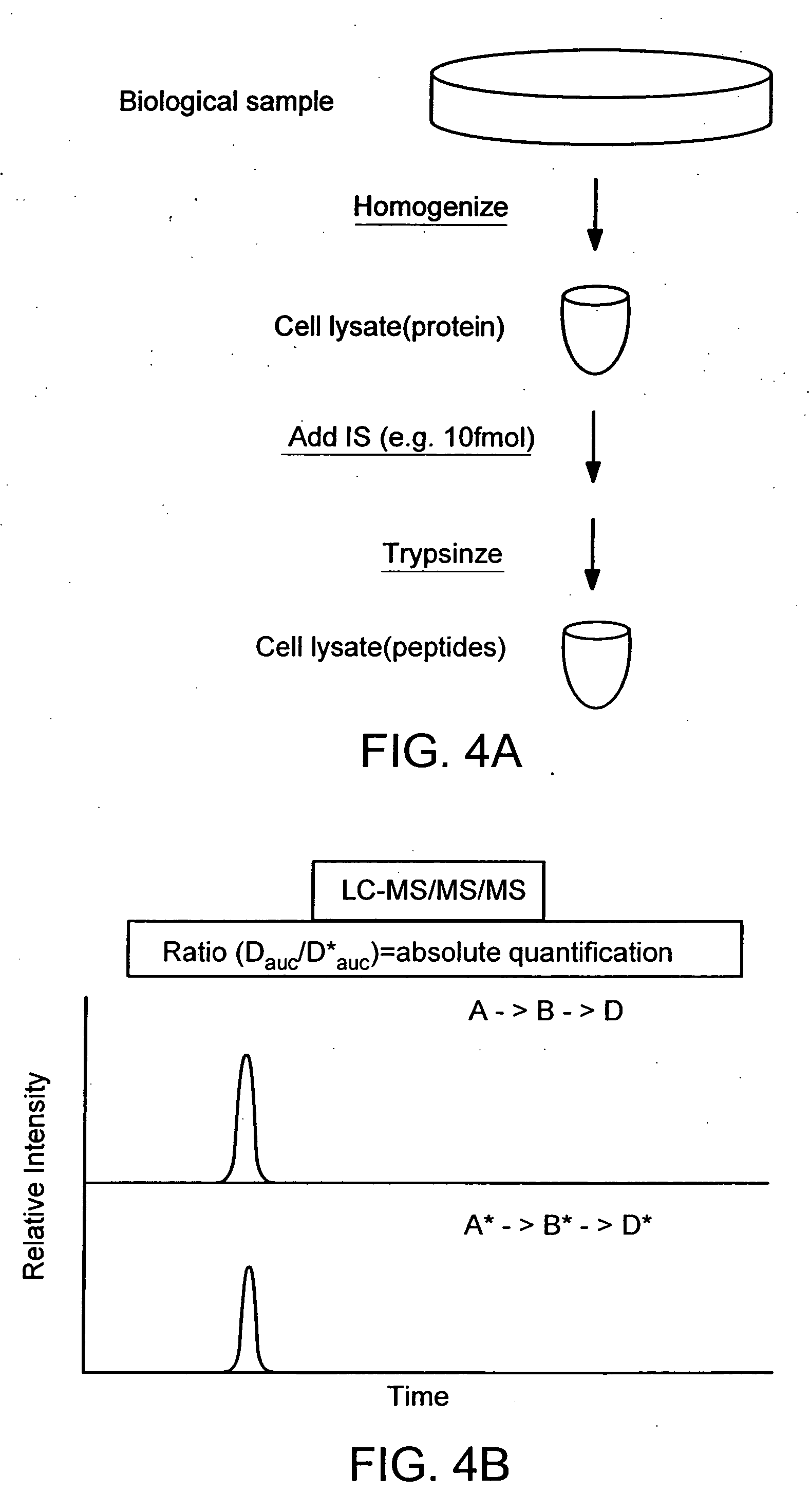
The instant invention provides an economical flow-through method for determining amount of target proteins in a sample. An antibody preparation (whether polyclonal or monoclonal, or any equivalent specific binding agent) is used to capture and thus enrich a specific monitor peptide (a specific peptide fragment of a protein to be quantitated in a proteolytic digest of a complex protein sample) and an internal standard peptide (the same chemical structure but including stable isotope labels). Upon elution into a suitable mass spectrometer, the natural (sample derived) and internal standard (isotope labeled) peptides are quantitated, and their measured abundance ratio used to calculate the abundance of the monitor peptide, and its parent protein, in the initial sample
Owner:ANDERSON FORSCHUNG GROUP
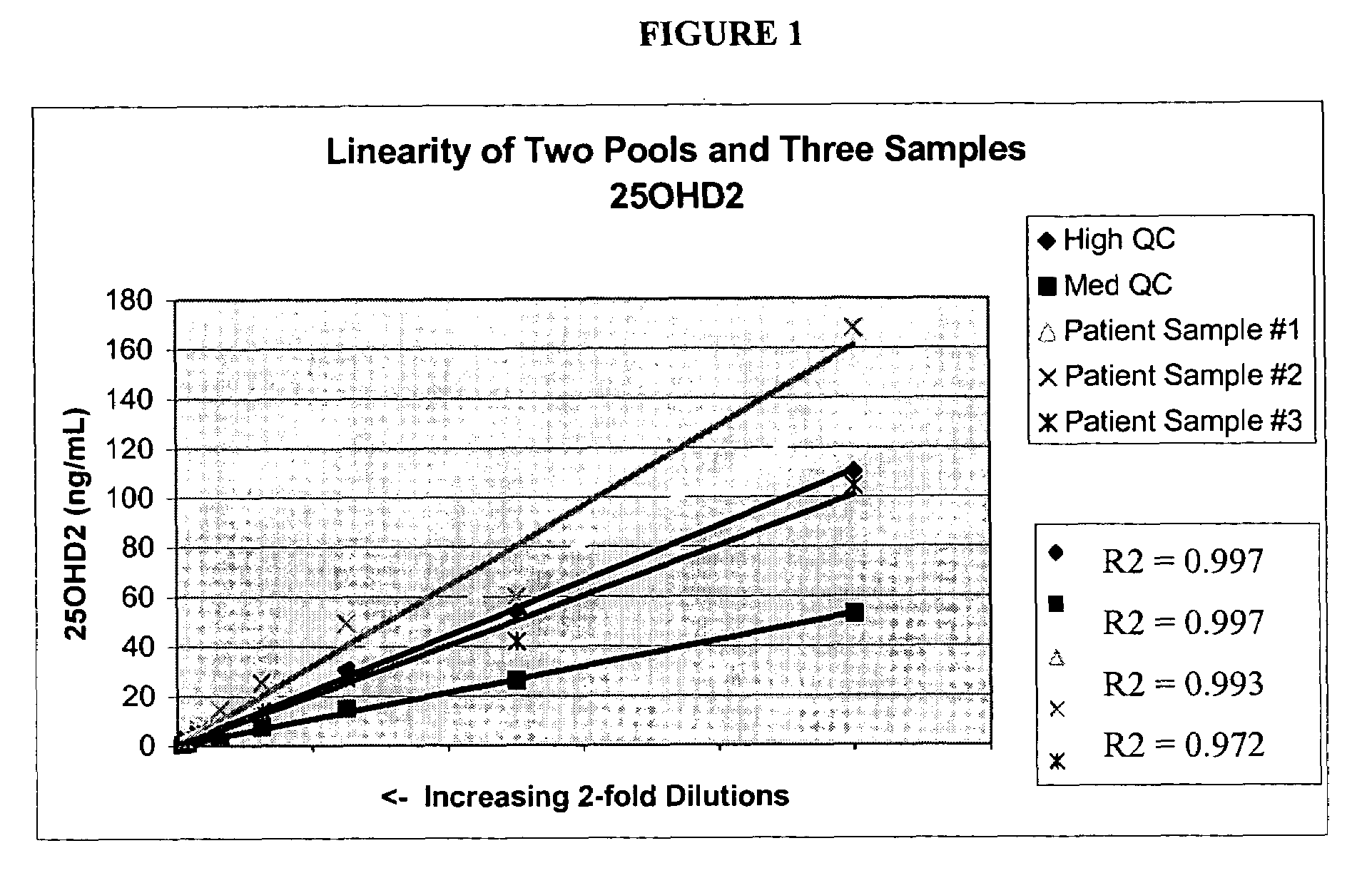
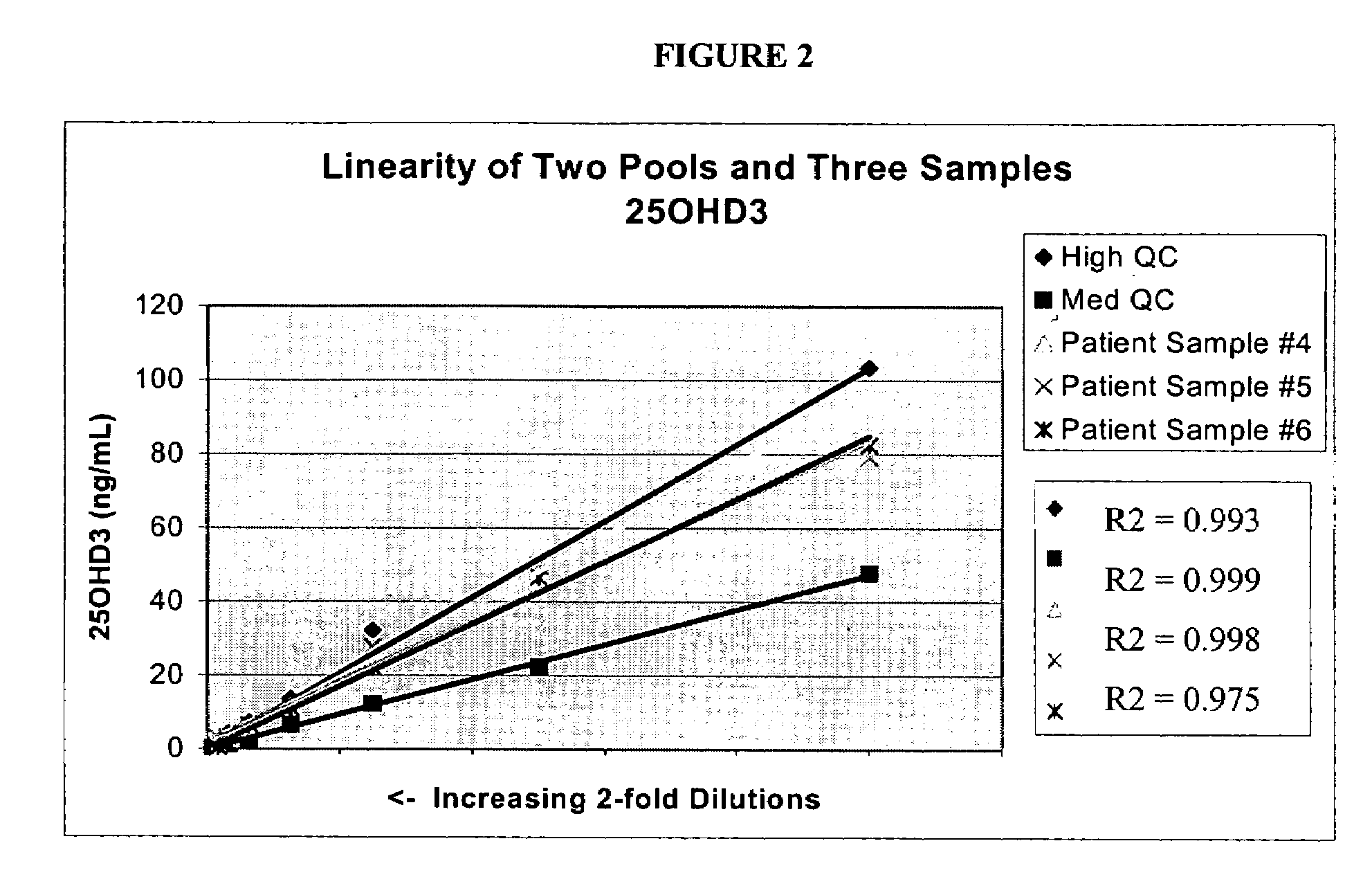
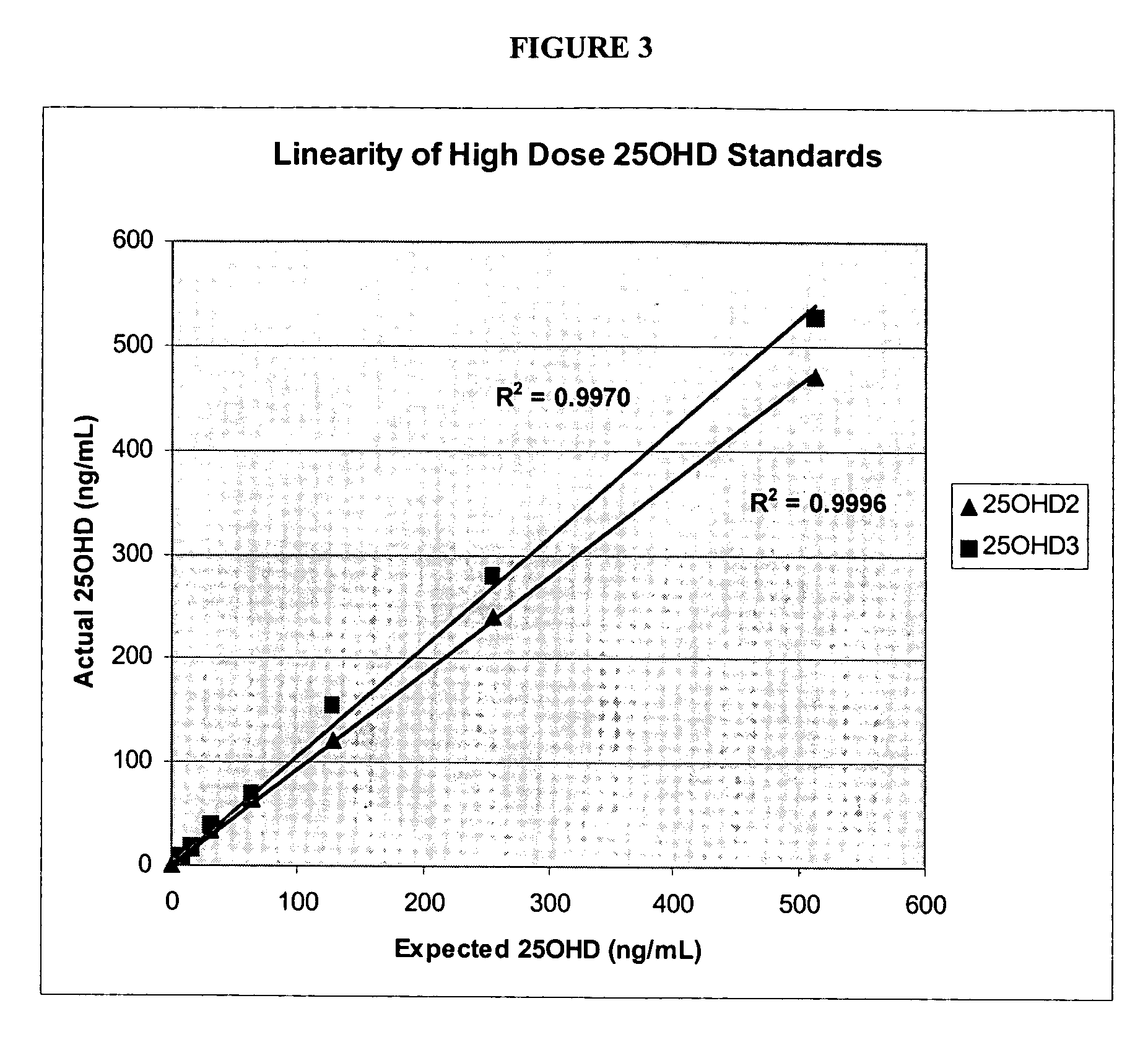
Methods for detecting vitamin D metabolites
ActiveUS20060228808A1Increased turbulenceEasy to separateComponent separationMass spectrometric analysisMetaboliteMass Spectrometry-Mass Spectrometry
Provided are methods of detecting the presence or amount of a vitamin D metabolite in a sample using mass spectrometry. The methods generally comprise ionizing a vitamin D metabolite in a sample and detecting the amount of the ion to determine the presence or amount of the vitamin D metabolite in the sample. Also provided are methods to detect the presence or amount of two or more vitamin D metabolites in a single assay.
Owner:QUEST DIAGNOSTICS INVESTMENTS INC
High sensitivity quantitation of peptides by mass spectrometry
InactiveUS7632686B2Reduce complexityOverwhelm resolutionSamplingComponent separationChemical structureProtein target
The instant invention provides an economical flow-through method for determining amount of target proteins in a sample. An antibody preparation (whether polyclonal or monoclonal, or any equivalent specific binding agent) is used to capture and thus enrich a specific monitor peptide (a specific peptide fragment of a protein to be quantitated in a proteolytic digest of a complex protein sample) and an internal standard peptide (the same chemical structure but including stable isotope labels). Upon elution into a suitable mass spectrometer, the natural (sample derived) and internal standard (isotope labeled) peptides are quantitated, and their measured abundance ratio used to calculate the abundance of the monitor peptide, and its parent protein, in the initial sample.
Owner:ANDERSON FORSCHUNG GROUP
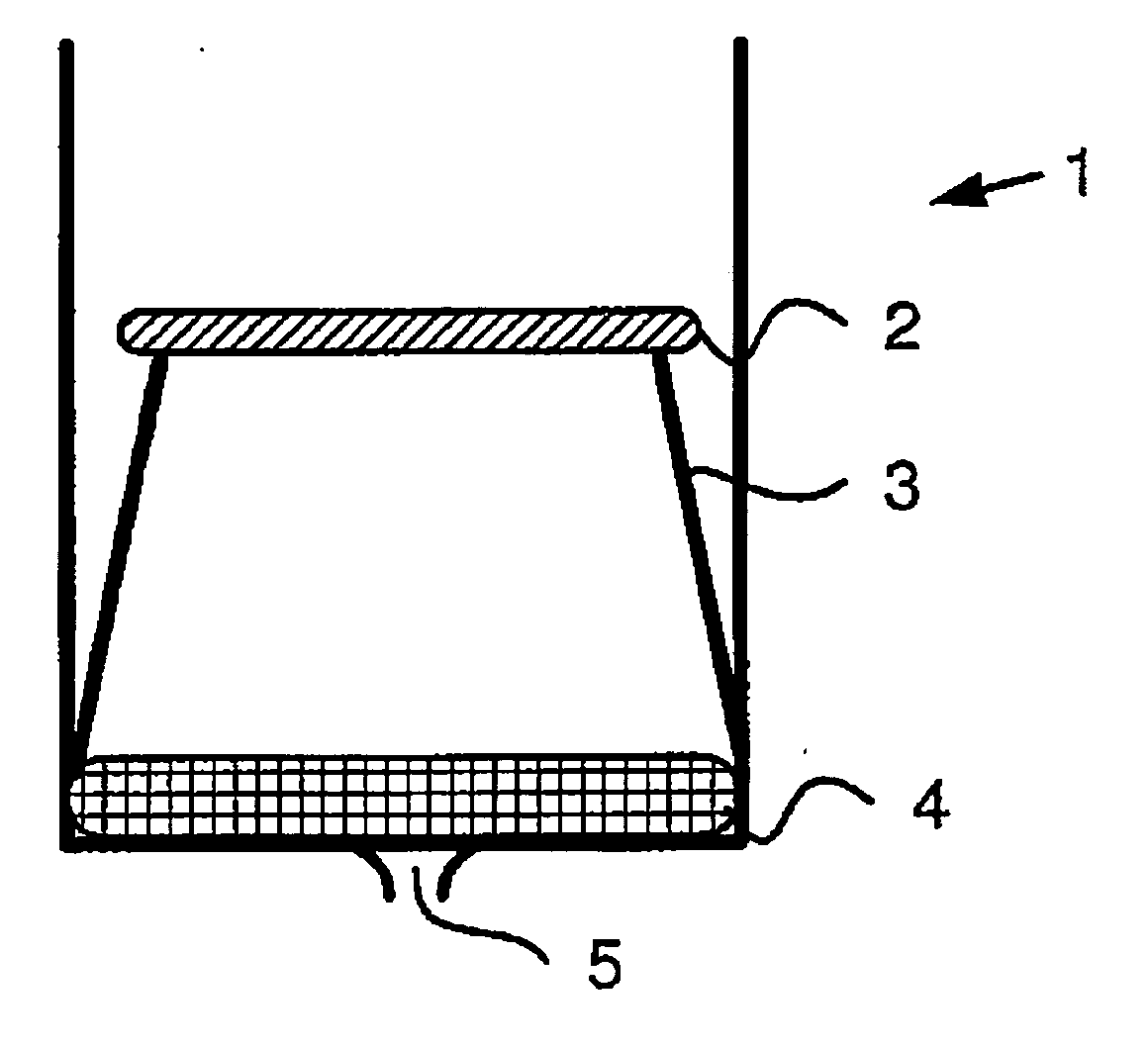
Device for quantitative analysis of a drug or metabolite profile
ActiveUS20070003965A1Improve efficiencyImprove reliabilityBioreactor/fermenter combinationsBiological substance pretreatmentsMedicineInternal standard
A device, in particular a sample preparation device for the quantitative analysis of a drug and / or metabolite profile in a biological sample, which includes an insert for such a device being impregnated with at least one internal standard, to the internal standard itself, and to a kit comprising the device. Further, the invention also relates to an apparatus containing the device, and to a method for the quantitative analysis of a drug and / or metabolite profile in a biological sample employing the device.
Owner:BIOCRATES LIFE SCIENCES AG
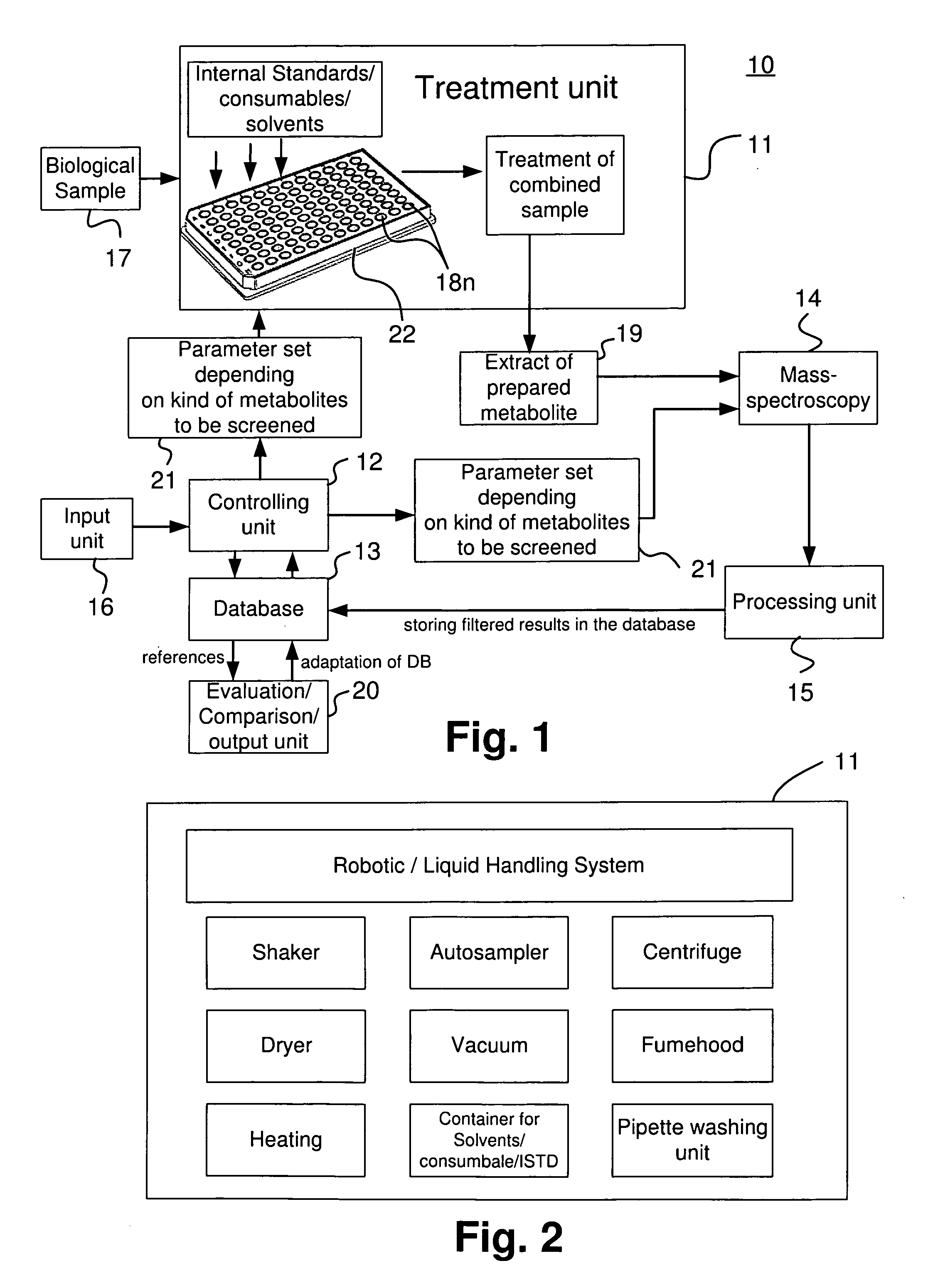
Apparatus and method for analyzing a metabolite profile
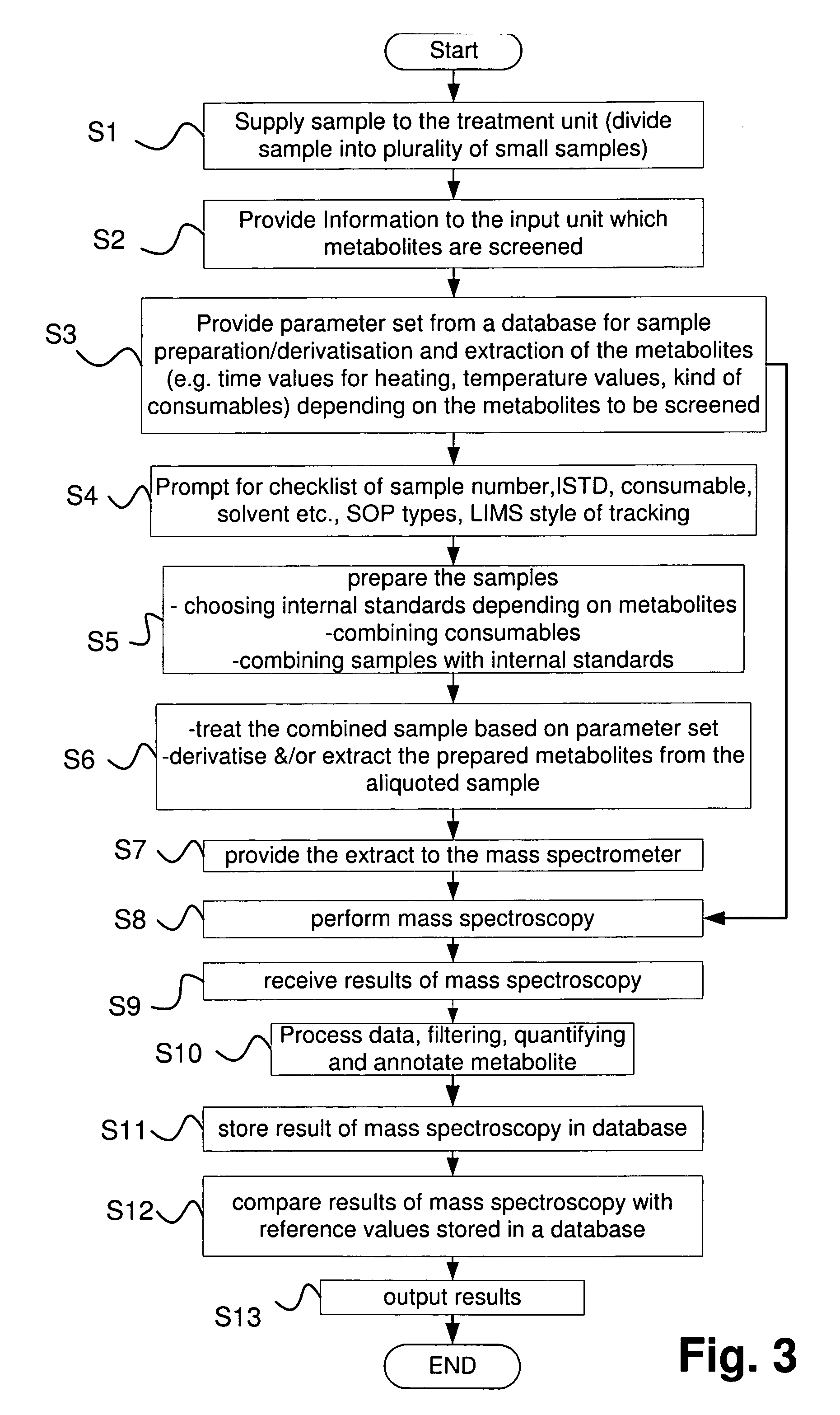
ActiveUS20070004044A1Reduce the amount requiredEasy to analyze resultsParticle spectrometer methodsMass spectrometric analysisMedicineMass Spectrometry-Mass Spectrometry
An apparatus and method analyzes a metabolic profile in a biological sample. The apparatus includes an input unit for inputting the drugs and / or metabolites to be screened; a controlling unit for determining a parameter set for metabolites preparation and for mass spectrometry analysis depending on the input of the kind of metabolites to be screened; a treatment unit for preparing the metabolites to be screened depending on the determined parameter set; a mass spectrometer for performing mass spectrometry analysis on prepared metabolites depending on the parameter set; a database for storing results of analyzing and parameter sets for metabolite preparation and for mass spectrometry analyses; and an evaluation unit for evaluating the results of mass spectrometry by use of reference results stored in the database to output an analysis of the metabolites profile.
Owner:BIOCRATES LIFE SCIENCES AG
Selected Reaction Monitoring Assays
ActiveUS20130203096A1Minimal interferenceEarly detectionMicrobiological testing/measurementMass spectrometric analysisMass Spectrometry-Mass SpectrometrySelected reaction monitoring
Provided herein are methods for developing selected reaction monitoring mass spectrometry (LC-SRM-MS) assays.
Owner:BIODESIX
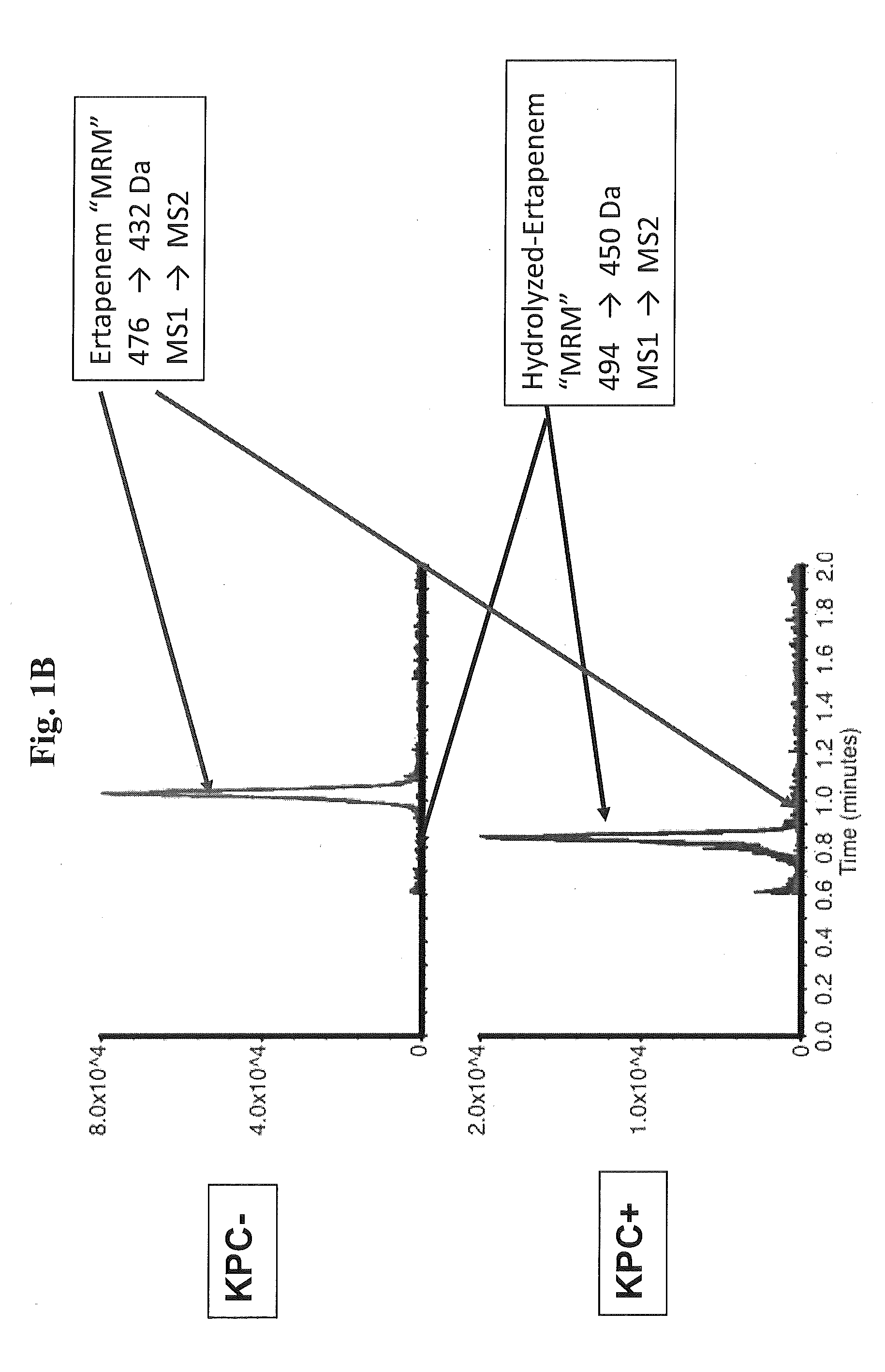
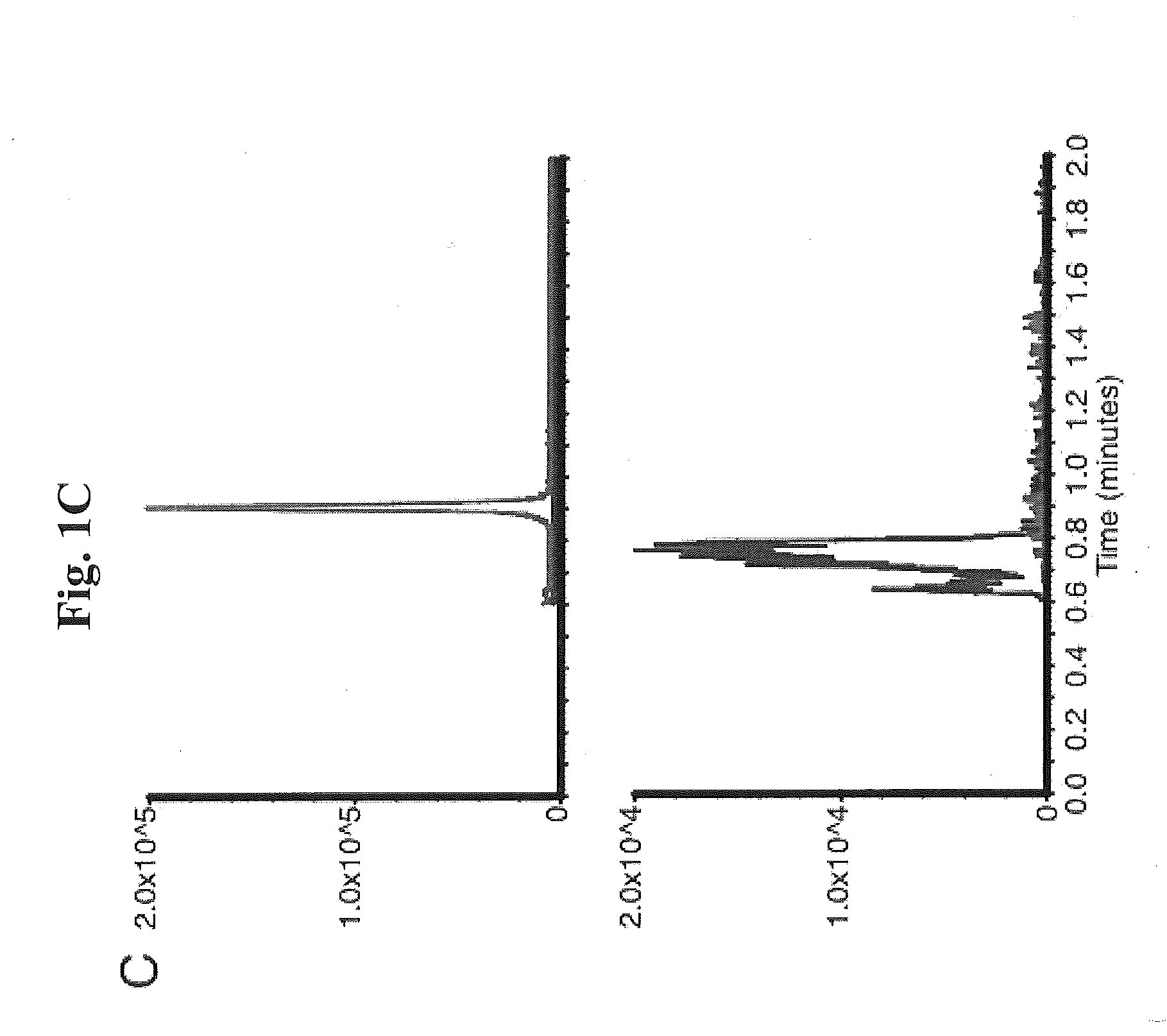
Methods and Kits for Detection of Antibiotic Resistance
InactiveUS20120196309A1Promotes bacterial resistanceMicrobiological testing/measurementMass spectrometric analysisResistant bacteriaMass Spectrometry-Mass Spectrometry
The present invention relates to a method of detecting antibiotic resistant bacteria in a sample. The method includes the steps of analyzing a sample derived from bacteria via mass spectrometry to produce a data set, and determining from the data set the presence or absence of a covalently modified antibiotic compound in the sample, wherein the presence of a covalently modified antibiotic compound in the sample is indicative that the bacteria are resistant to the antibiotic. The present invention also relates to a kit for determining the presence or absence of antibiotic resistant bacteria in a sample. The kit includes reagents for preparing and performing the assay, and instructions for the set-up, performance, monitoring, and interpretation of the assay.
Owner:YALE UNIV
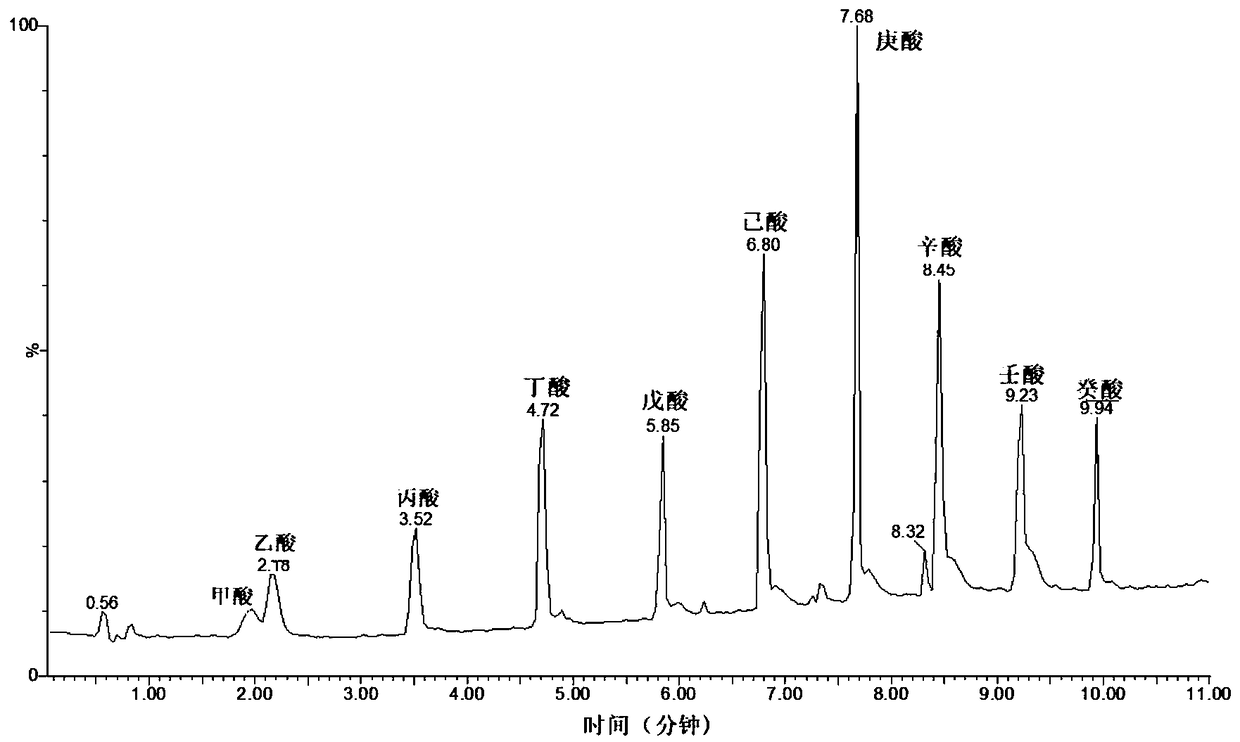
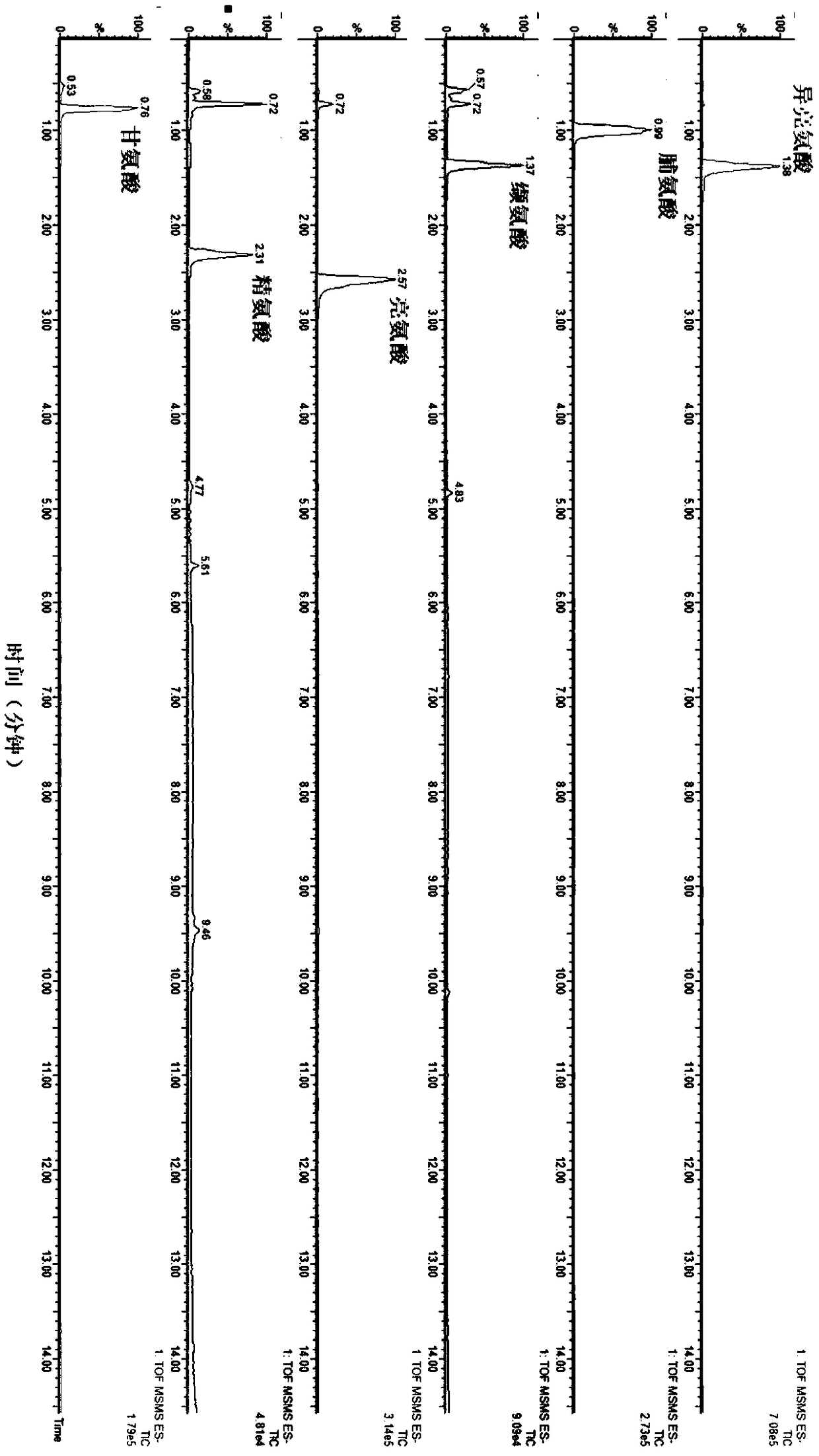
Quantitative detecting method for various metabolites in biological sample, and metabolic chip
ActiveCN109298115AFast contentContent quicklyComponent separationMass spectrometric analysisMetaboliteDerivatization
The invention discloses a quantitative detecting method for various metabolic components in a biological sample, and a metabolic chip used for the quantitative detecting method. The quantitative detecting method is characterized in that the biological sample is subjected to derivatization treatment, and the biological sample subjected to derivatization is detected through liquid chromatography andmass spectrometry combination; and during derivatization treatment, 3-nitrophenylhydrazine serves as a derivatization reagent, and 1-(3-dimethylaminopropyl)-3-ethly carbodiimide serves as a derivatization reaction catalyst. According to the quantitative detecting method, high-sensitivity detection can be achieved, and a plurality of level-crossing metabolic components can be covered during detection, and the quantitative detecting method is simple, fast and suitable for being applied to clinical and scientific research testing. The metabolic chip comprises a chip carrier microtitration plateand related reagents, and thus the multiple level-crossing metabolites such as amino acid, phenols, phenyl or benzyl derivatives, indole, organic acid, fatty acid, sugar and bile acid in the biological samples can be subjected to quantitative detection on the same microtitration plate.
Owner:麦特绘谱生物科技(上海)有限公司
Detection and quantification of modified proteins
ActiveUS20060148093A1Rapid and high throughput analysisQuantitative precisionMicrobiological testing/measurementMass spectrometric analysisInternal standardData file
The invention provides a method detecting and quantifying proteins by mass spectrophotometric analysis using peptide internal standards and provides a highly sensitive way of detecting protein modifications. In one aspect, the invention provides a method for determining a site of ubiquitination in a polypeptide and for evaluating ubiquitination targets in a population of polypeptides. In this way, a proteome ubiquitination map can be obtained which comprises information relating to the ubiquitination states of a plurality of cellular polypeptides. Maps can be obtained for a variety of different types of cells and cell states. For example, ubiquitination targets in normal and diseased cells can be evaluated. Preferably, the map is stored as data files in a database. Individual ubiquitinated polypeptides identified can be used to generate molecular probes diagnostic of a cell state and / or can serve as targets for agents that modulate one or more cellular processes.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
Methods for detecting vitamin D metabolites
ActiveUS7745226B2Component separationMass spectrometric analysisMass Spectrometry-Mass SpectrometryVitamin D metabolism
Provided are methods of detecting the presence or amount of a vitamin D metabolite in a sample using mass spectrometry. The methods generally comprise ionizing a vitamin D metabolite in a sample and detecting the amount of the ion to determine the presence or amount of the vitamin D metabolite in the sample. Also provided are methods to detect the presence or amount of two or more vitamin D metabolites in a single assay.
Owner:QUEST DIAGNOSTICS INVESTMENTS INC
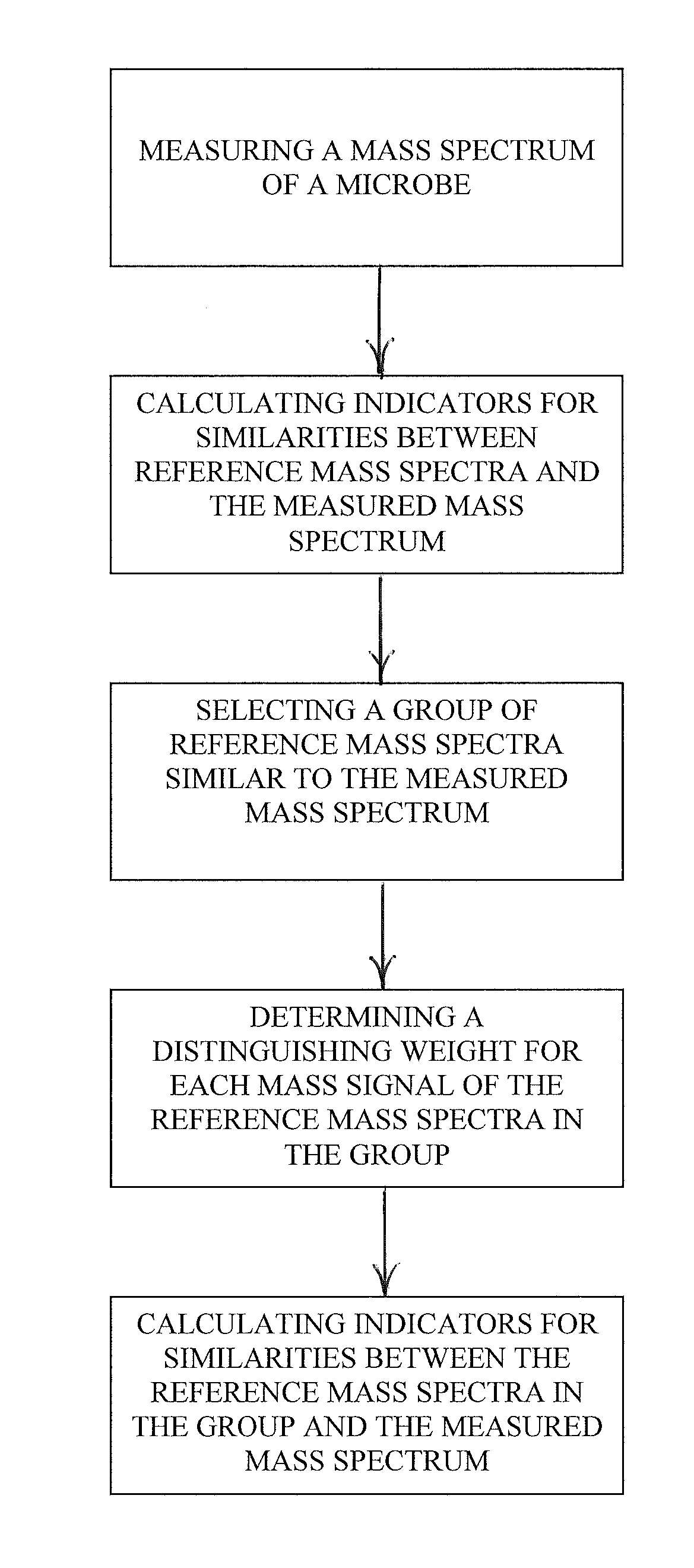
Spectrophotometric identification of microbe subspecies
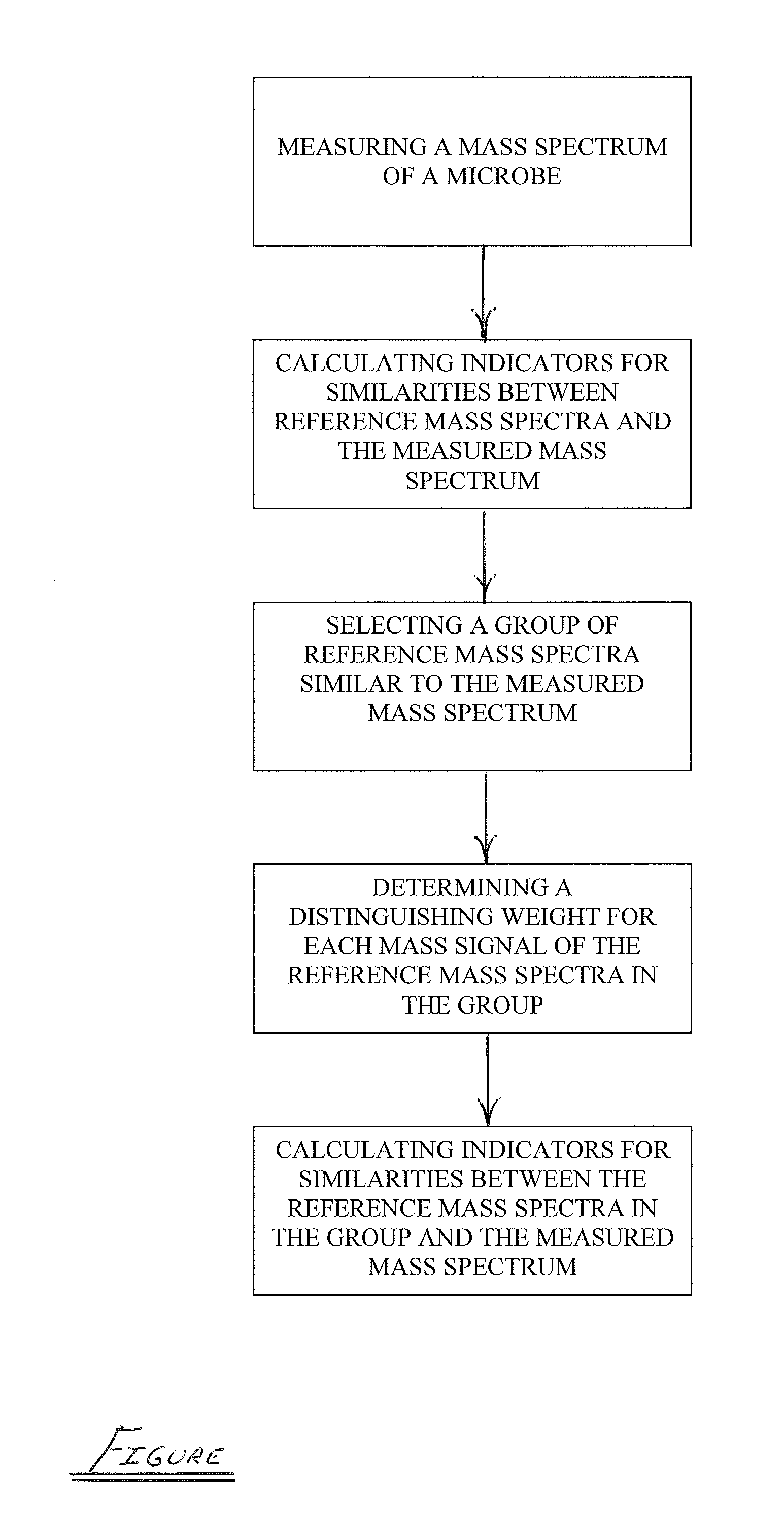
ActiveUS20110012016A1Particle separator tubesMicrobiological testing/measurementMicroorganismDual stage
A dual-stage method is provided for identifying a microbe by, for example, its species or its subspecies. The method includes measuring a mass spectrum of the microbe using a mass spectrometer, calculating indicators for similarities between reference mass spectra in a library and the measured mass spectrum, selecting a group of reference mass spectra similar to the measured mass spectrum, determining a distinguishing weight for each mass signal of the reference mass spectra in the group, where the distinguishing weights emphasize differences between the reference mass spectra in the group, and calculating indicators for similarities between the reference mass spectra in the group and the measured mass spectrum as a function of the distinguishing weights.
Owner:BRUKER DALTONIK GMBH & CO KG
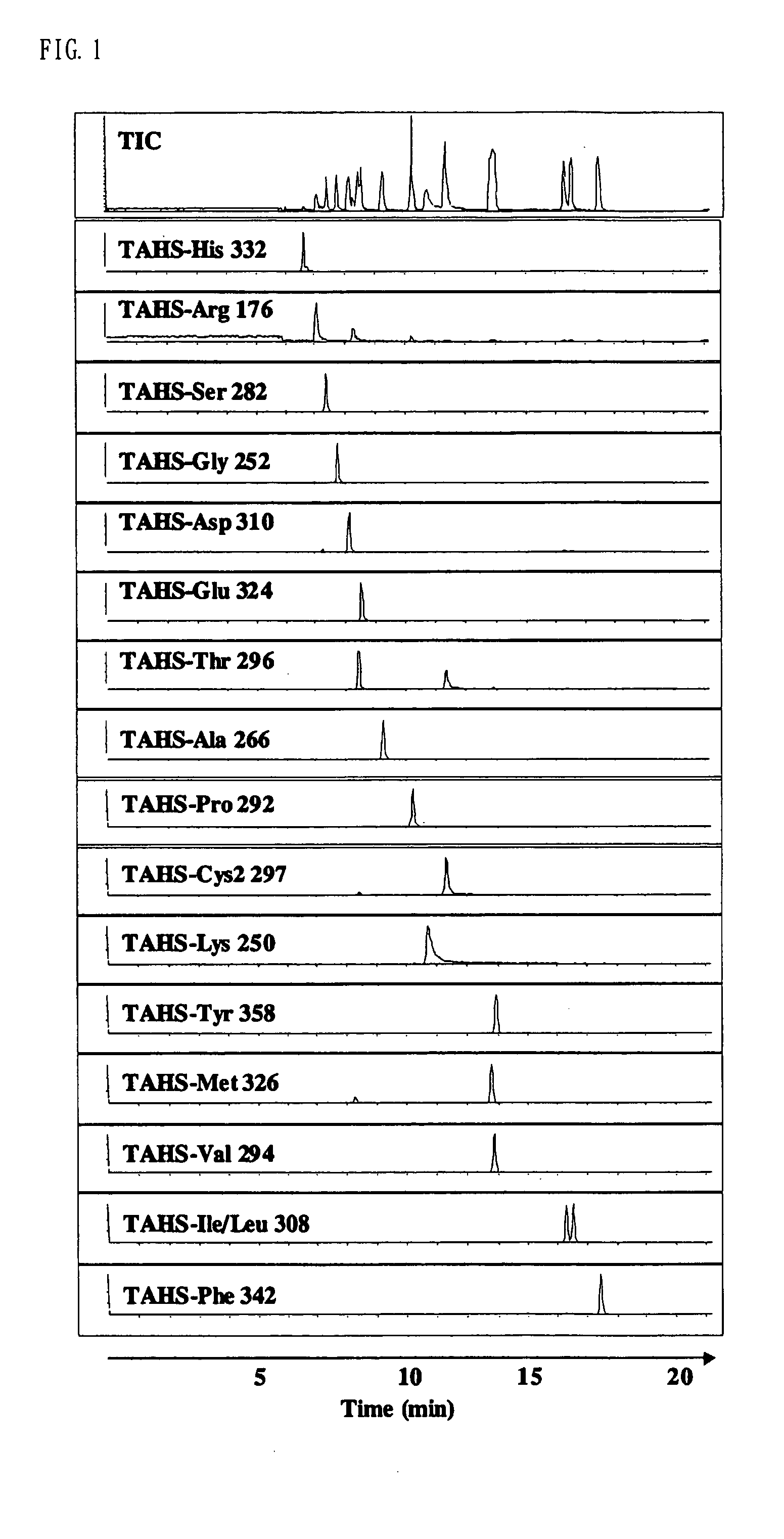
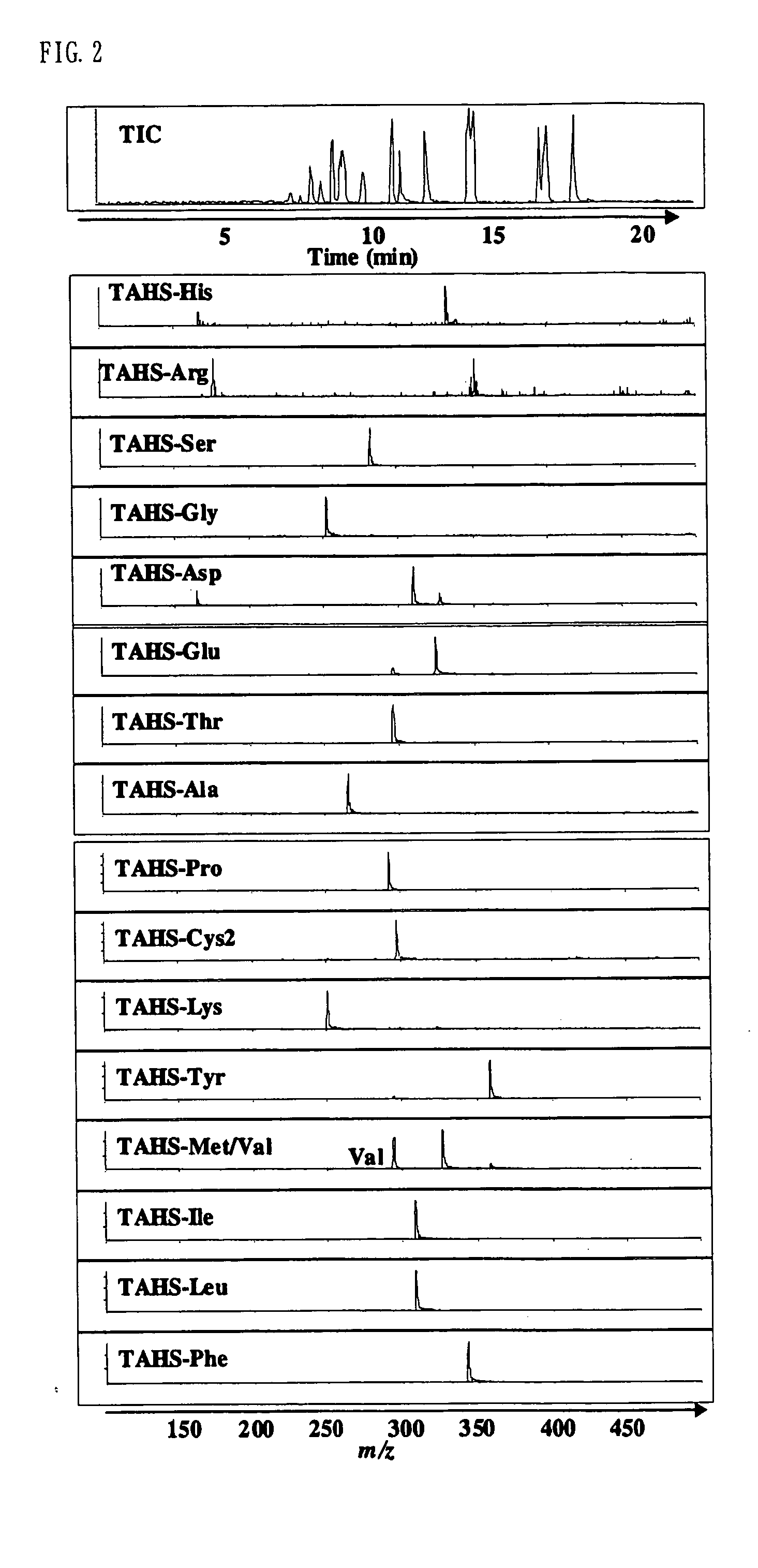
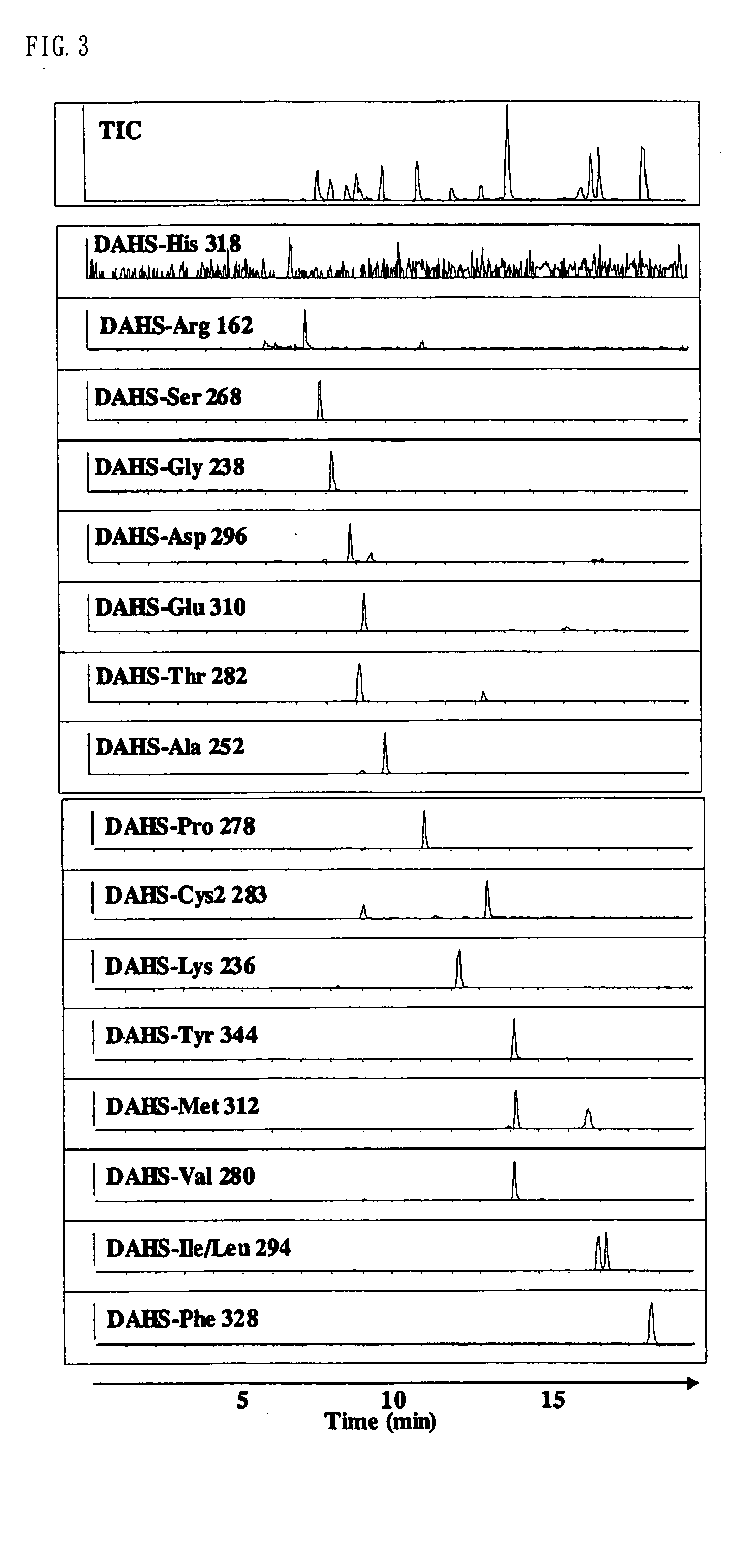
Method for analysis of compounds with amino group and analytical reagent therefor
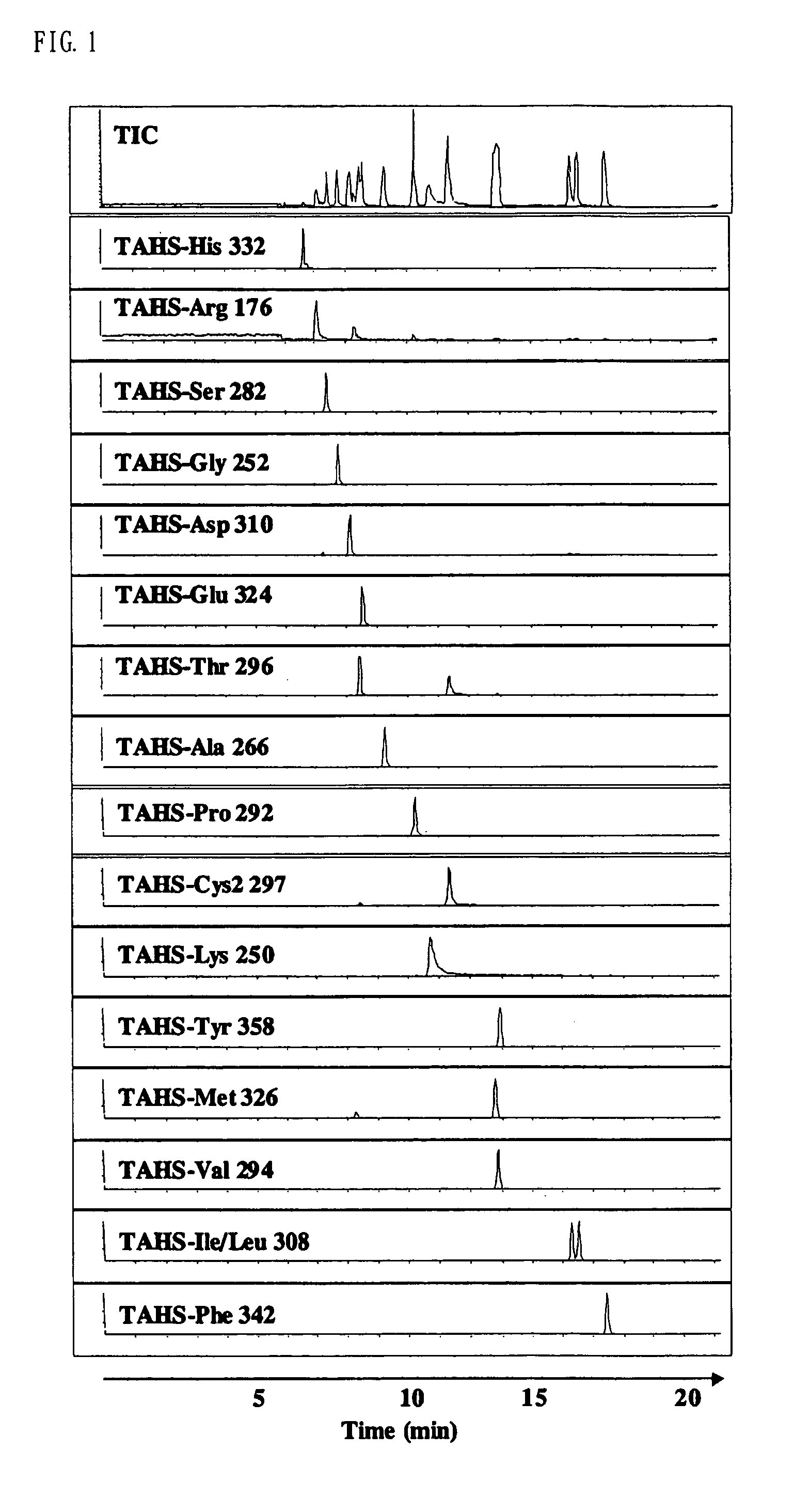
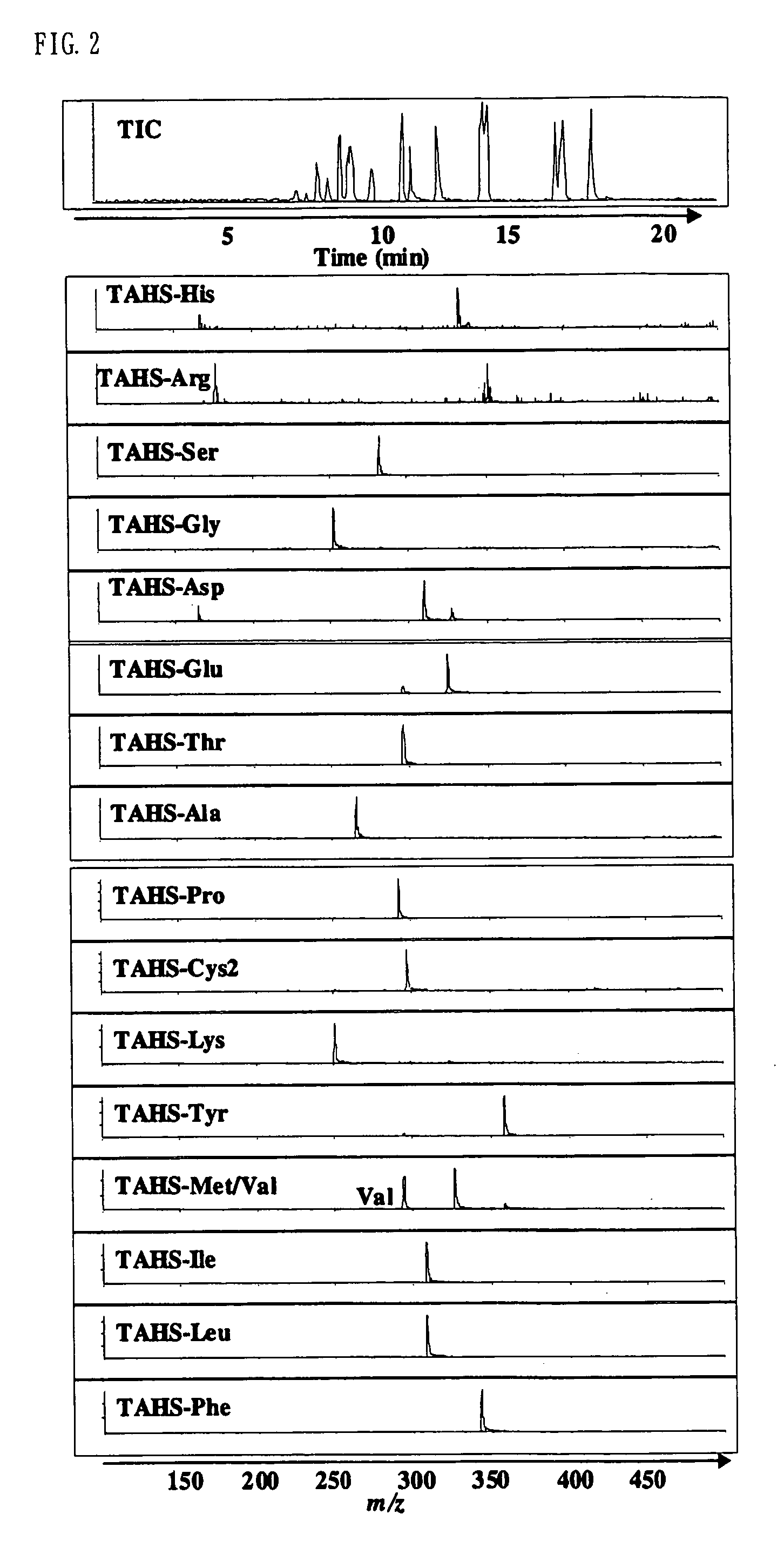
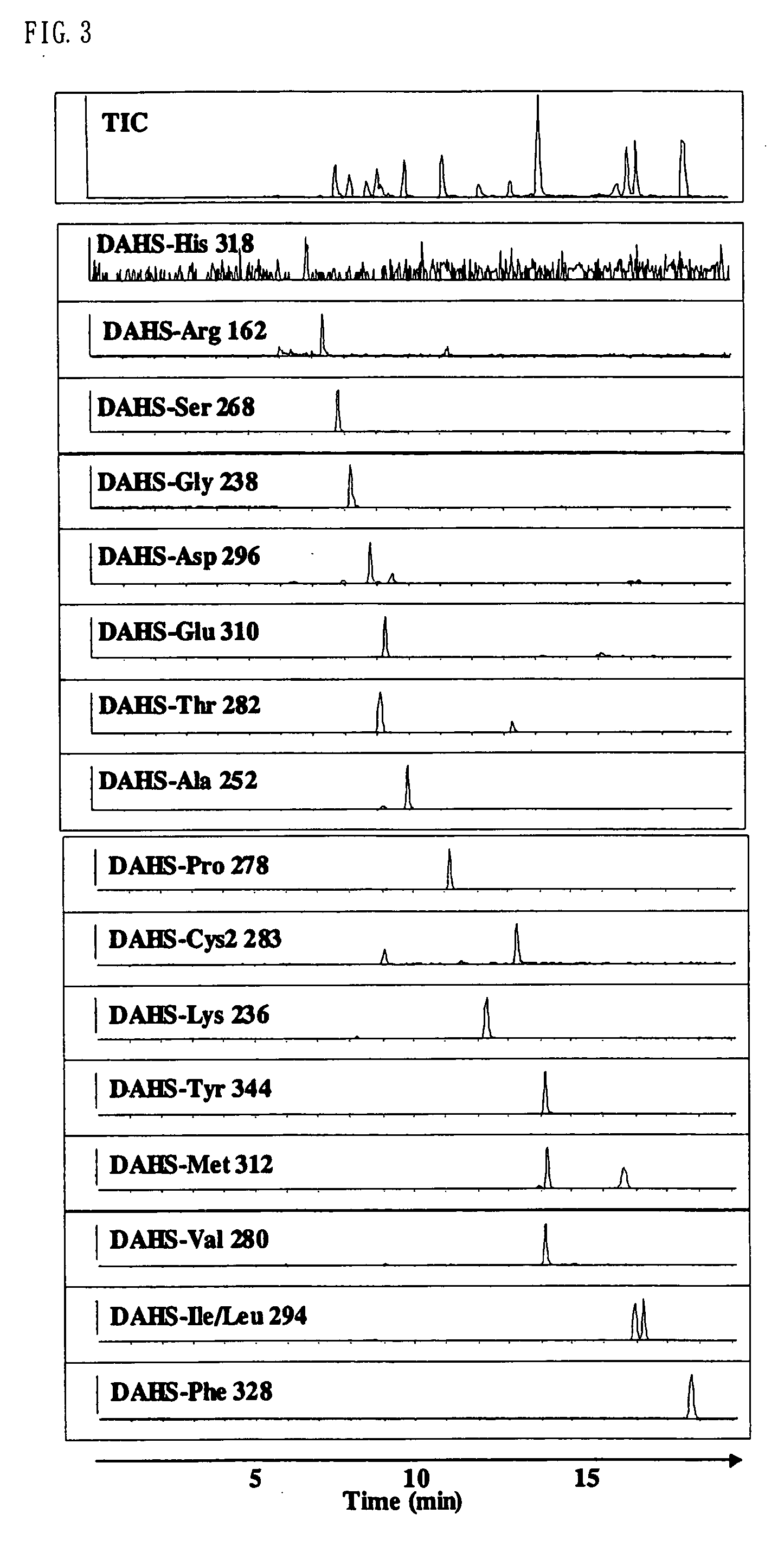
InactiveUS7148069B2Convenient and highly and selectiveMass spectrometric analysisPeptide preparation methodsCarbamateChemical compound
The present invention provides a method for the analysis of a compound with amino group (e.g., an amino acid or peptide) contained in a sample and convenient manner with a high sensitivity. The compound with amino group in a sample containing the compound with amino group is labeled with a specific carbamate compound such as p-trimethylammonium anilyl-N-hydroxysuccinimidyl carbamate iodide to enhance the selectivity and sensitivity. The present invention is preferably used in conjunction with mass spectrometry such as MS / MS method to facilitate quantitative analysis. The present invention further provides labeling reagents for mass spectrometry.
Owner:AJINOMOTO CO INC
Method for analysis of compounds with amino group and analytical reagent therefor
InactiveUS20050079624A1Convenient and highly and selectiveMass spectrometric analysisPeptide preparation methodsCarbamateN-Hydroxysuccinimide
The present invention provides a method for the analysis of a compound with amino group (e.g., an amino acid or peptide) contained in a sample and convenient manner with a high sensitivity. The compound with amino group in a sample containing the compound with amino group is labeled with a specific carbamate compound such as p-trimethylammonium anilyl-N-hydroxysuccinimidyl carbamate iodide to enhance the selectivity and sensitivity. The present invention is preferably used in conjunction with mass spectrometry such as MS / MS method to facilitate quantitative analysis. The present invention further provides labeling reagents for mass spectrometry.
Owner:AJINOMOTO CO INC
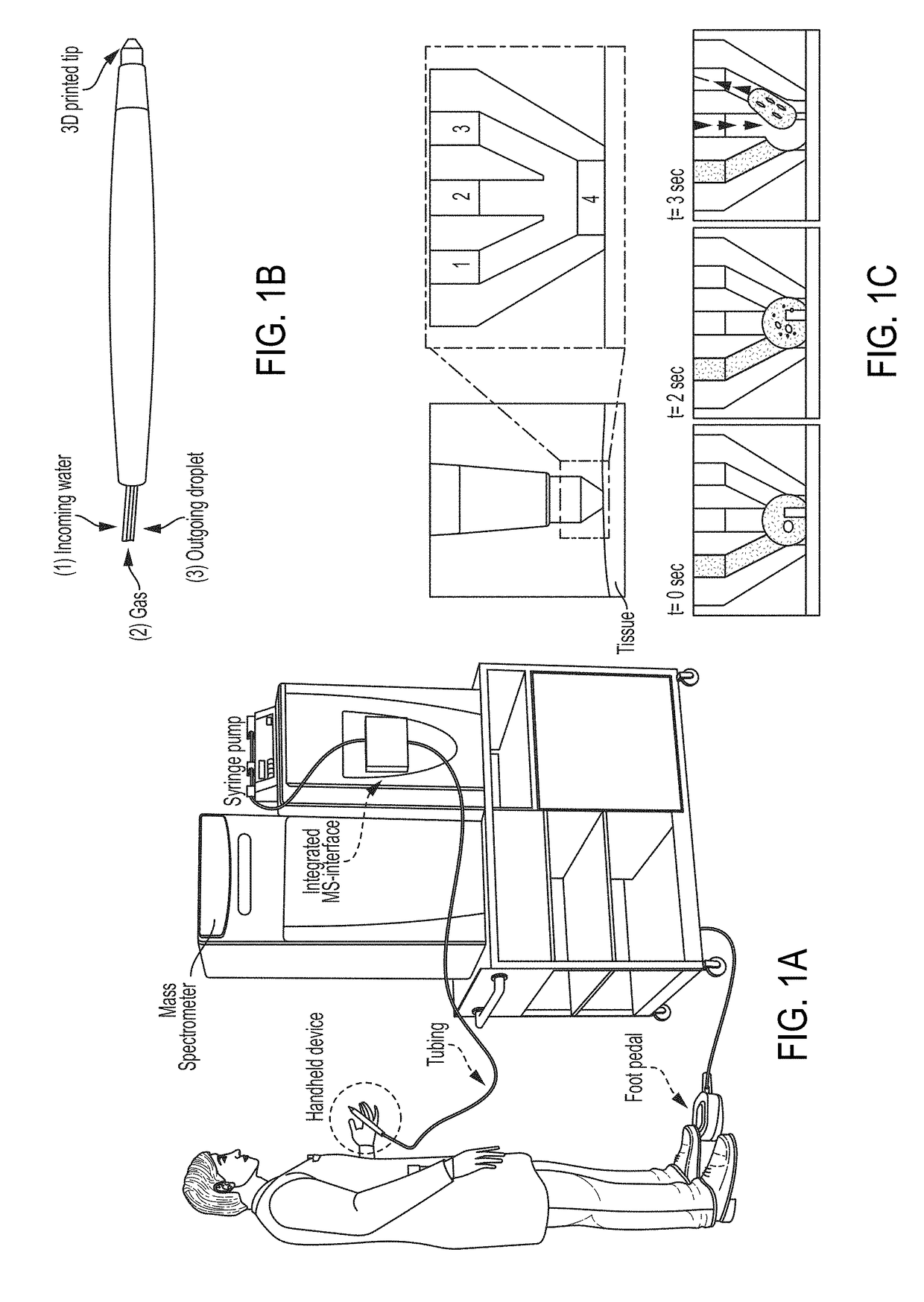
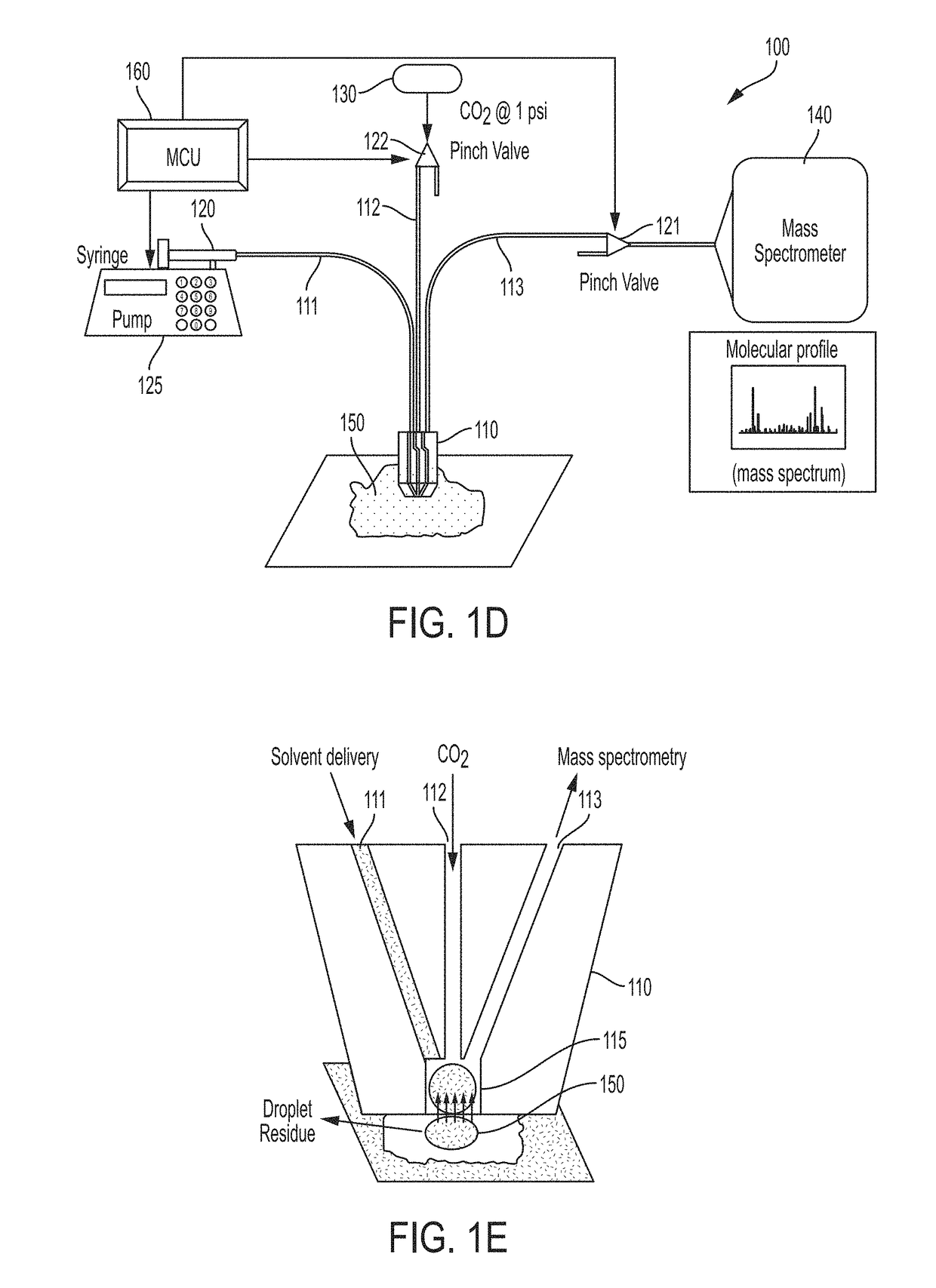
Collection probe and methods for the use thereof
ActiveUS20180158661A1Easy to identifyEasy to readComponent separationSamples introduction/extractionMolecular analysisFluid tissues
Method and devices are provided for assessing tissue samples from a plurality of tissue sites in a subject using molecular analysis. In certain aspects, devices of the embodiments allow for the collection of liquid tissue samples and delivery of the samples for mass spectrometry analysis.
Owner:BOARD OF REGENTS
Mass-Spectral Method for Selection, and De-Selection, of Cancer Patients for Treatment with Immune Response Generating Therapies
ActiveUS20130344111A1Raise the possibilityFast conductionParticle separator tubesMass spectrometric analysisYeastMass Spectrometry-Mass Spectrometry
A method and system for predicting in advance of treatment whether a cancer patient is likely, or not likely, to obtain benefit from administration of a yeast-based immune response generating therapy, which may be yeast-based immunotherapy for mutated Ras-based cancer, alone or in combination with another anti-cancer therapy. The method uses mass spectrometry of a blood-derived patient sample and a computer configured as a classifier using a training set of class-labeled spectra from other cancer patients that either benefitted or did not benefit from an immune response generating therapy alone or in combination with another anti-cancer therapy. Also disclosed are methods of treatment of a cancer patient, comprising administering a yeast-based immune response generating therapy, which may be yeast-based immunotherapy for mutated Ras-based cancer, to a patient selected by a test in accordance with predictive mass spectral methods disclosed herein, in which the class label for the spectra indicates the patient is likely to benefit from the yeast-based immunotherapy.
Owner:GLOBE IMMUNE INC
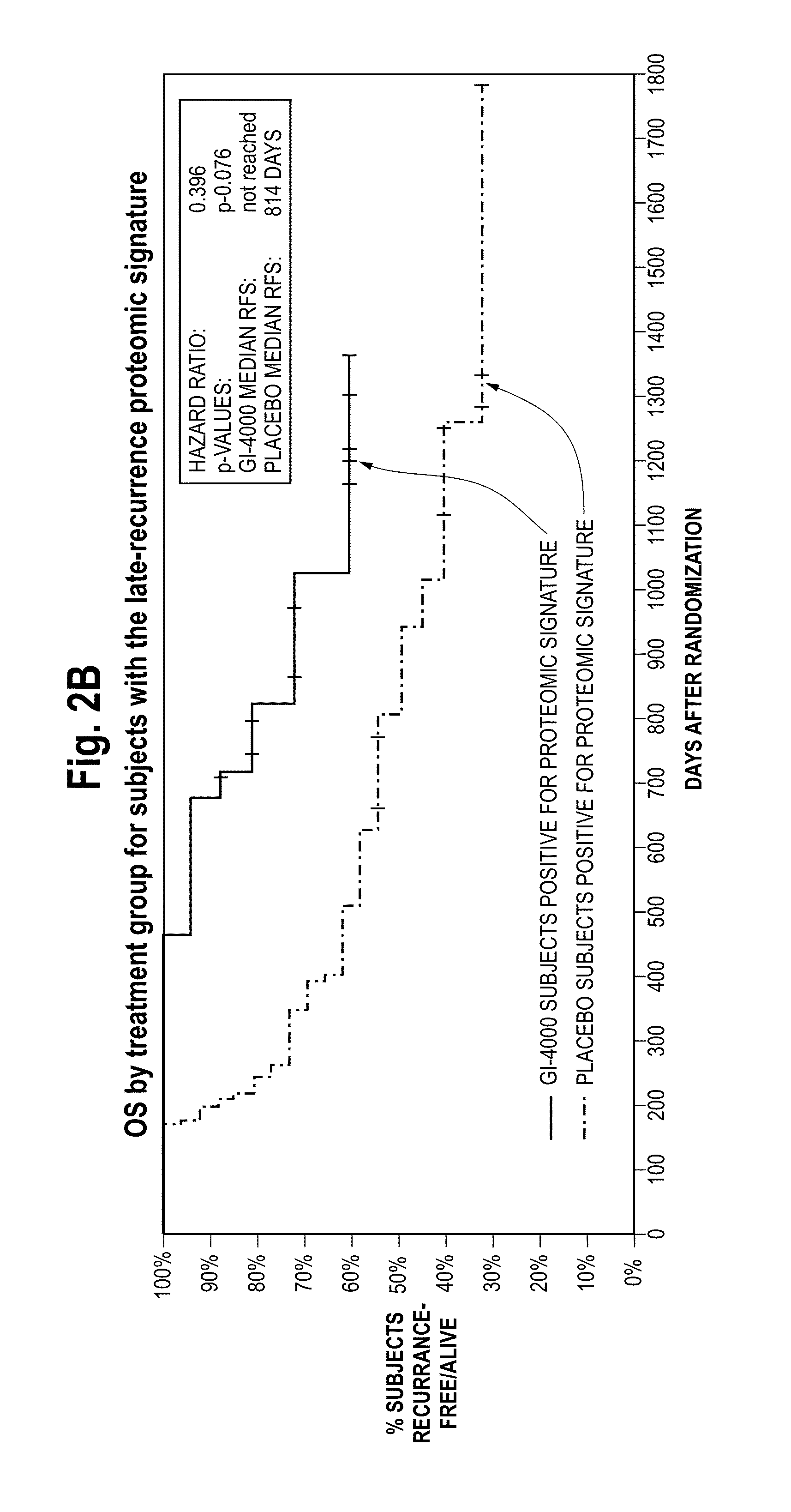
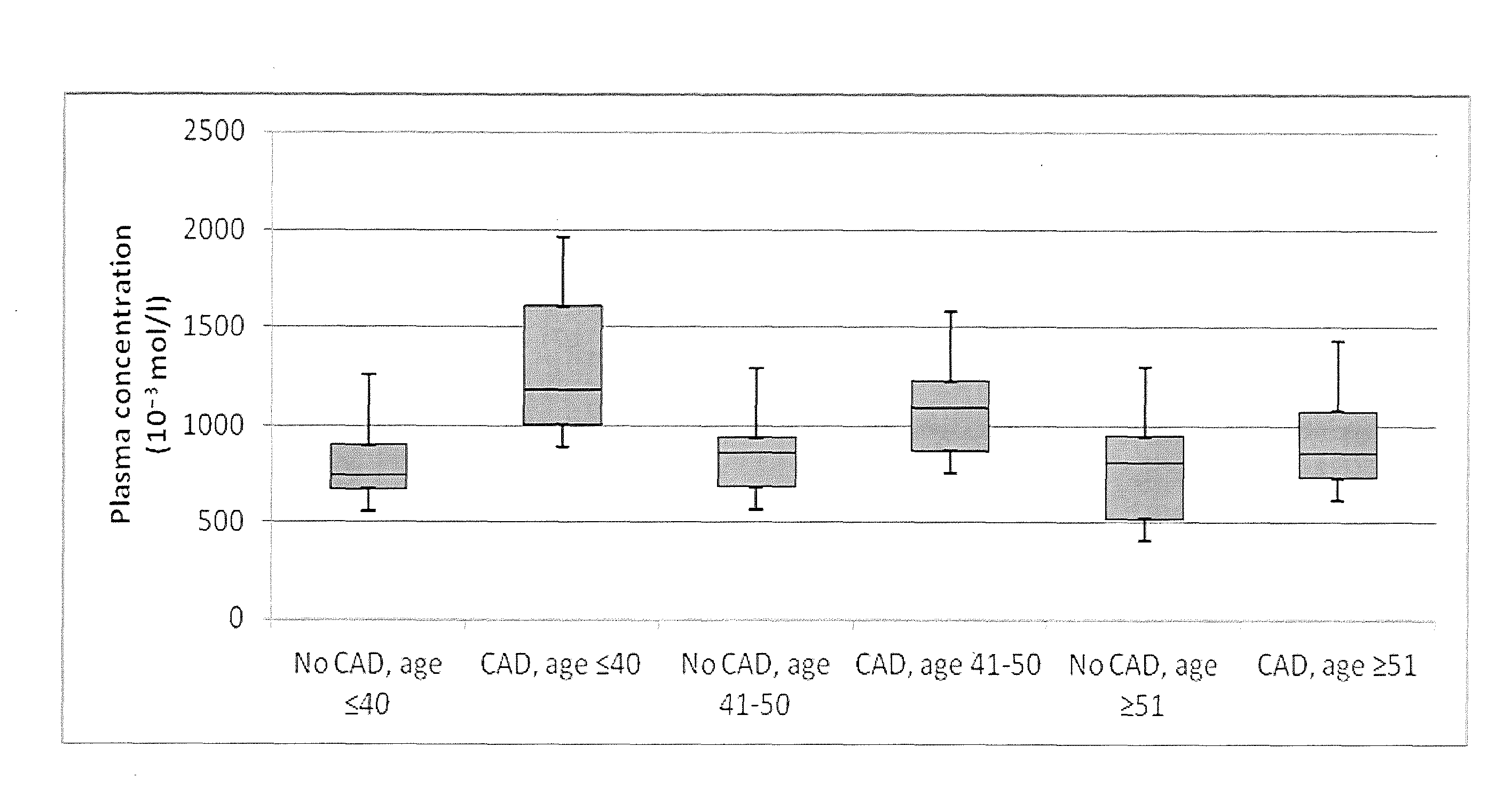
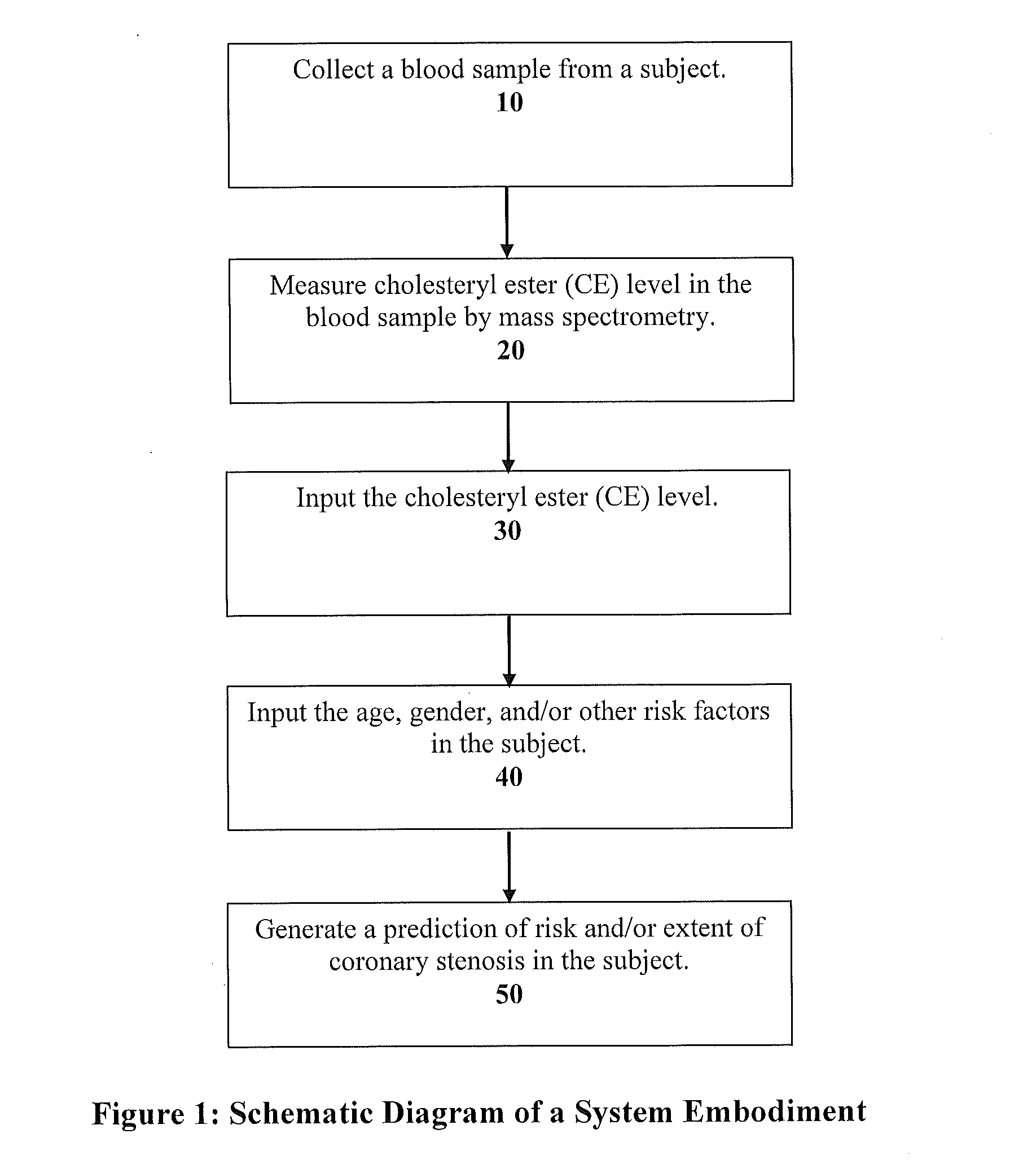
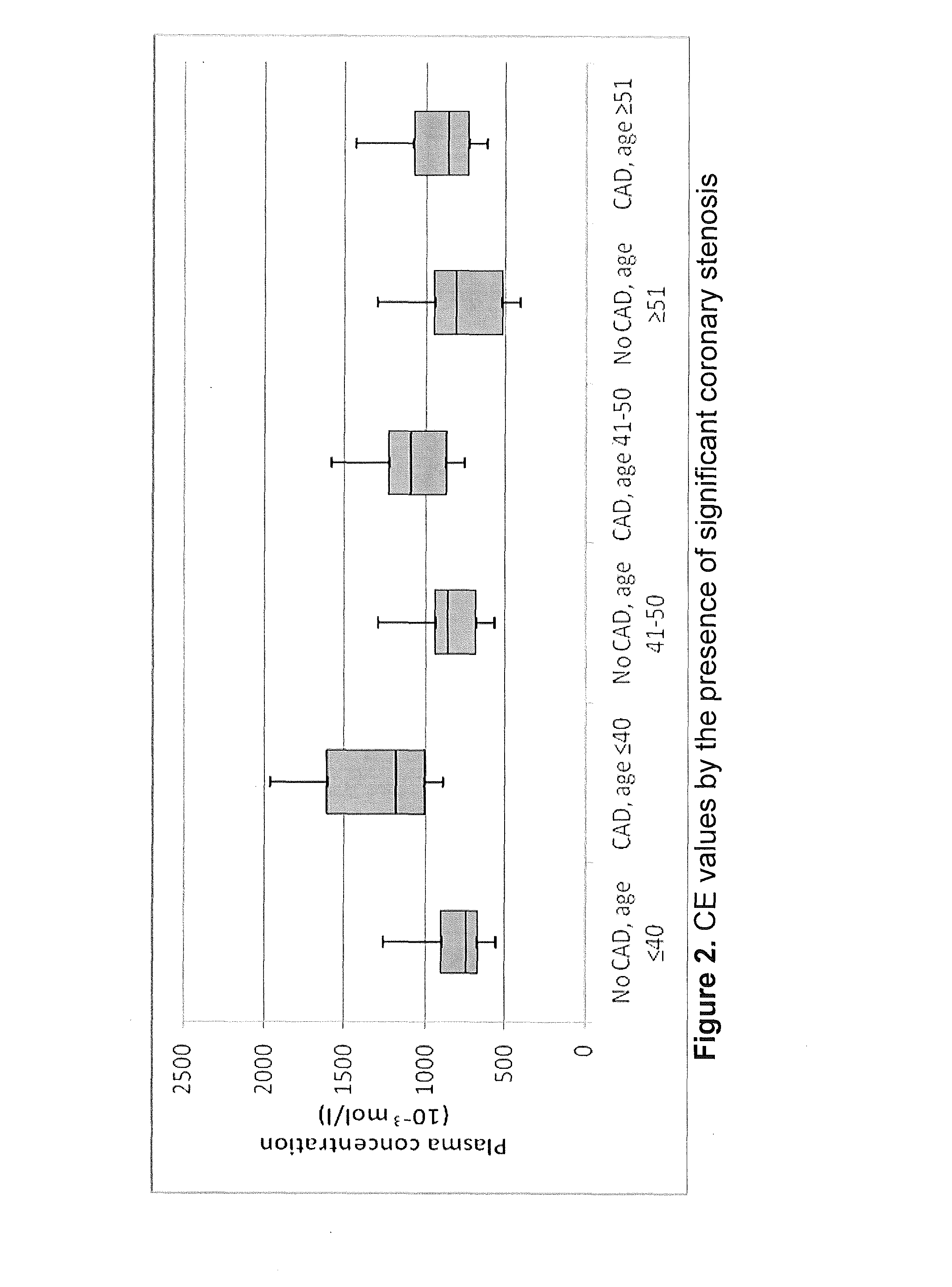
Systems and apparatus for indicating risk of coronary stenosis
ActiveUS20140134654A1Increased riskHealth-index calculationMicrobiological testing/measurementAge and genderMass Spectrometry-Mass Spectrometry
A computer-based method for determining a prediction of risk and / or an indication of extent of coronary stenosis in a human subject, comprises the steps of: (a) inputting the level of at least one cholesteryl ester measured in a blood sample collected from said subject; and then (b) inputting the age and gender of said subject; and then (c) generating in said computer from said cholesteryl ester level input, said age input and said gender input a prediction of risk and / or an indication of extent of coronary stenosis blood sample by mass spectrometry.
Owner:WAKE FOREST UNIV HEALTH SCI INC
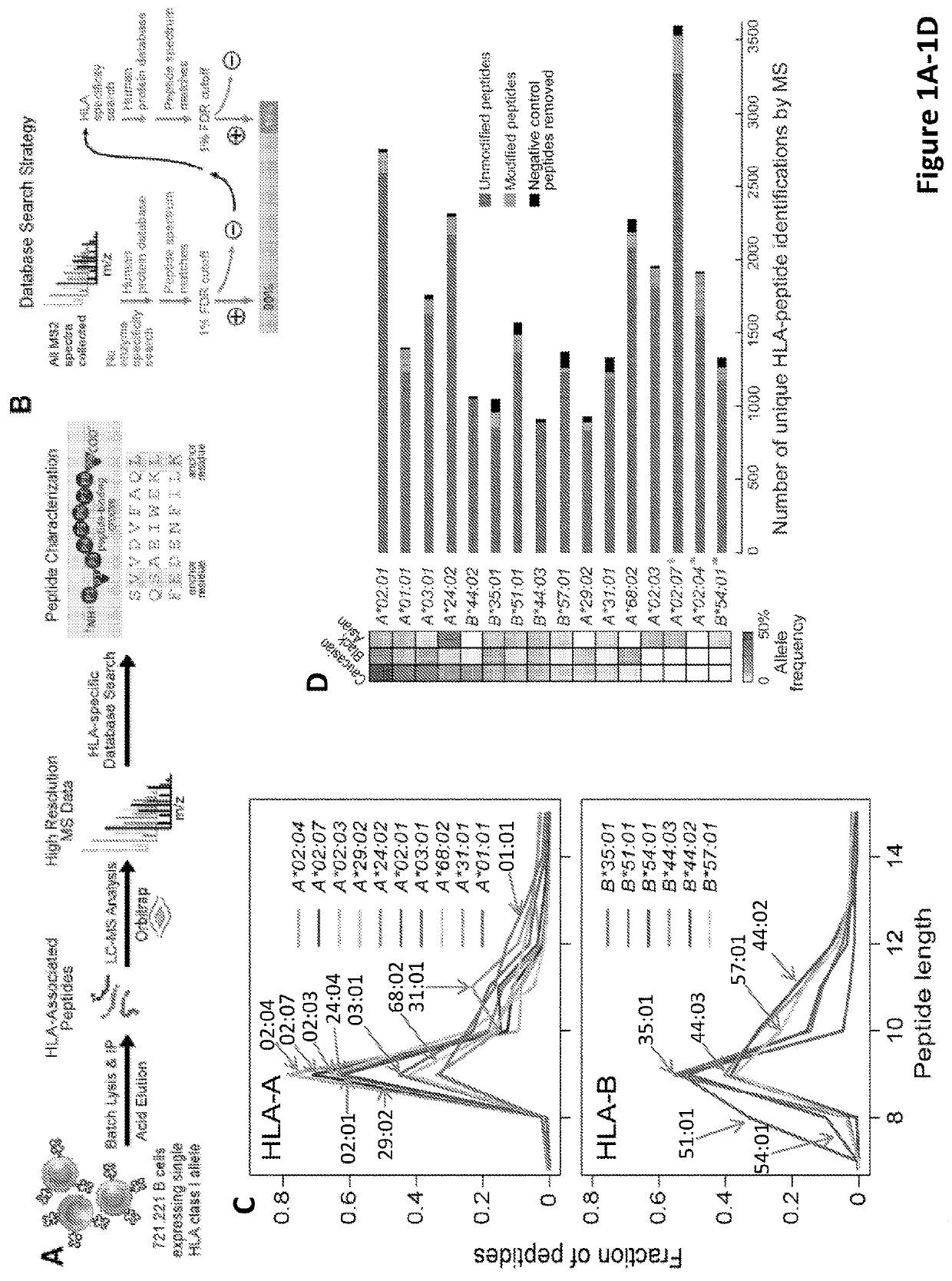
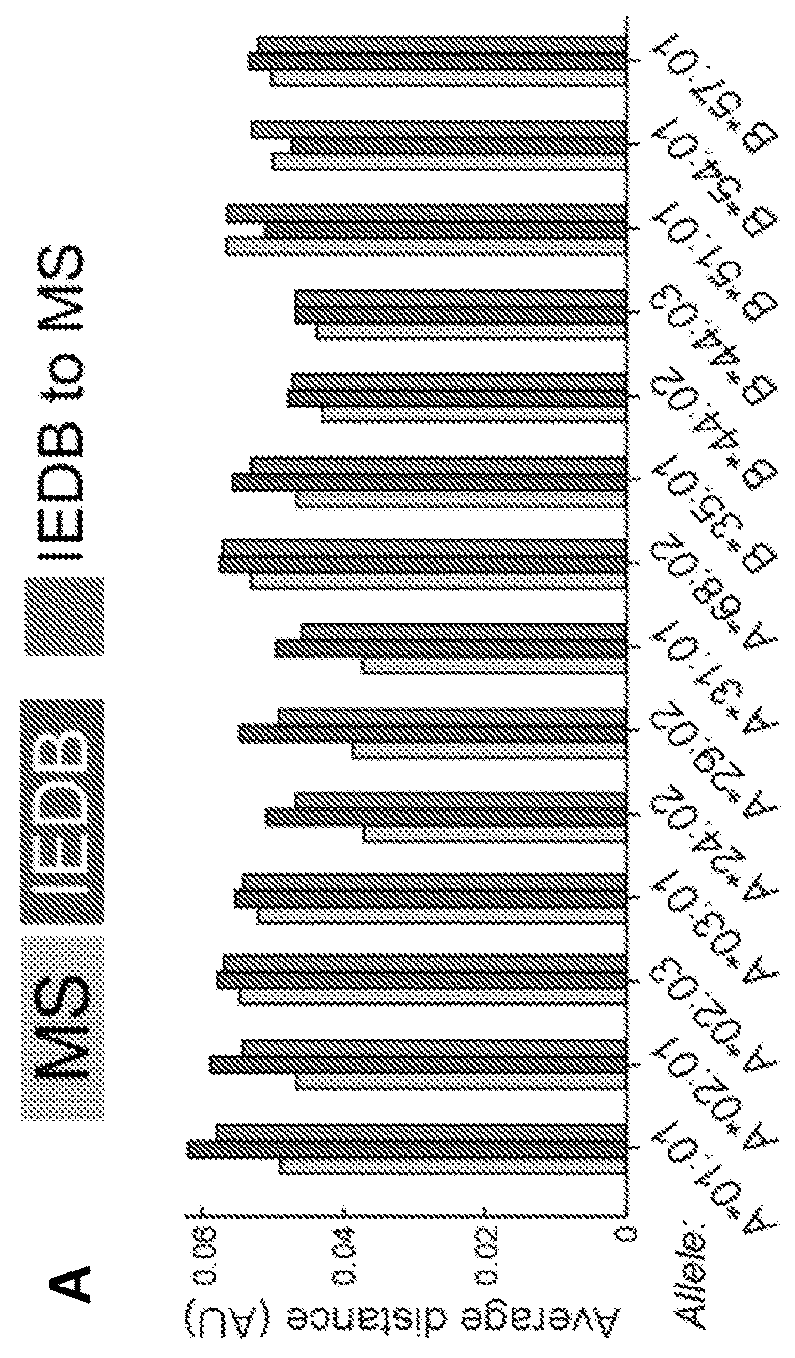
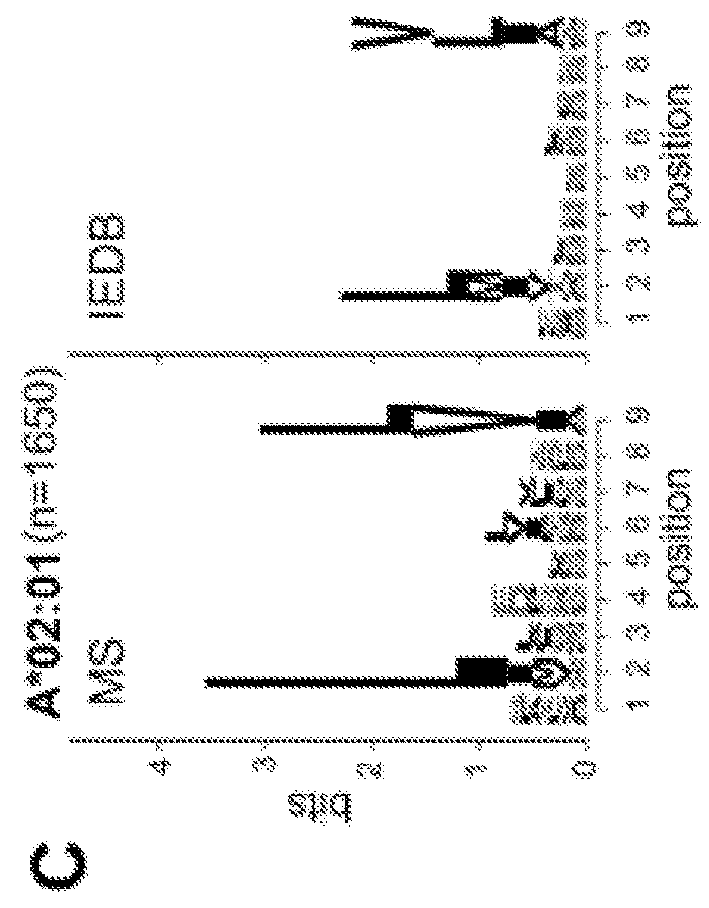
Improved HLA epitope prediction
PendingUS20190346442A1Microbiological testing/measurementMass spectrometric analysisT cellHuman health
Adaptive immune responses rely on the ability of cytotoxic T cells to identify and eliminate cells displaying disease-specific antigens on human leukocyte antigen (HLA) class I molecules. Investigations into antigen processing and display have immense implications in human health, disease and therapy. To extend understanding of the rules governing antigen processing and presentation, immunopurified peptides from B cells, each expressing a single HLA class I allele, were profiled using accurate mass, high-resolution liquid chromatography-mass spectrometry (LC-MS / MS). A resource dataset containing thousands of peptides bound to 28 distinct class I HLA-A, -B, and -C alleles was generated by implementing a novel allele-specific database search strategy. Applicants discovered new binding motifs, established the role of gene expression in peptide presentation and improved prediction of HLA-peptide binding by using these data to train machine-learning models. These streamlined experimental and analytic workflows enable direct identification and analysis of endogenously processed and presented antigens.
Owner:THE BROAD INST INC +4
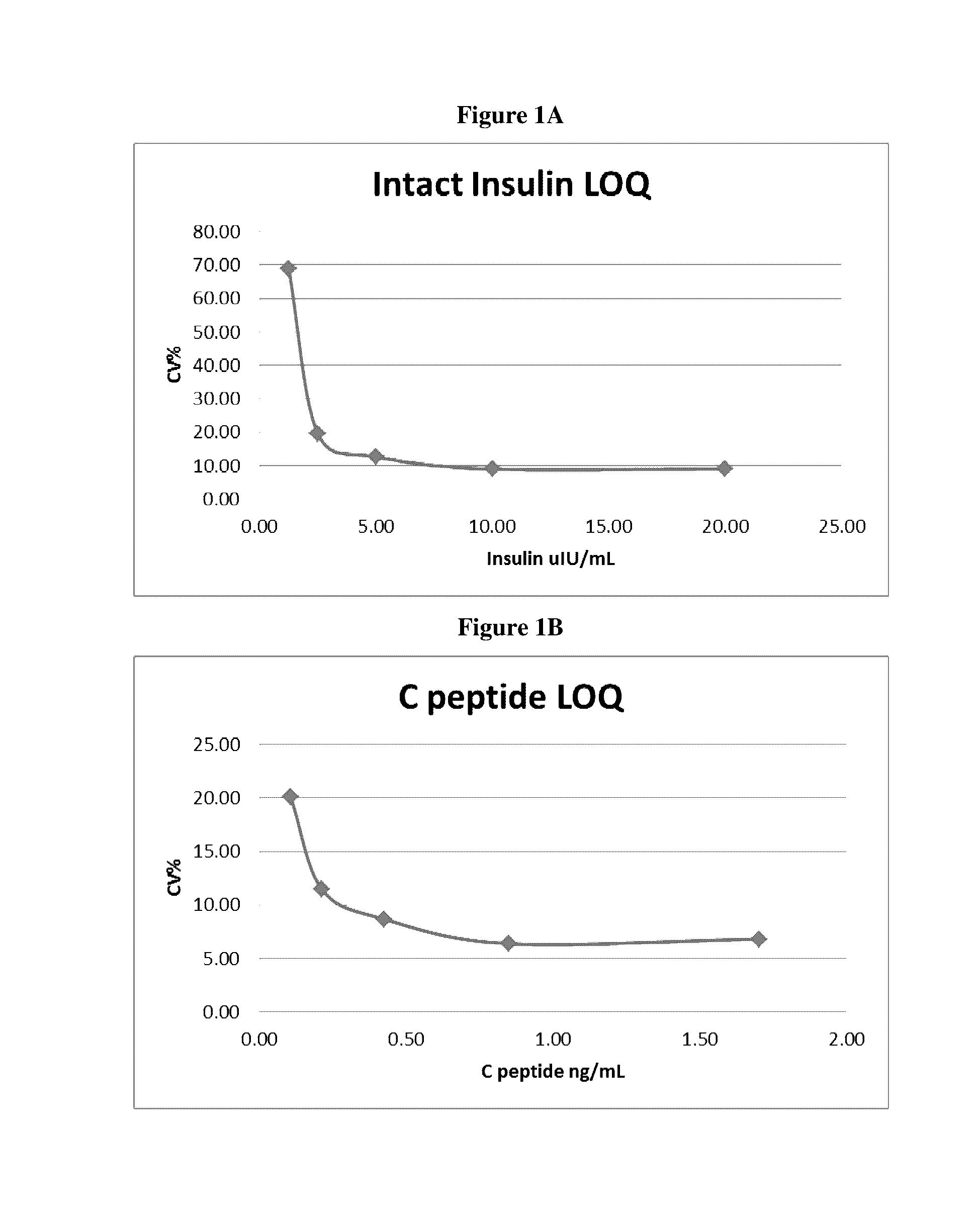
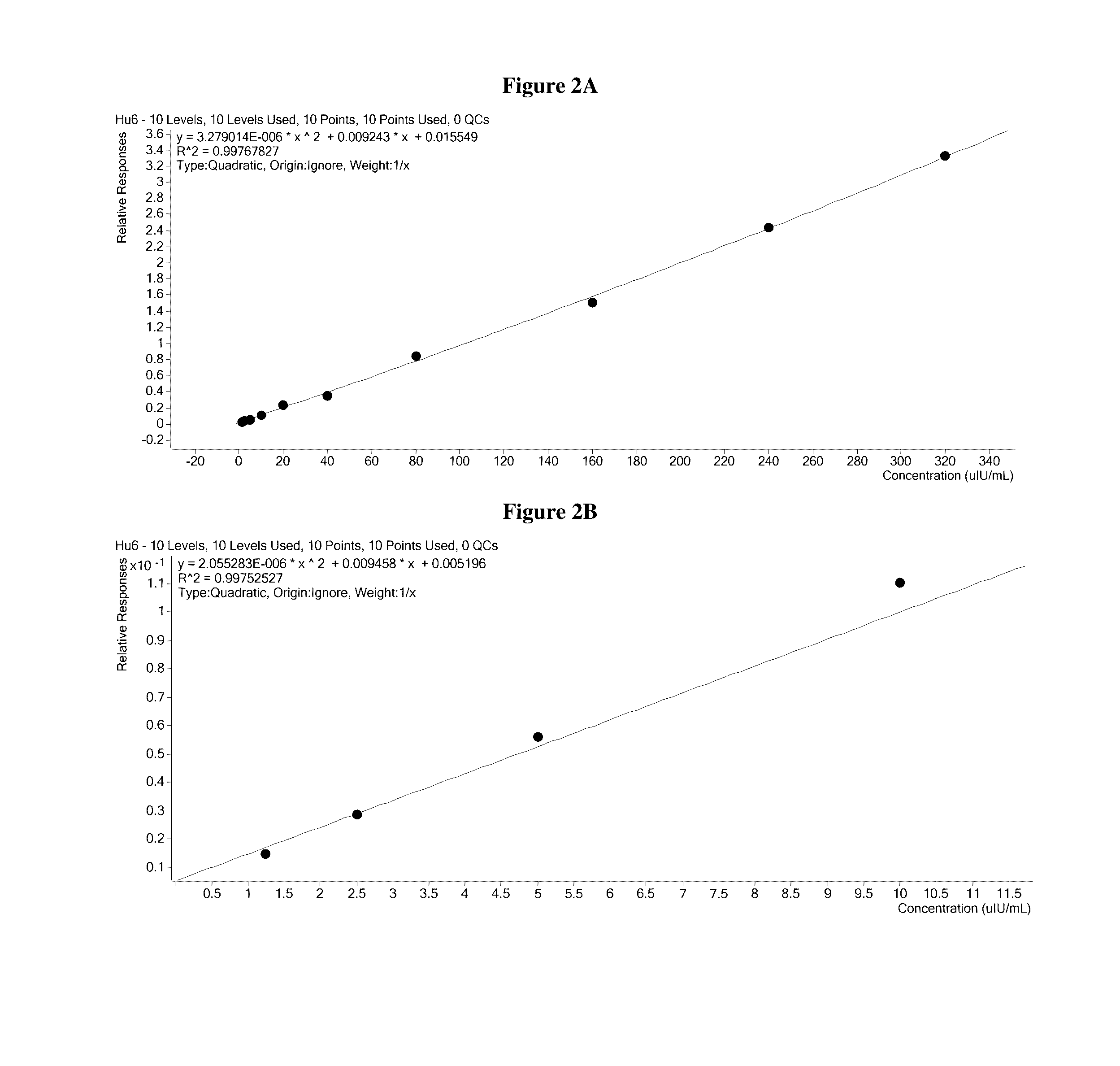
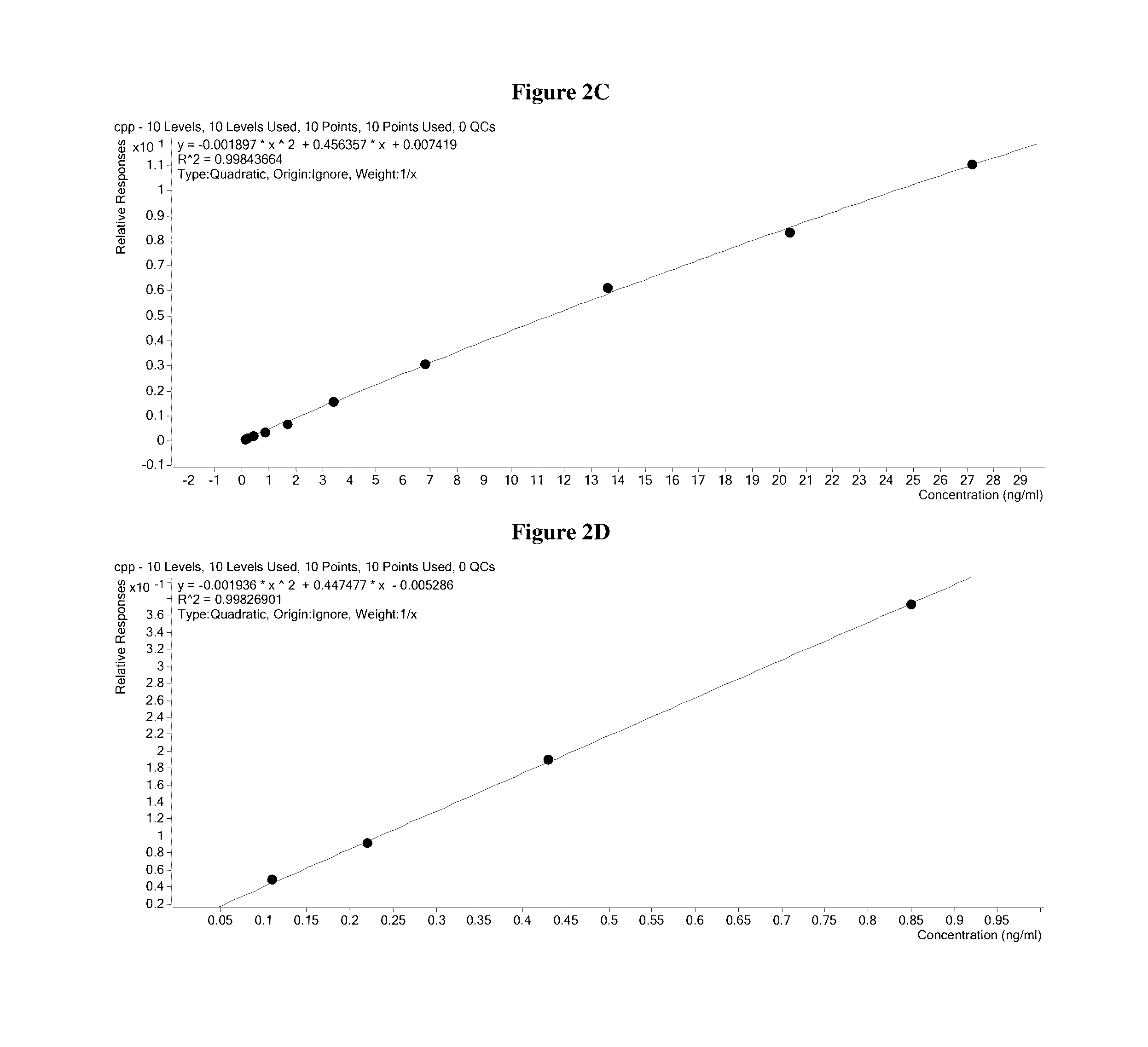
Methods for quantitation of insulin levels by mass spectrometry
ActiveUS20160282328A1Improve accuracyComponent separationMicrobiological testing/measurementPurification methodsLevel insulin
Methods are described for determining the amount of insulin in a sample. Provided herein are mass spectrometric methods for detecting and quantifying insulin and C-peptide in a biological sample utilizing enrichment and / or purification methods coupled with tandem mass spectrometric or high resolution / high accuracy mass spectrometric techniques. Also provided herein are mass spectrometric methods for detecting and quantifying insulin and b-chain in a biological sample utilizing enrichment and / or purification methods coupled with tandem mass spectrometric or high resolution / high accuracy mass spectrometric techniques.
Owner:QUEST DIAGNOSTICS INVESTMENTS INC
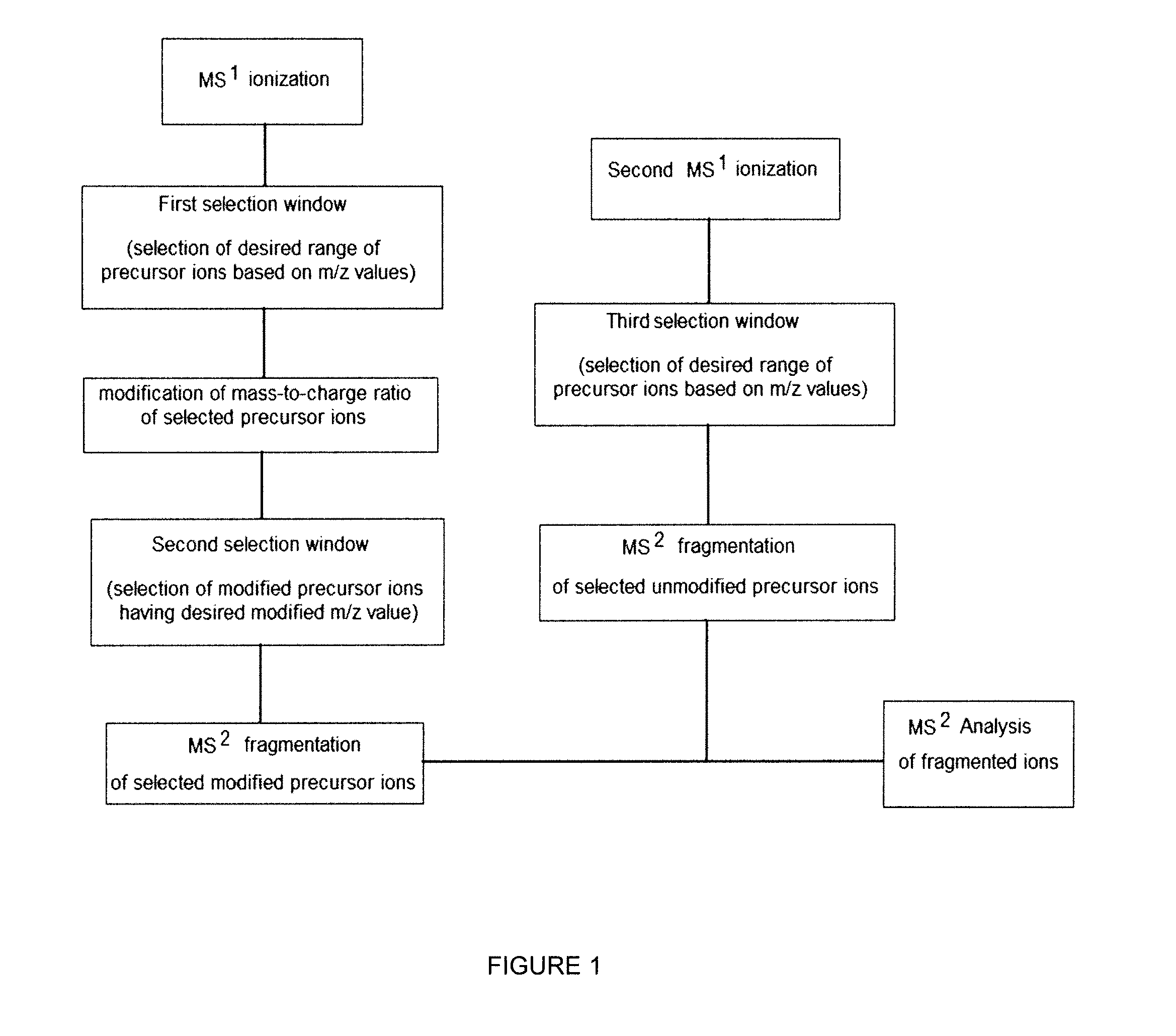
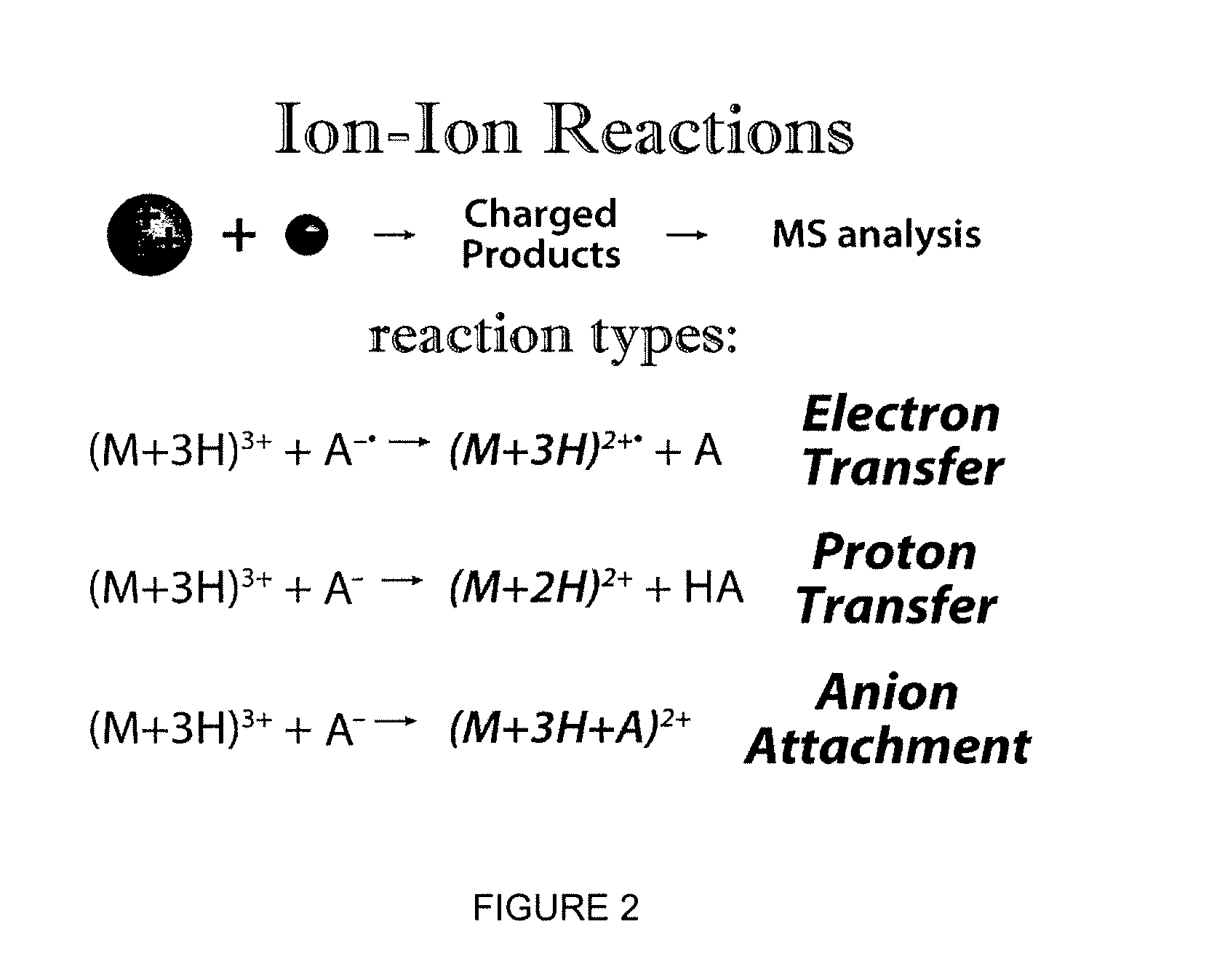
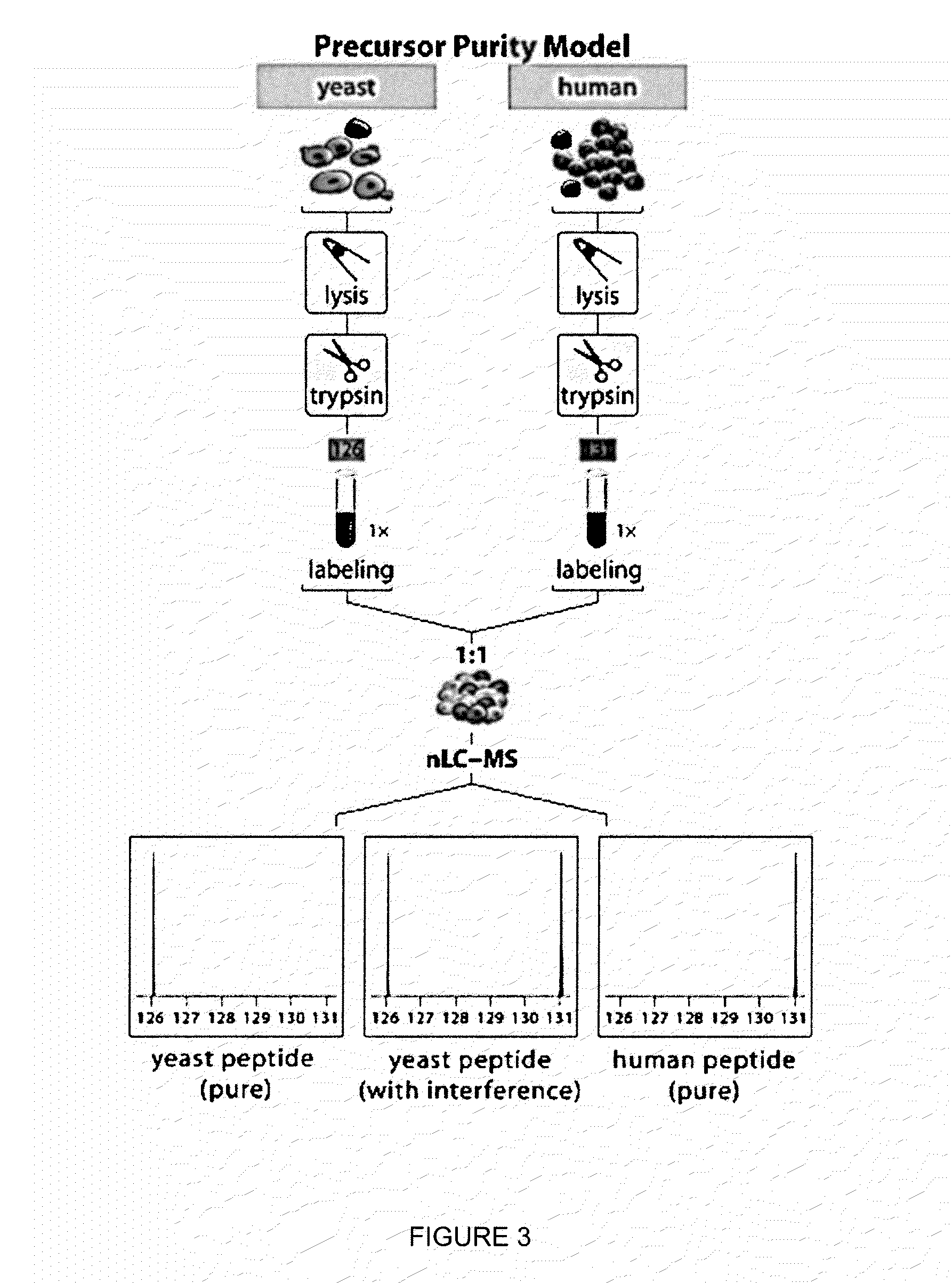
Gas-phase purification for accurate isobaric tag-based quantification
ActiveUS20130084645A1Improve mass spectrometry analysisEasy to quantifySamples introduction/extractionMass spectrometric analysisGas phaseMass Spectrometry-Mass Spectrometry
Described herein are mass spectrometry systems and methods which improve the accuracy of isobaric tag-based quantification by alleviating the pervasive problem of precursor interference and co-isolation of impurities through gas-phase purification. During the gas-phase purification, the mass-to-charge ratios of precursor ions within at least a selected range are selectively changed allowing ions having similar unmodified mass-to-charge ratios to be separated before further isolation, fragmentation or analysis.
Owner:WISCONSIN ALUMNI RES FOUND
Methods and means for characterizing antibiotic resistance in microorganisms
ActiveUS20130244230A1Rapid diagnosisEarly detectionBioreactor/fermenter combinationsBiological substance pretreatmentsMicroorganismAntibiotic resistance
The present invention relates to a method for characterizing the antibiotic resistance of a microorganism, said method comprising the steps of (a) providing a reference mass spectrum of an antimicrobial compound, its enzymatic modification product, its molecular target, or of a substrate compound of a its modifying enzyme; (b) exposing a microorganism, a cell lysate thereof, or a growth medium supernatant thereof, to said antimicrobial compound or said substrate compound in aqueous liquid to thereby provide an exposed sample; (c) acquiring a mass spectrum of the exposed sample; (d) comparing the mass spectrum acquired in step c) with the reference mass spectrum of step (a), and (e) determining from said comparison whether modification of said antimicrobial compound, its modification product or its molecular target or of said substrate has occurred following said exposure, and establishing that said microorganism is potentially resistant to said antimicrobial compound when said modification is observed.
Owner:ERASMUS UNIV MEDICAL CENT ROTTERDAM ERASMUS MC
Transgenic plants for nitrogen fixation
InactiveCN105874070ALow artificial nitrogen application requirementsMicrobiological testing/measurementClimate change adaptationGenetic MaterialsNutrient deficiency
Provided are genetic material and nucleic acid sequences useful of increasing yield, biomass, growth rate, vigor, nitrogen use efficiency and / or abiotic stress tolerance, preferably tolerance to nutrient deficiency of a plant. Specifically, the improvement of nitrogen fixation properties in cultivated plants is described.
Owner:UNIV OF BREMEN
Methods for analysis of free and autoantibody-bound biomarkers and associated compositions, devices, and systems
ActiveUS9140695B2Analysis using chemical indicatorsMass spectrometric analysisAnalyteAutoantibody production
The present invention provides methods, compositions, and kits associated with analyzing, enriching, and / or isolating a biomarker or analyte in a biological sample. In one aspect, for example, a method for determining a concentration of a biomarker in a biological sample can include binding any unbound biomarker with an antibody specific for the biomarker to form antibody-bound biomarker, enriching the antibody-bound biomarker and any endogenous autoantibody-bound biomarker to form an enriched fraction, identifying the biomarker in the enriched fraction, and determining the concentration of the biomarker in the biological sample. In one aspect, the concentration of the biomarker is derived from initially unbound biomarker and autoantibody-bound biomarker in the biological sample.
Owner:UNIV OF UTAH RES FOUND
Method, composition, isolation and identification of a plaque particle and related biomarker
ActiveUS20150276771A1Improve suppression propertiesAvoid assemblyMicrobiological testing/measurementMass spectrometric analysisDiseaseMedicine
The disclosure relates to an in vitro technology of detecting presence of a plaque particle, isolating the plaque particle followed by its composition analysis in several diseases states or before the disease sets in. A mechanism and a process leading to plaque formation, identifying a component in the mechanism of plaque formation, an identification of a biomarker for diagnosis / early diagnosis of plaque associated disease is described. A method of screening a candidate agent as an anti-plaque agent using flow cytometer, mass spectrometer and specific biomarkers is performed. Provided also are kits for use in practicing embodiment of the methods. A plaqueproteome database is also generated with novel protein sequences for diagnosis and specific antibodies for specific proteins are also disclosed.
Owner:PLAXGEN +1
Method for the direct detection and/or quantification of at least one compound with a molecular weight of at least 200
ActiveUS9612250B2Enhanced advantageGood reproducibilitySugar derivativesMass spectrometric analysisCulture cellRetention time
Owner:SANIFIT THERAPEUTICS SA
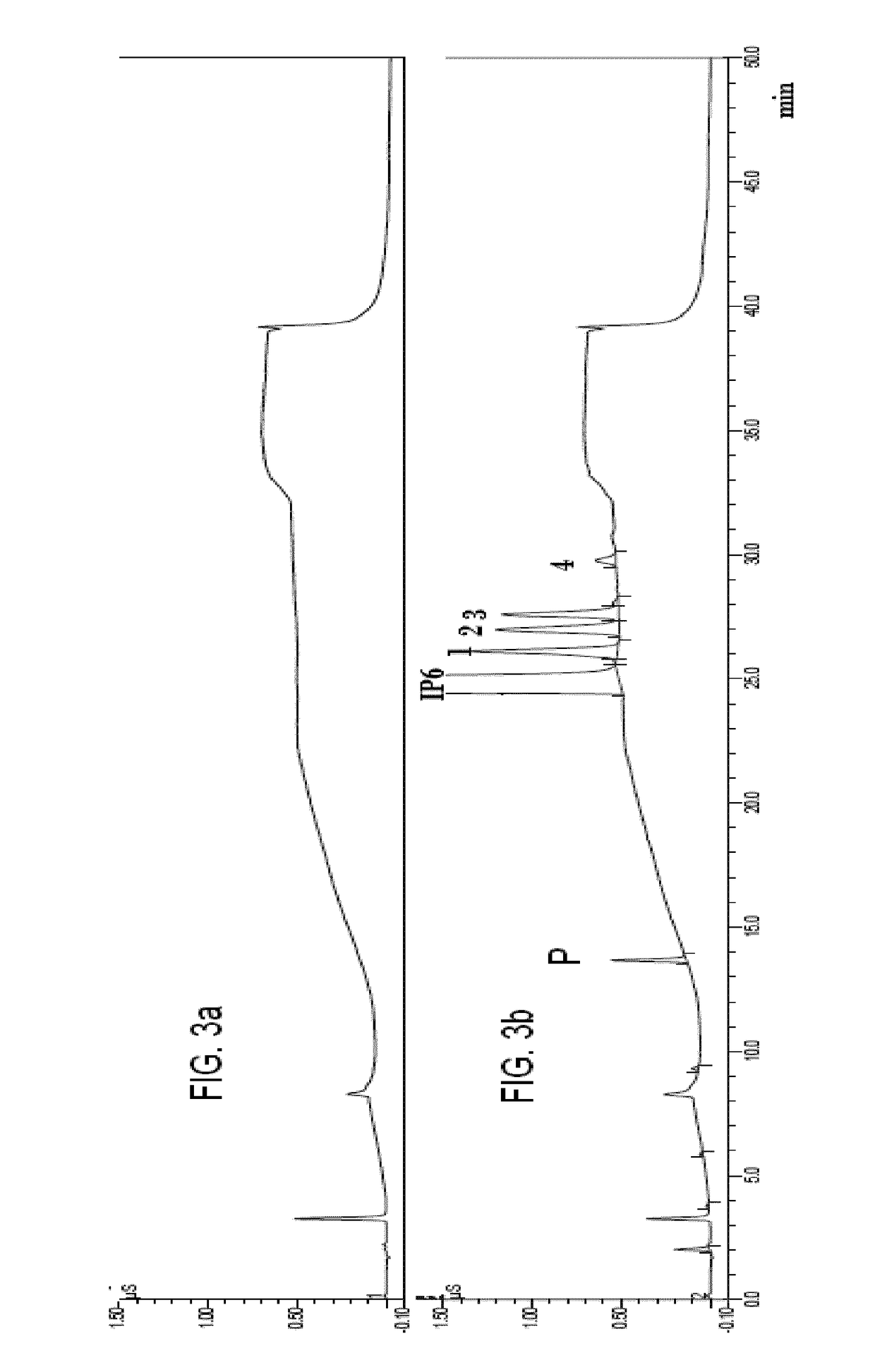
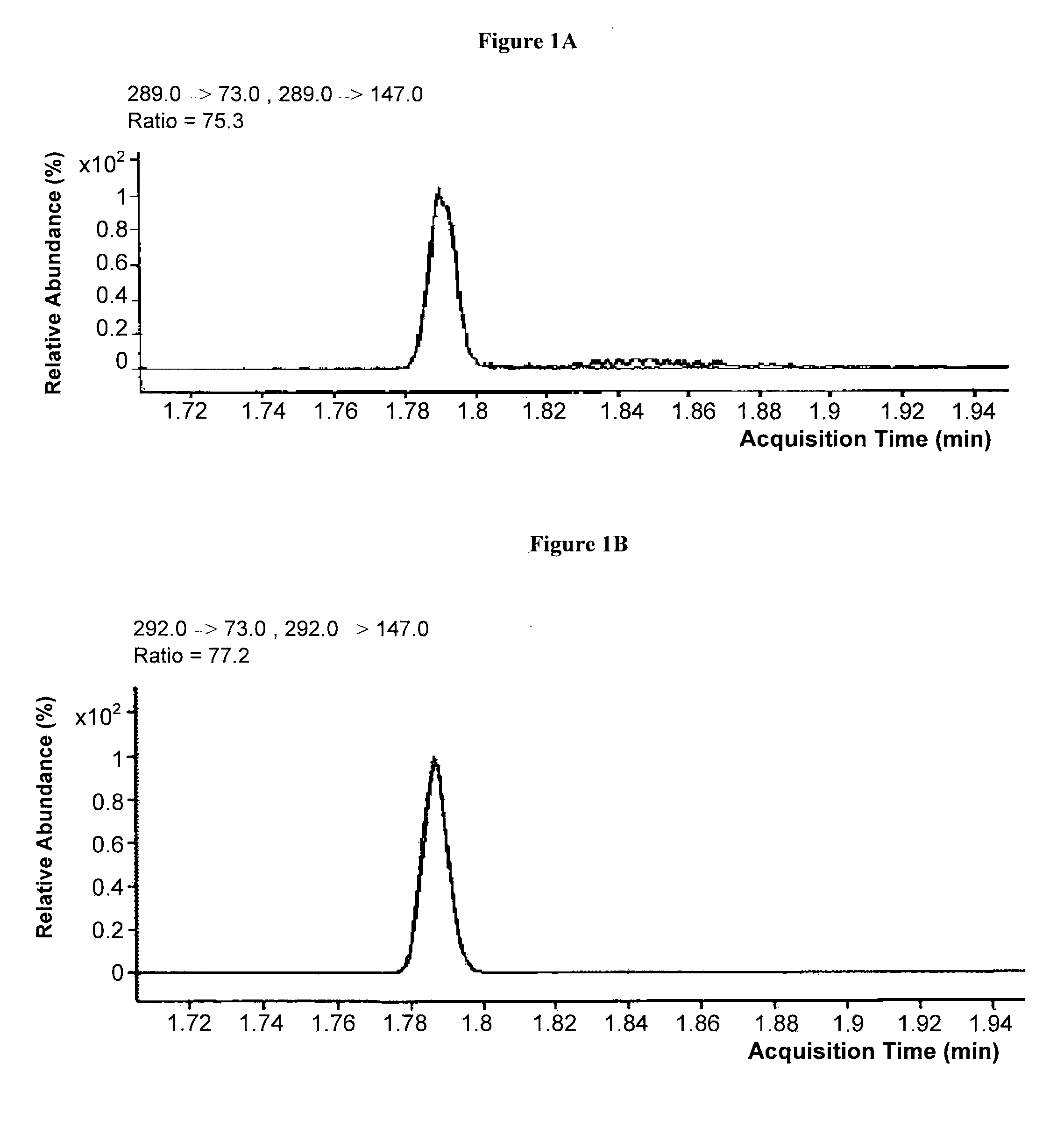
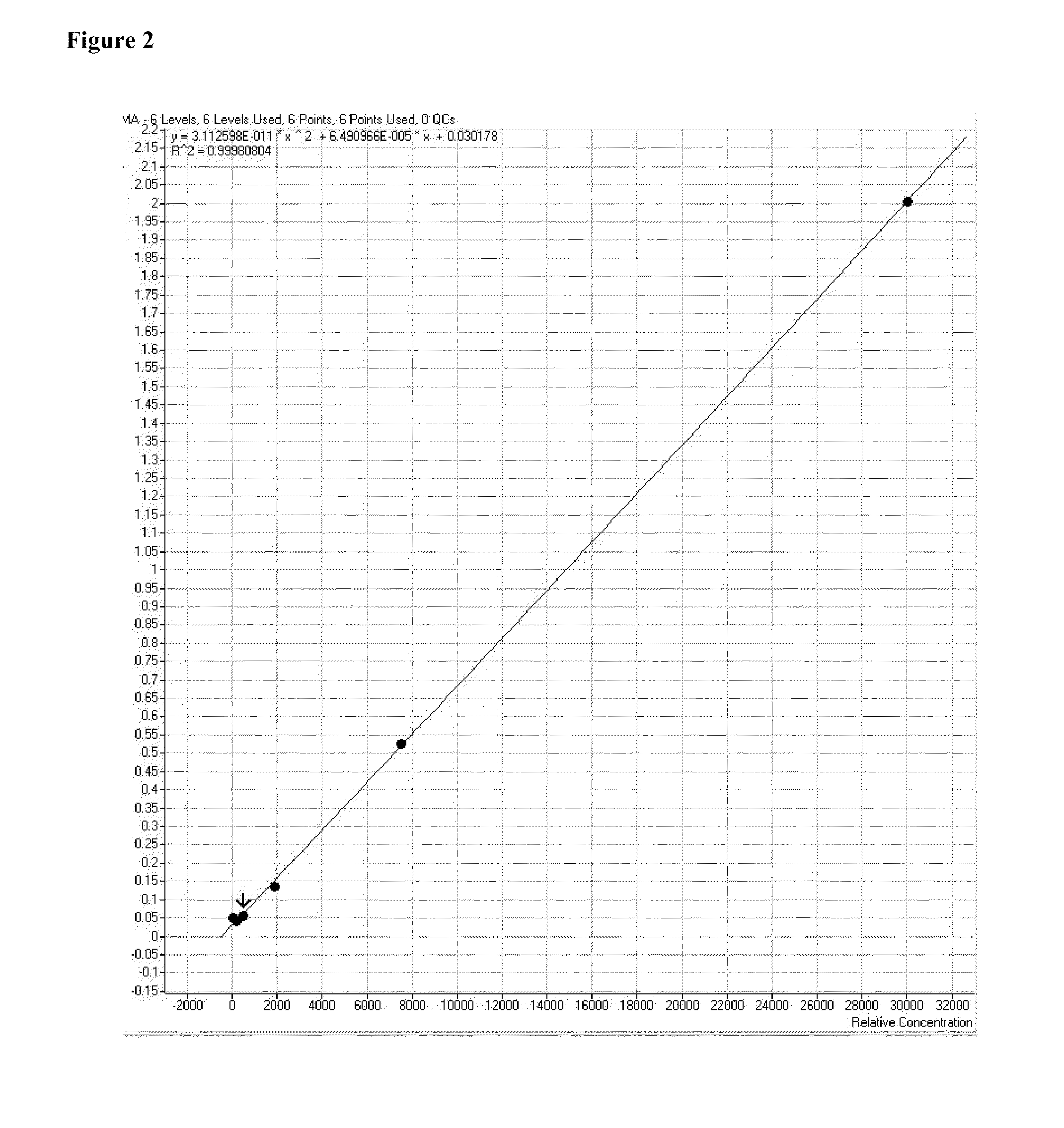
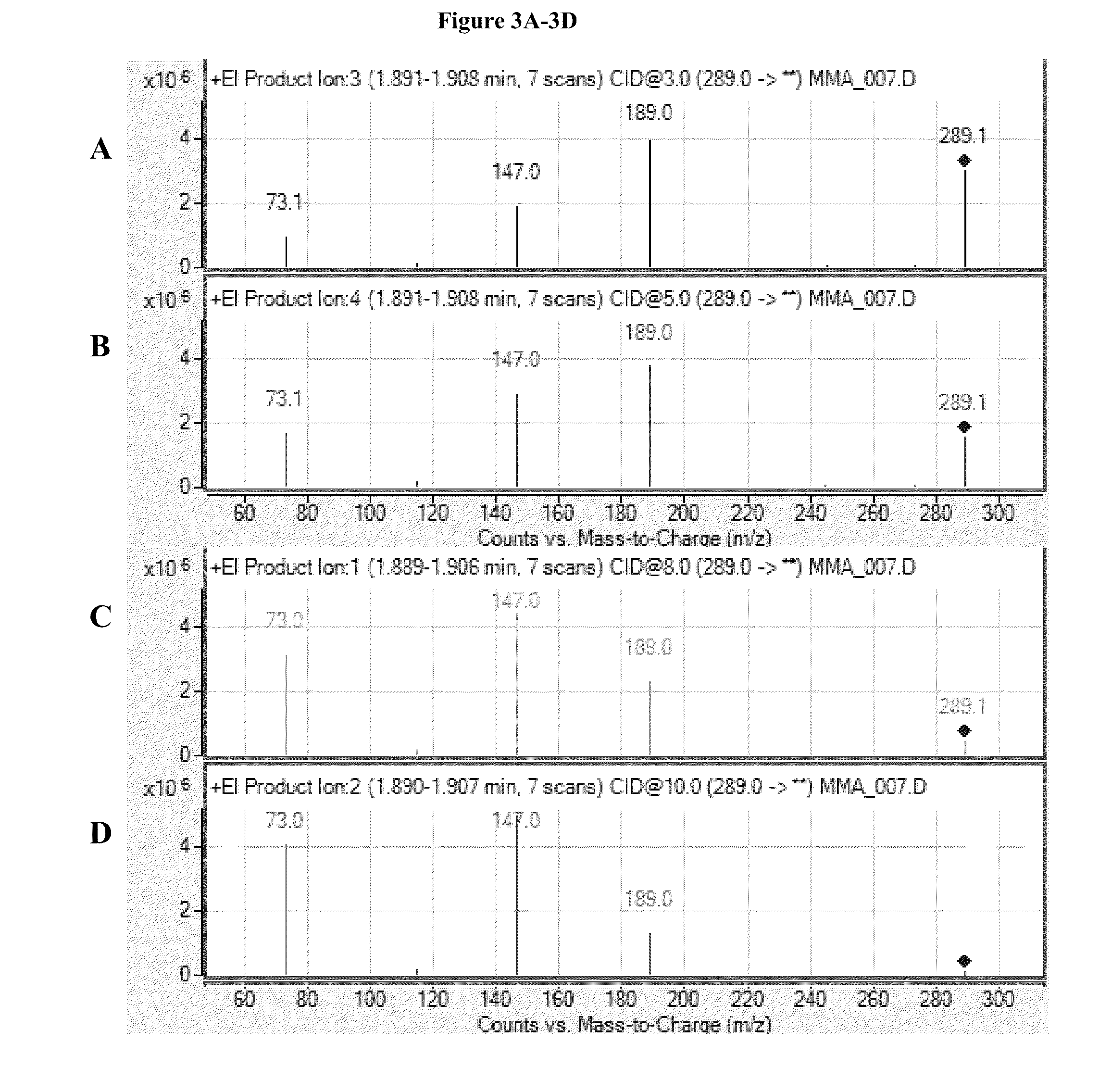
Mass spectrometric determination of derivatized methylmalonic acid
ActiveUS20110306144A1Avoid condensationSolvent is evaporatedComponent separationMass spectrometric analysisMethylmalonic acidMass Spectrometry-Mass Spectrometry
The invention relates to the detection of methylmalonic acid (MMA). In a particular aspect, the invention relates to methods for detecting derivatized methylmalonic acid (MMA) by mass spectrometry.
Owner:QUEST DIAGNOSTICS INVESTMENTS INC
Detection and quantification method for post-translational modification proteomics
ActiveCN108627647AIncrease the total abundanceImprove the odds of analysisPreparing sample for investigationMass spectrometric analysisInternal standardProteomic Profile
Owner:SICHUAN UNIV
Solid Phase Extraction, Derivatization with Crown Ethers, and Mass Spectrometry, Methods, Reagents and Kits
InactiveUS20160282371A1Similar accuracySimilar sensitivityMass spectrometric analysisPeptide preparation methodsAnalyteMass Spectrometry-Mass Spectrometry
The present disclosure is directed to methods reagents and kits for solid phase extraction, derivatization with crown ether containing derivatizing agents, and mass spectrometry of the derivatized analytes.
Owner:TECAN SP INC
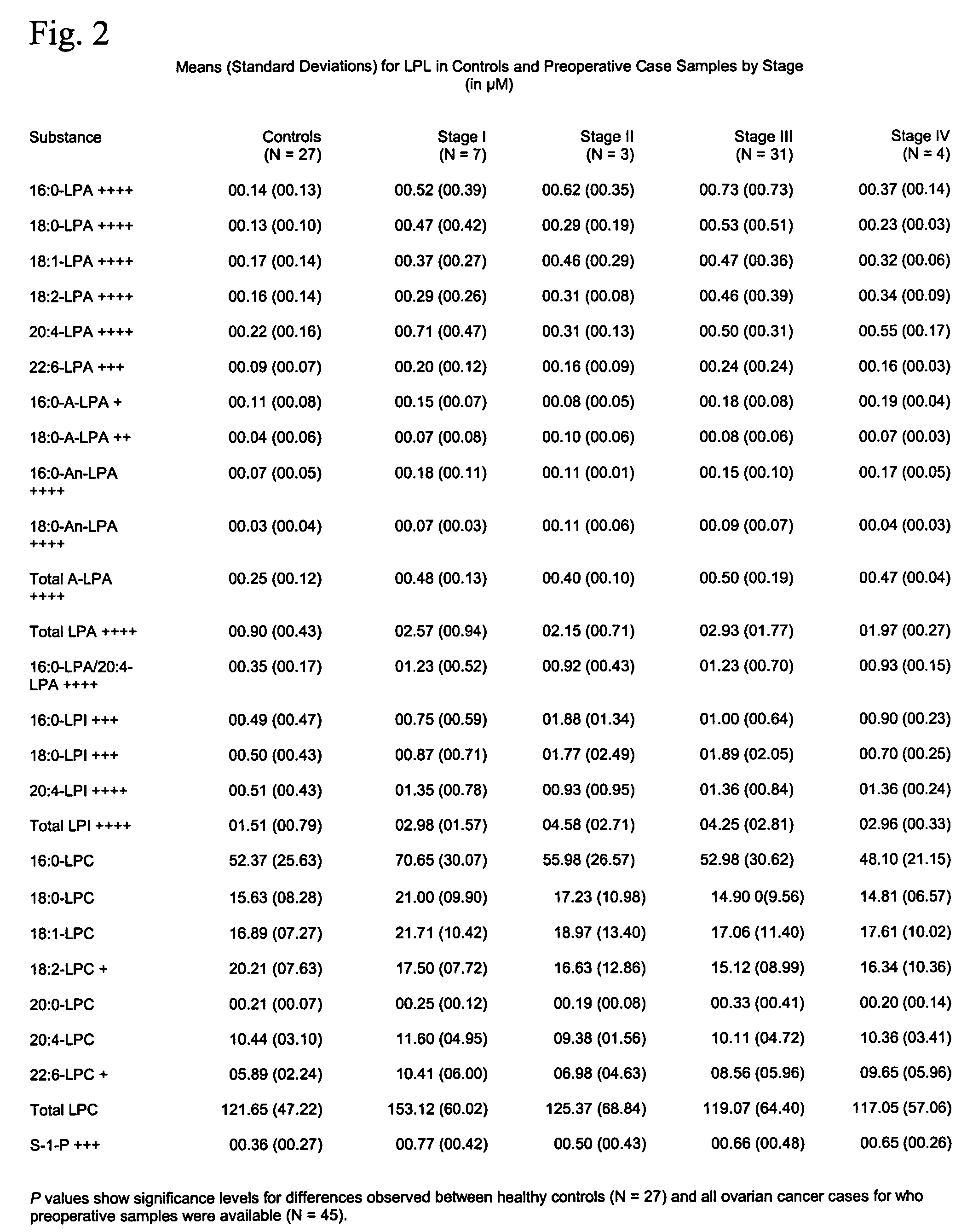
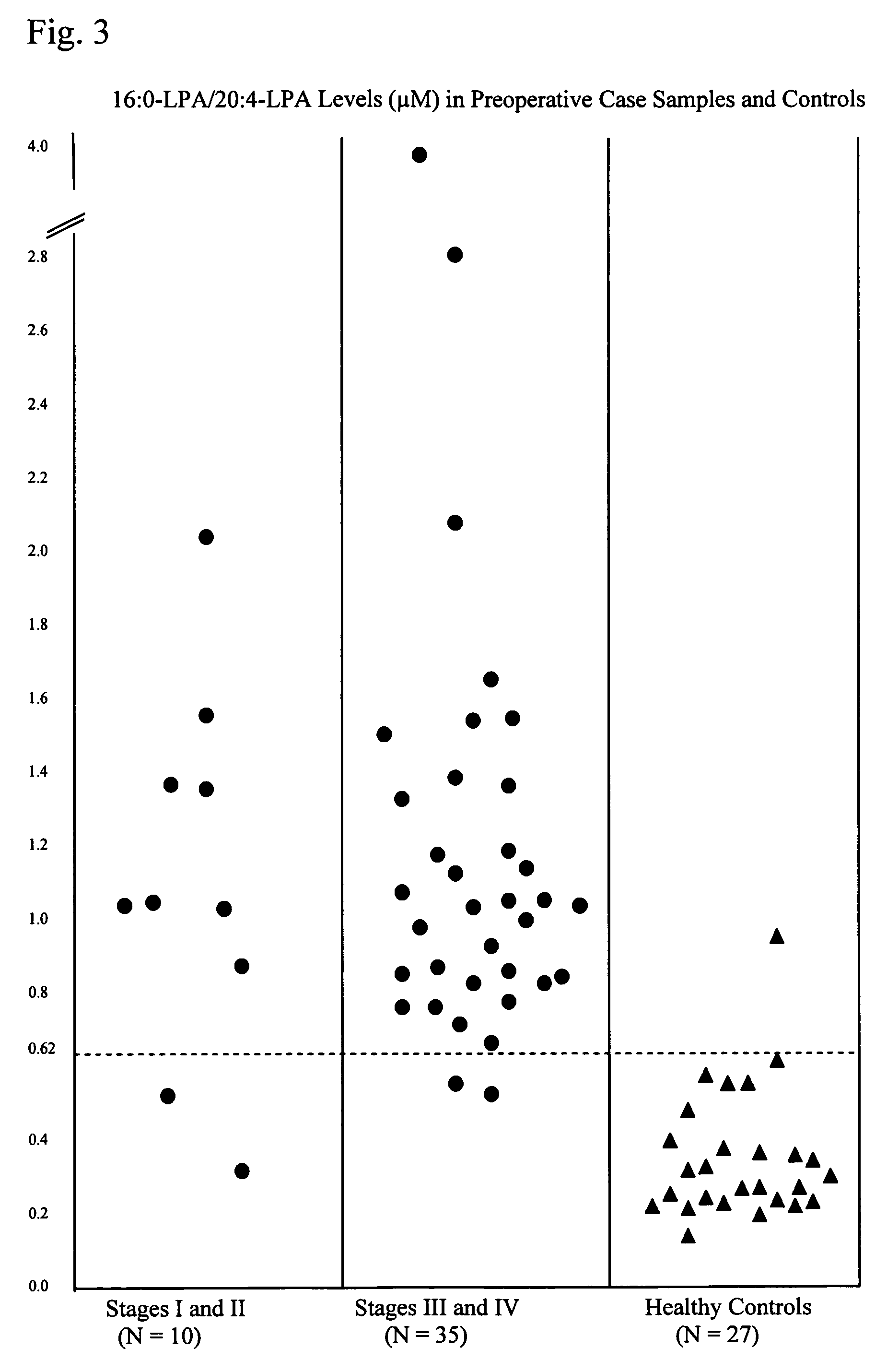
Lysophospholipids as biomarkers of ovarian cancer
InactiveUS7964408B1Improve survivalIncreased mortalityMicrobiological testing/measurementMass spectrometric analysisPhosphateSubspecies
A method of using a bioactive lysophospholipid (LL) as a biomarker for detecting the presence and recurrence of ovarian cancer. Subspecies of LL, such as lysophosphatidic acid (LPA), lysophosphatidylinositol (LPI), lysophosphatidylcholine (LPC), and lysosphingolipid sphinsosine-1-phosphate (S1P), are used alone or in conjunction to increase the specificity and sensitivity of the assay.
Owner:UNIV OF SOUTH FLORIDA
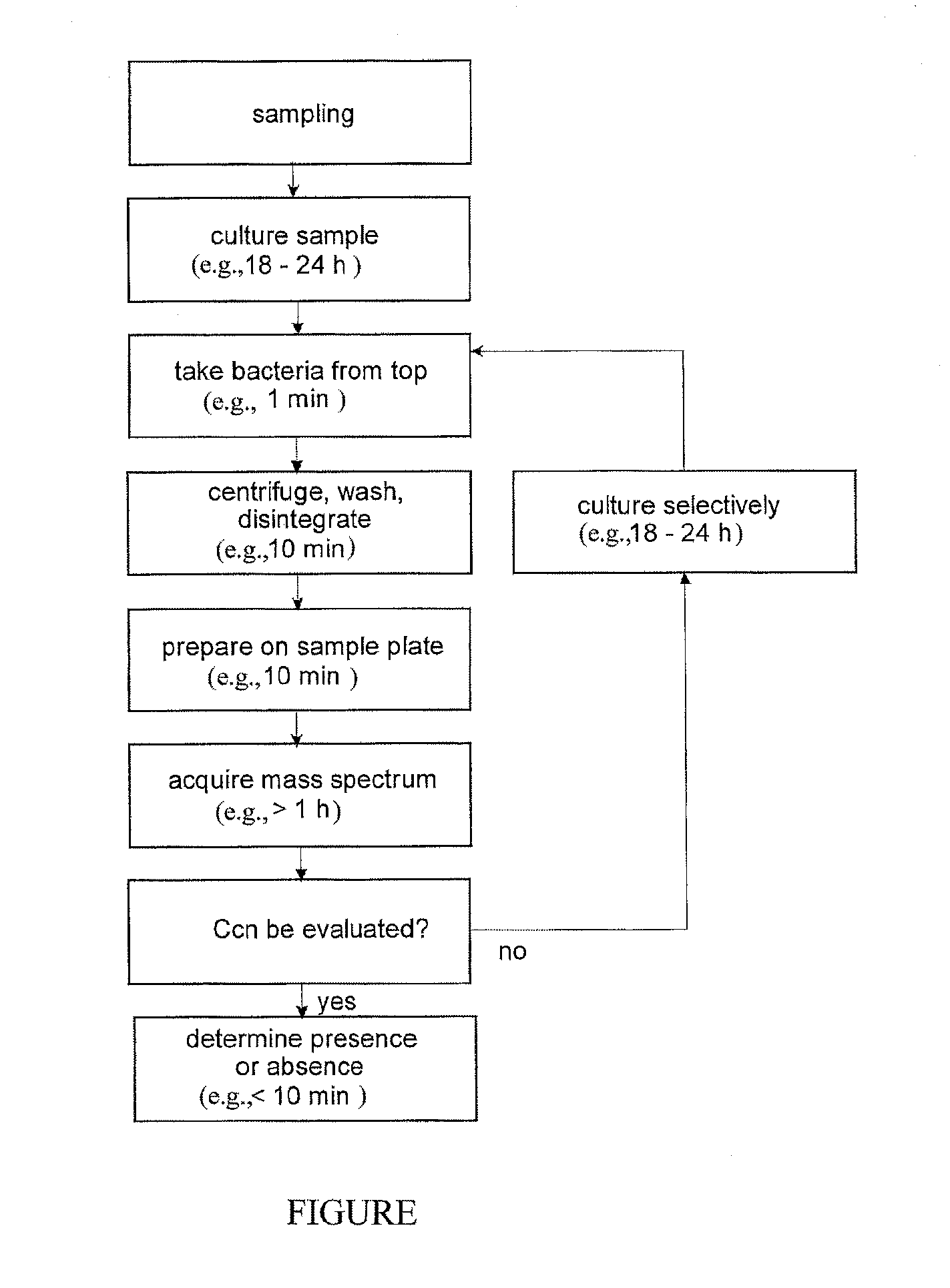
Mass spectrometric rapid detection of salmonella
ActiveUS20120107864A1Microbiological testing/measurementMass spectrometric analysisMicroorganismFeces
The invention relates to the detection of specified, flagellated bacteria, particularly Salmonella, in food and stool. A single culturing period of about 12 to 24 hours in a liquid nutrient medium without agitation is combined with a position-selective sampling of the flagellated microbes from the liquid of the culture, after which a mass spectrometric detection method is used which recognizes the target bacteria in mixtures. A second culture step is only necessary in exceptional cases. A species-selective or genus-selective culture medium is advantageous. Positional selection becomes possible because these bacteria use their flagella to counteract sedimentation by chemotaxis, and they collect near the surface. This provides a low-cost detection method that is several days faster than conventional methods
Owner:BRUKER DALTONIK GMBH & CO KG
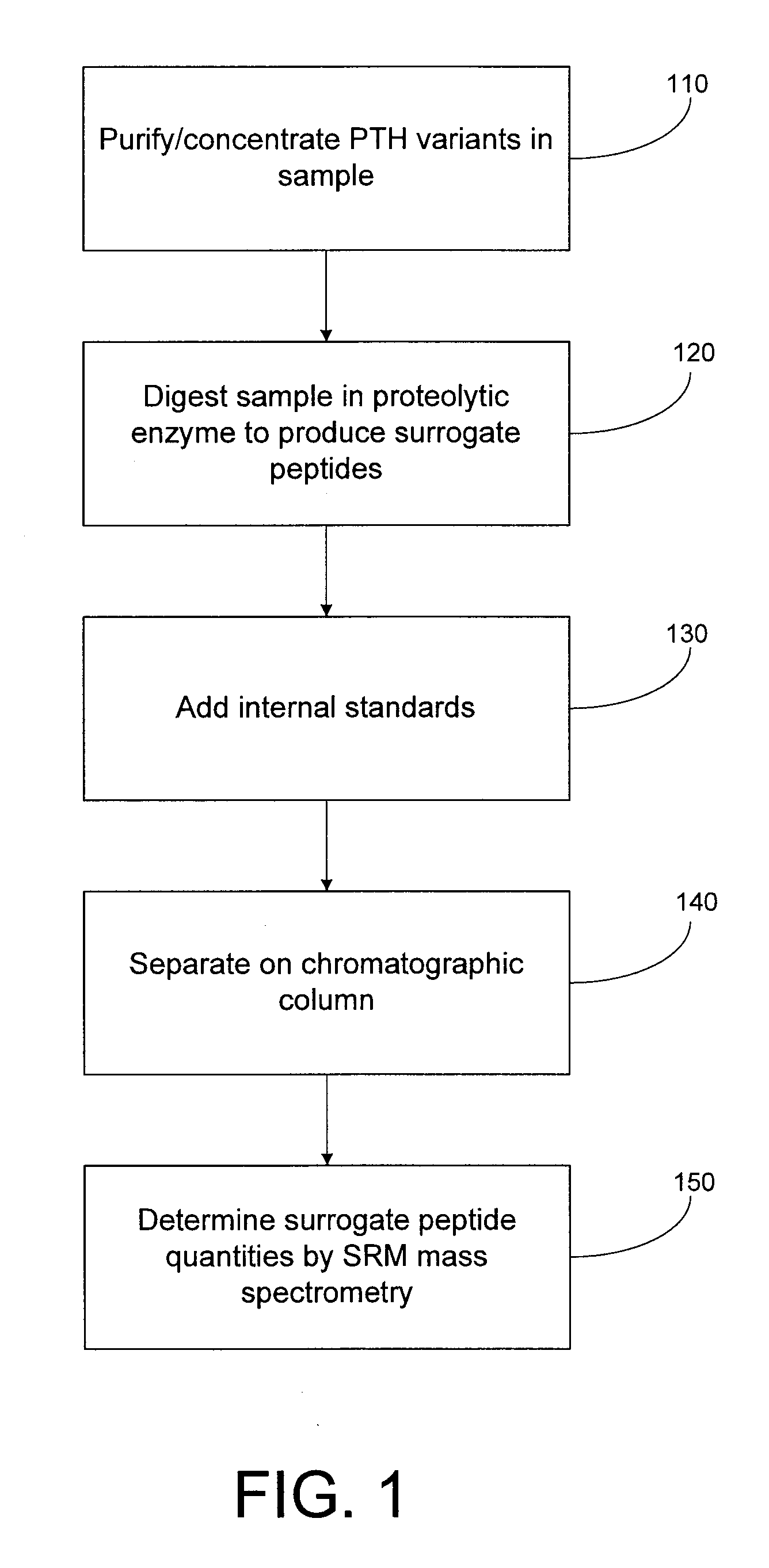
Assay for monitoring parathyroid hormone (PTH) variants by tandem mass spectrometry
InactiveUS8383417B2Mass spectrometric analysisMaterial analysis by electric/magnetic meansMass Spectrometry-Mass SpectrometryProtein mass spectrometry
Methods are described for monitoring the amounts of PTH variants in a biological sample by digesting the sample to produce surrogate peptides specific to the targeted PTH variants, and detecting and quantifying the surrogate peptides by selective reaction monitoring (SRM) mass spectrometry, using a set of precursor-to-product ion transitions optimized for sensitivity and selectivity. The PTH variants, or a portion thereof, may be concentrated in the sample by means of immunoaffinity capture or other suitable technique. The mass spectrometric method described herein enables the concurrent measurement of peptides representative of a plurality of targeted PTH variants in a single assay.
Owner:INTRINSIC BIOPROBES +1
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com