Transgenic plants for nitrogen fixation
A technology for nitrogen-fixing microorganisms and plants, applied in plant peptides, plant products, genetic engineering, etc., can solve the problems of reducing the annual yield of crops and transferring nitrogen-fixing organic matter.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
[0117] It is to be noted that the term "a" or "an" entity refers to one or more of that entity; for example, "a plant cell" is understood to mean one or more plant cells. Likewise, the terms "a" (or "an"), "one or more" and "at least one" are used interchangeably herein.
[0118] Cultivars and cultivars, respectively, are systematic groups of cultivated plants whose characteristics are distinct, uniform and stable and which, when propagated by appropriate means, retain these characteristics. A cultivar group is a group of appropriately named cultivars based on one or more criteria. A cultivated plant is a plant whose origin or selection is primarily attributable to the purposive activities of man. Such plants may be produced by deliberate or unintentional crossing of cultivars, or by selection from existing cultivars, or such plants may be the result of selection from a few variants in wild populations and produced only by deliberate Continuous propagation of cultivated plan...
Embodiment 1
[0167] Example 1: Gene Mining
[0168] The following description summarizes the process of how to define putative plant candidate genes involved in high nitrogen fixation potential in Oryza longistamina.
[0169] Abbreviations: BNF—biological nitrogen fixation; EST—expressed sequence tag;
[0170] nifH - one of the structural genes encoding nitrogenase, the key bacterial enzyme for nitrogen fixation.
[0171] First RNA analysis :
[0172] Since there is currently no O. longista genome sequence available for gene mining, deep-sequencing of the Oryza longistamin root transcriptome was performed on plants adapted to high BNF rates in soil without nitrogen fertilization. In total, early next-generation sequencing (pyrosequencing) revealed 60,155 ESTs that could be aligned to the japonica or indica genome sequence, of which a remarkable 9.993 were genes that had not been detected to be expressed in rice. About 11,000 ESTs are potential novel sequence tags found only in our Ory...
Embodiment 2
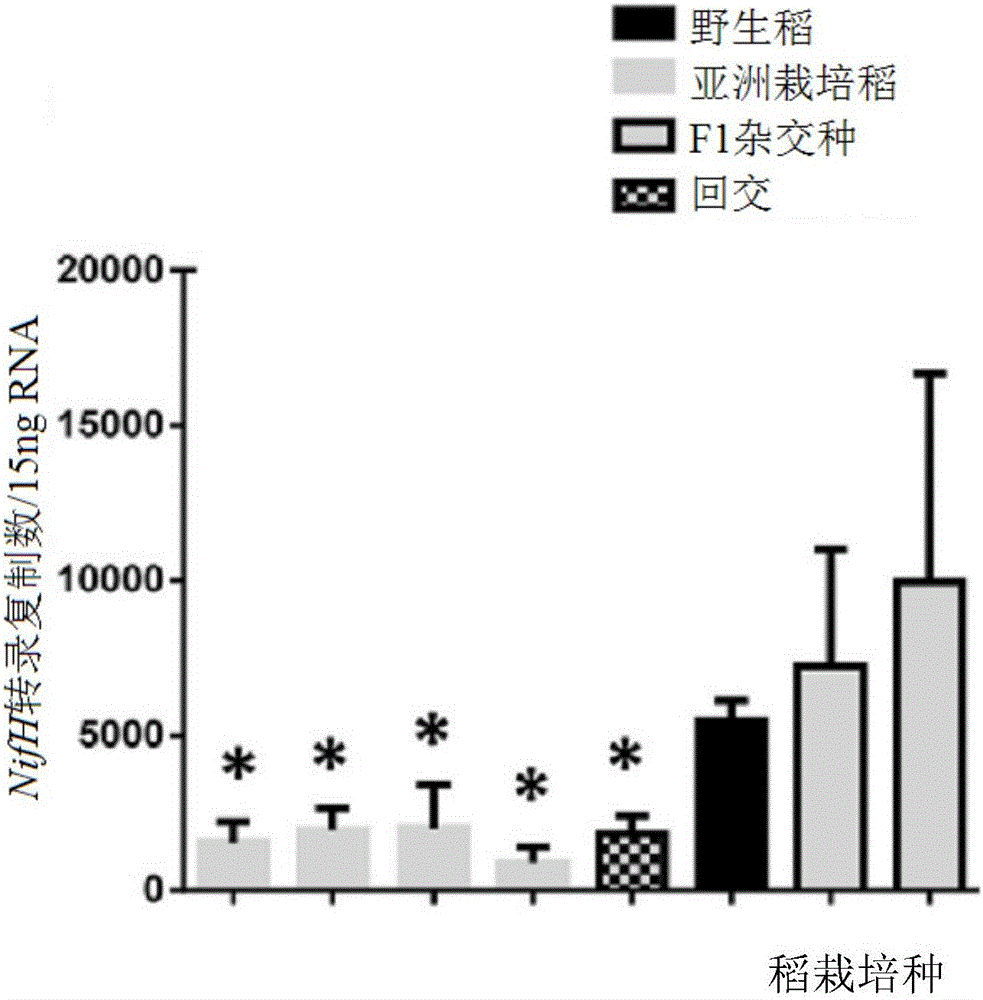
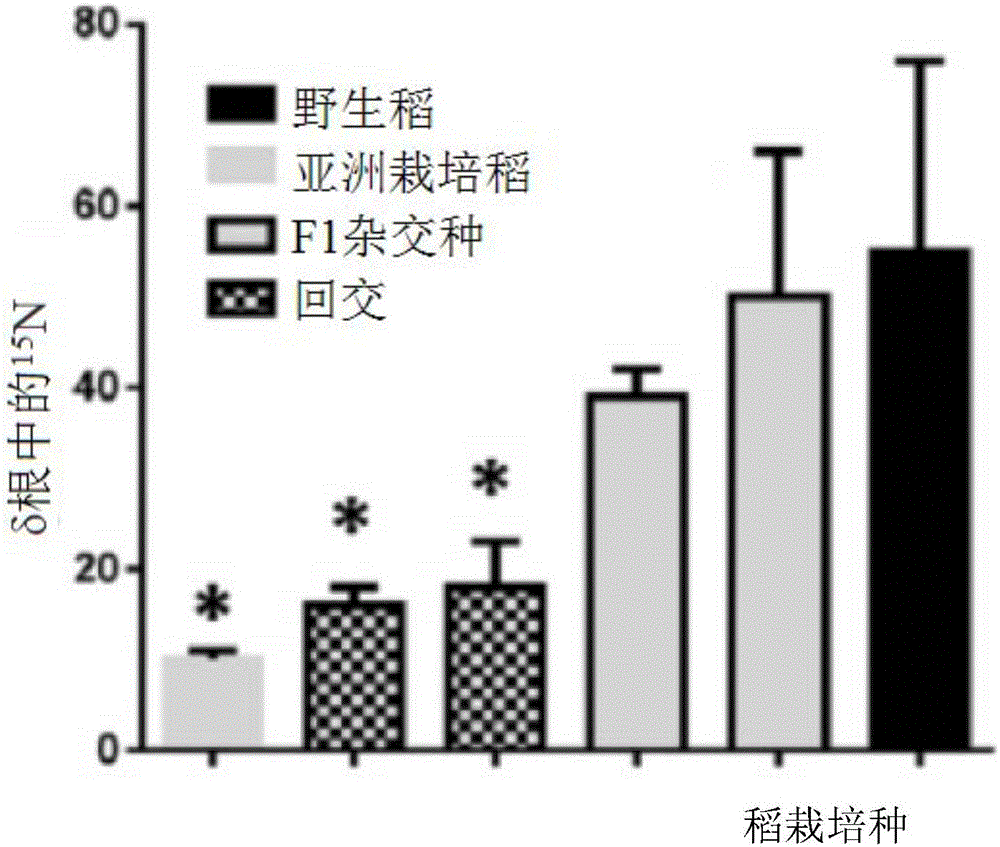
[0288] Example 2: Estimation of nifH transcript abundance
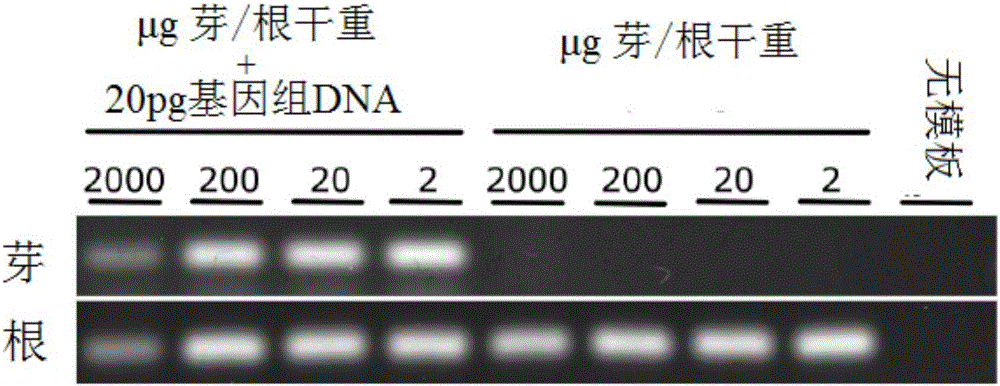
[0289] Levels of NifH transcripts associated with plant roots were assessed from total RNA extracts from plant roots as previously published (Hurek et al., 2002) and modified (Burbano et al., 2011, supra). figure 1 In, DNA amplification of the nifH fragment was performed with similar primers as in Hurek et al., 2002. for figure 2 , Amplification of the nifH fragment was obtained by the universal primer set in the nested reverse transcription (RT)-PCR method (Burbano et al., 2011, supra): A nested RT-PCR protocol was used. The RT step was performed at 42°C using primer nifH3. PCR amplification was performed with primers nifH3 and nifH4 using annealing 55°C. A second round of nested PCR amplification was performed with 1 μl of the first round product and primers ZehrF and ZehrR. For this second round, quantitative real-time PCR was used for quantification (Bio-Rad CFX 96). The actin gene was used to normalize quan...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com