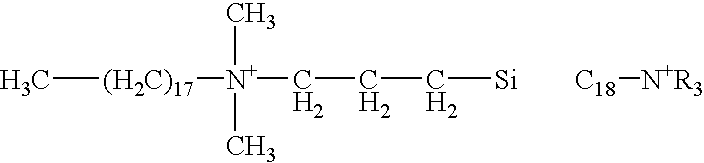
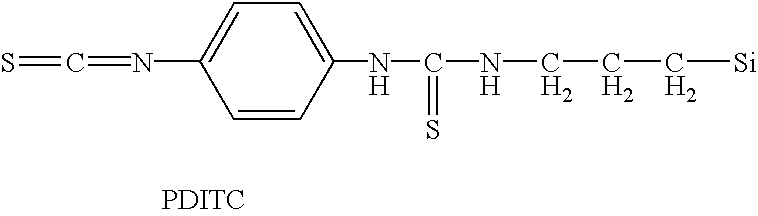
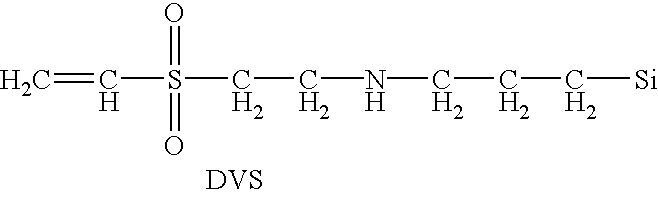
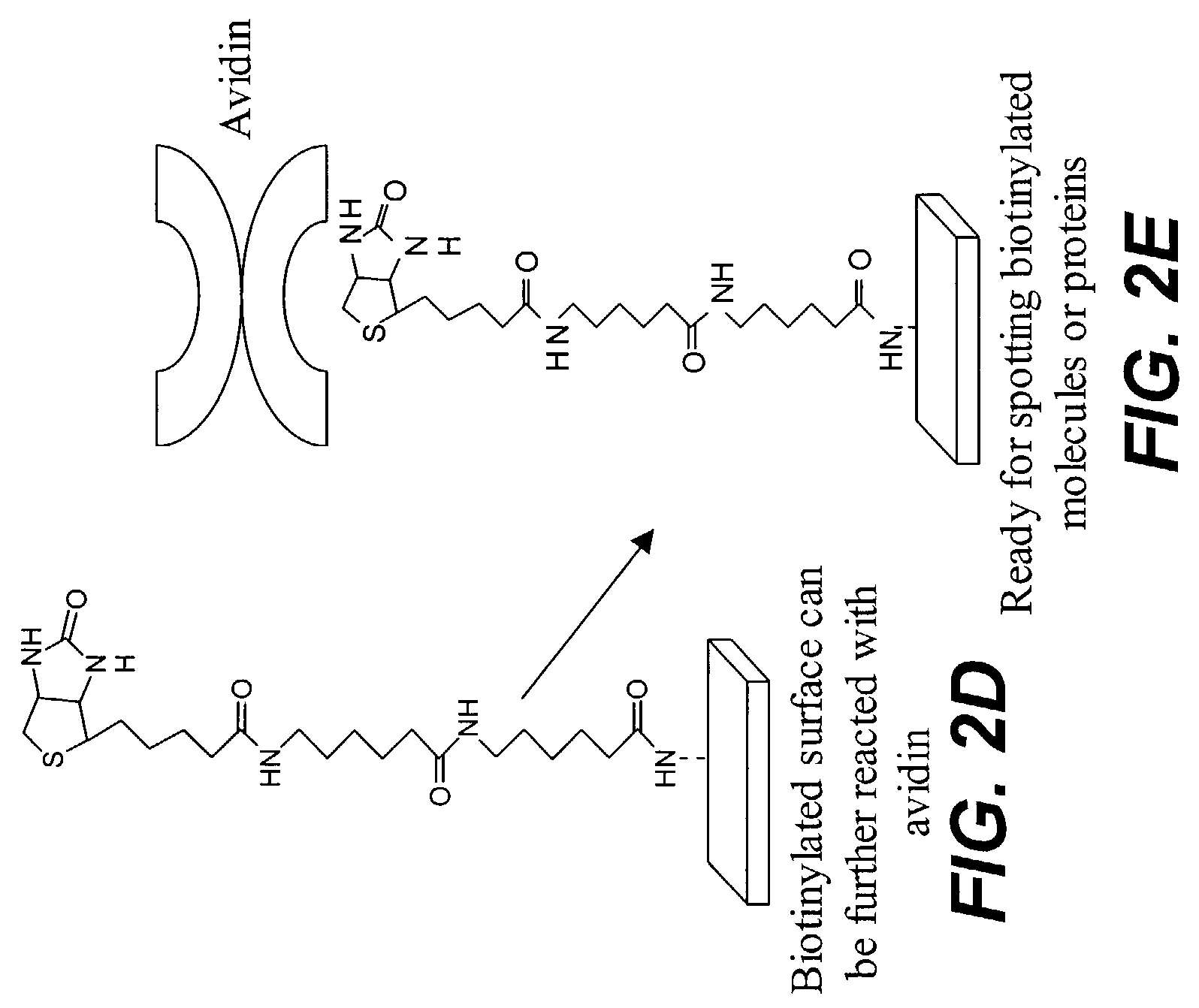
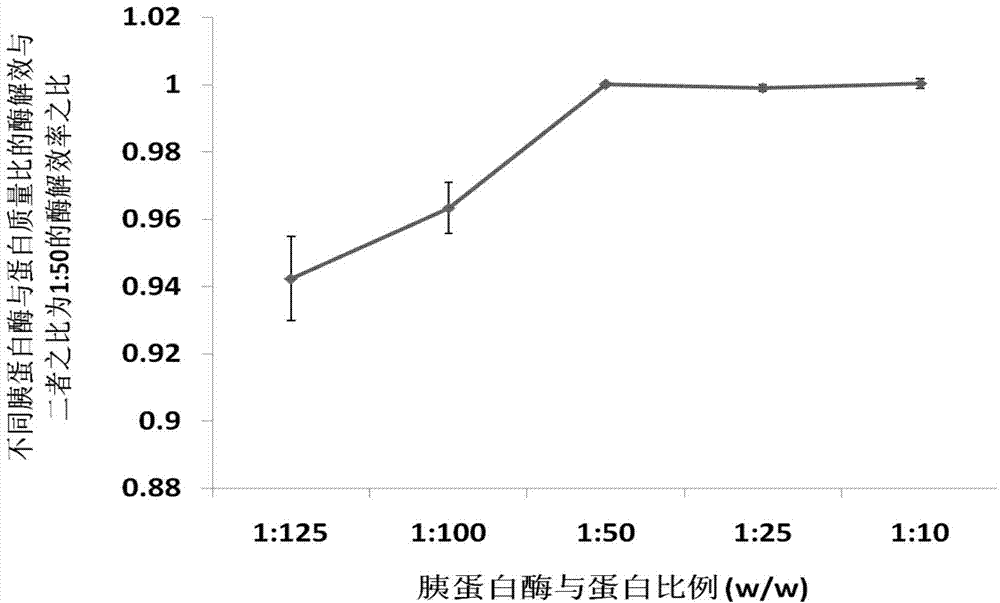
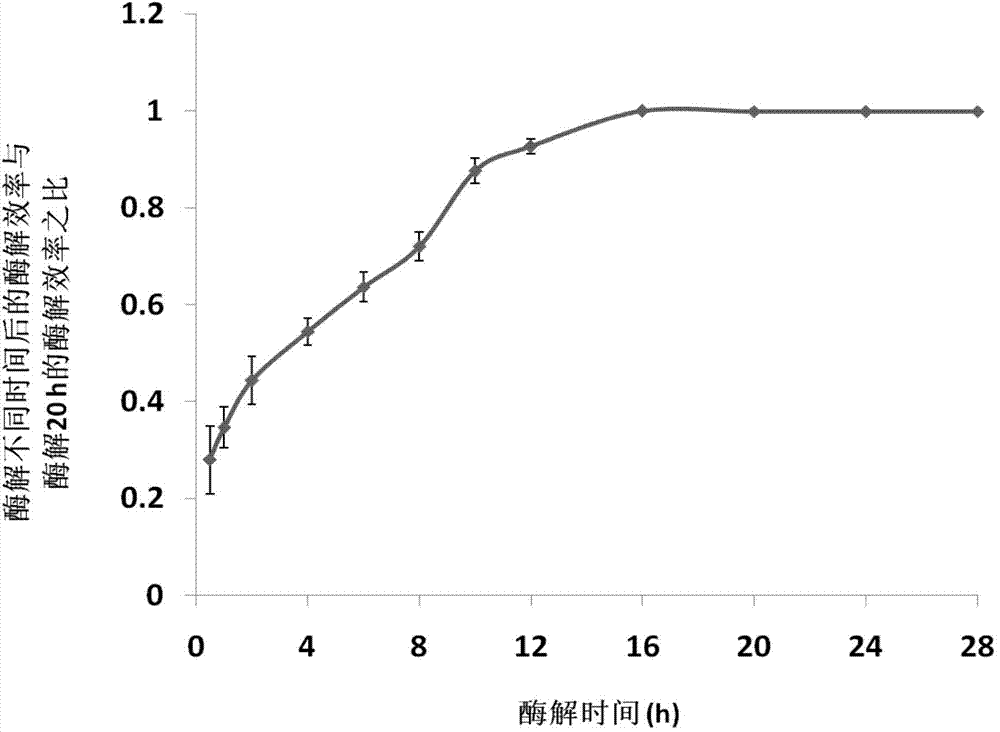
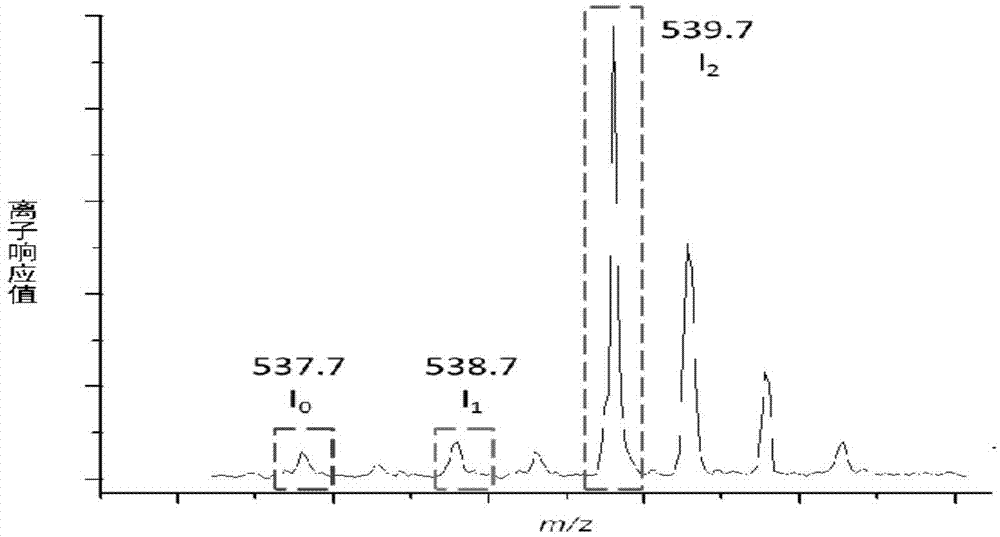
Patents
Literature
95 results about "Proteomic Profile" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Information about all proteins that are made in blood, other body fluids, or tissues, at certain times. A protein expression profile may be used to find and diagnose a disease or condition and to see how well the body responds to treatment.
Proteomic analysis of biological fluids
ActiveUS20070161125A1Eliminate needMicrobiological testing/measurementAnalogue computers for chemical processesDiseaseGynecology
The invention concerns the identification of proteomes of biological fluids and their use in determining the state of maternal / fetal conditions, including maternal conditions of fetal origin, chromosomal aneuploidies, and fetal diseases associated with fetal growth and maturation. In particular, the invention concerns a comprehensive proteomic analysis of human amniotic fluid (AF) and cervical vaginal fluid (CVF), and the correlation of characteristic changes in the normal proteome with various pathologic maternal / fetal conditions, such as intra-amniotic infection, pre-term labor, and / or chromosomal defects. The invention further concerns the identification of biomarkers and groups of biomarkers that can be used for non-invasive diagnosis of various pregnancy-related disorders, and diagnostic assays using such biomarkers.
Owner:HOLOGIC INC
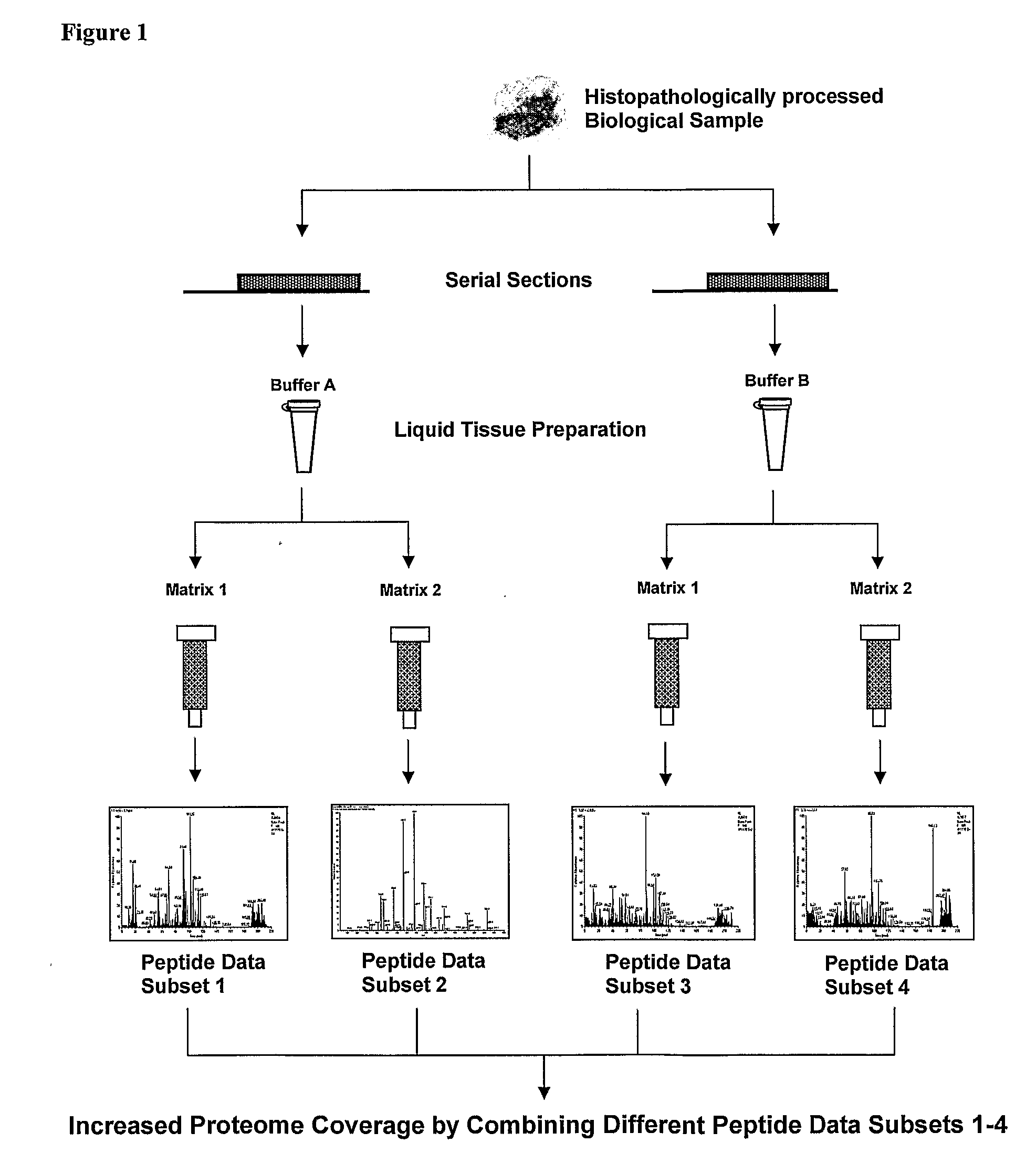
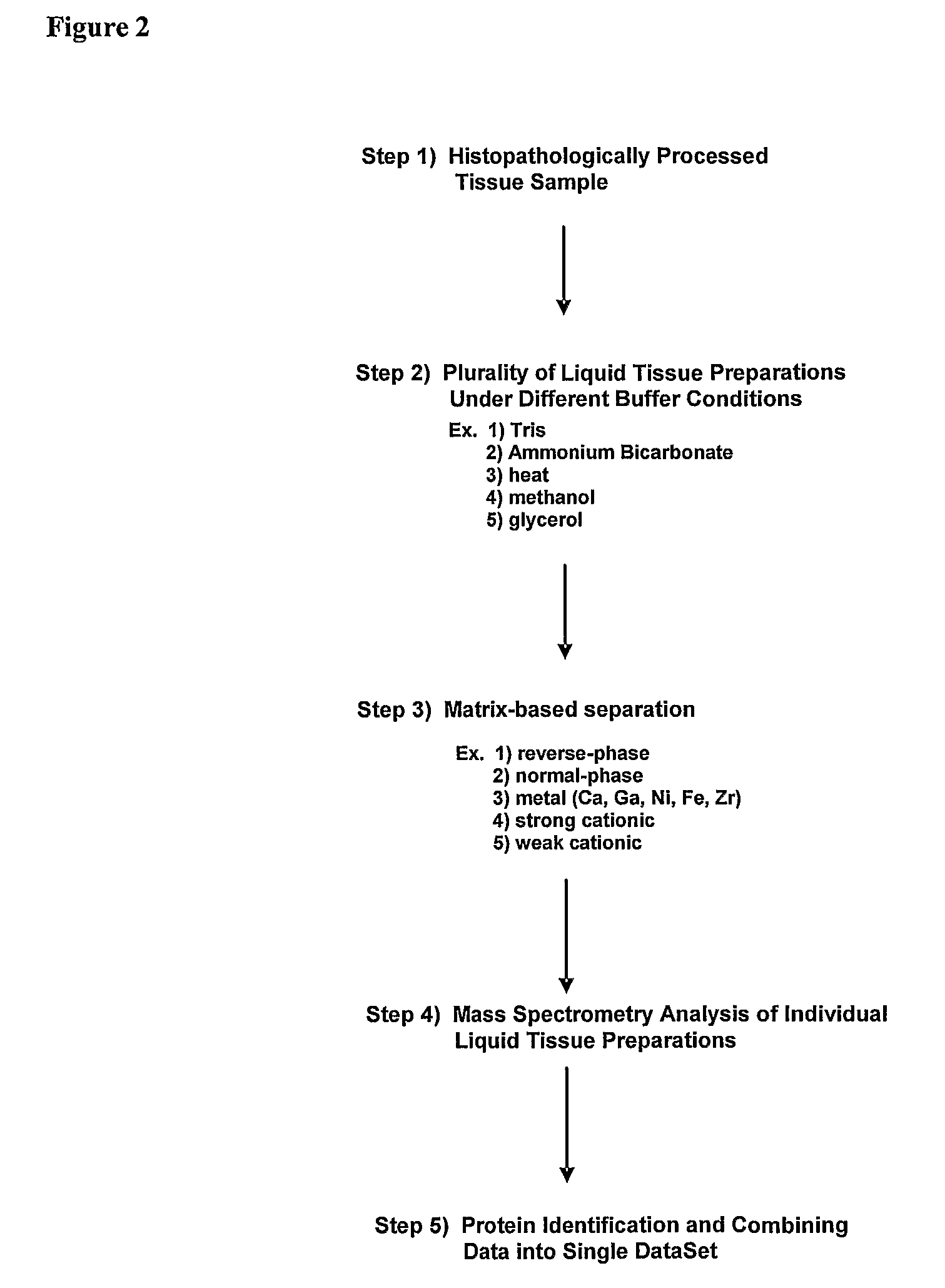
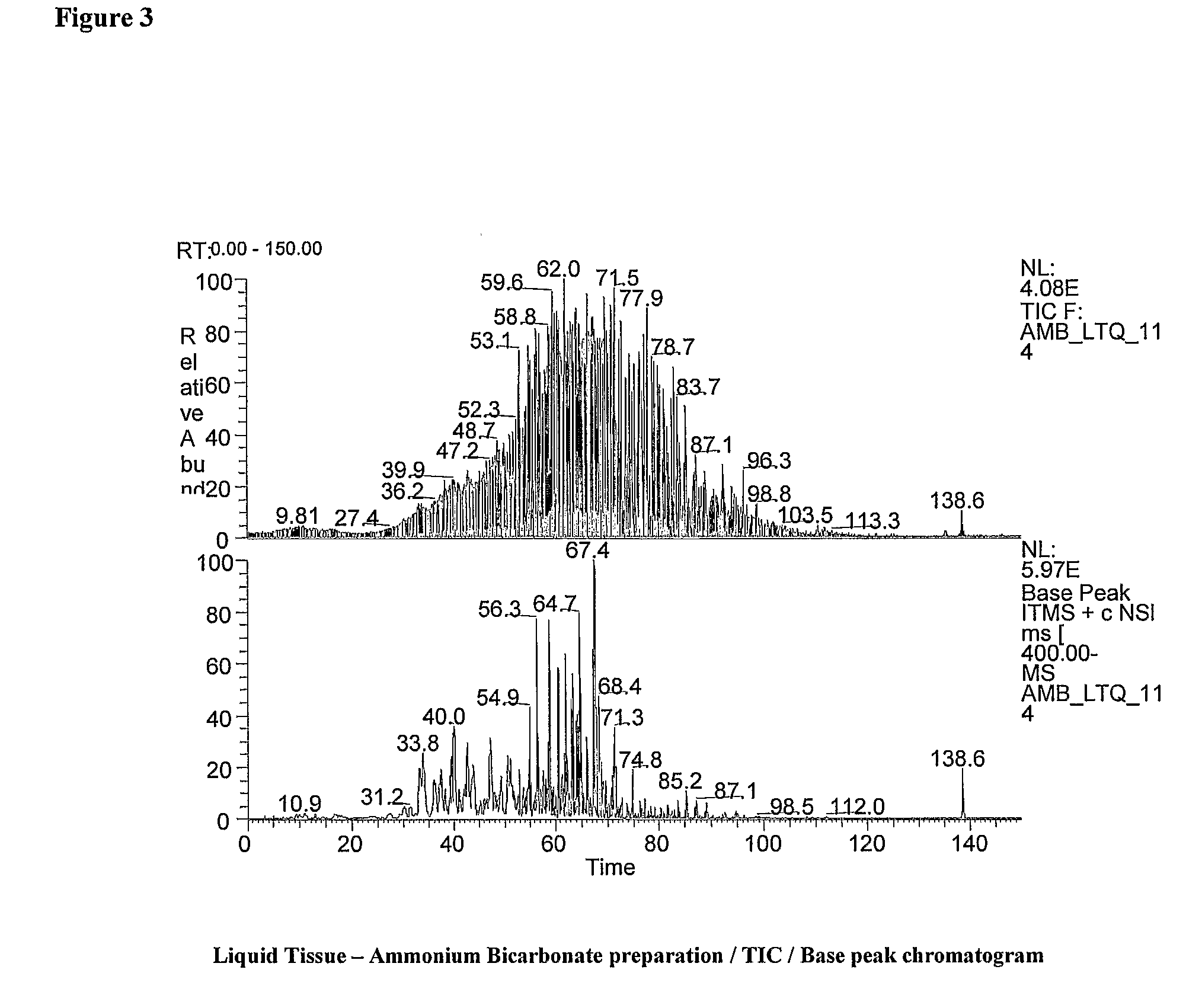
Multiplex liquid tissue method for increased proteomic coverage from histopathologically processed biological samples, tissues, and cells
ActiveUS20090136971A1Microbiological testing/measurementPreparing sample for investigationFluid tissuesProteomic Profile
The invention provides methods for multiplex analysis of biological samples of formalin-fixed tissue samples. The invention provides for a method to achieve a multiplexed, multi-staged plurality of Liquid Tissue preparations simultaneously from a single histopathologically processed biological sample, where the protocol for each Liquid Tissue preparation imparts a distinctive set of biochemical effects on biomolecules procured from histopathologically processed biological samples and which when each of the preparations is analyzed can render additive and complementary data about the same histopathologically processed biological sample.
Owner:EXPRESSION PATHOLOGY
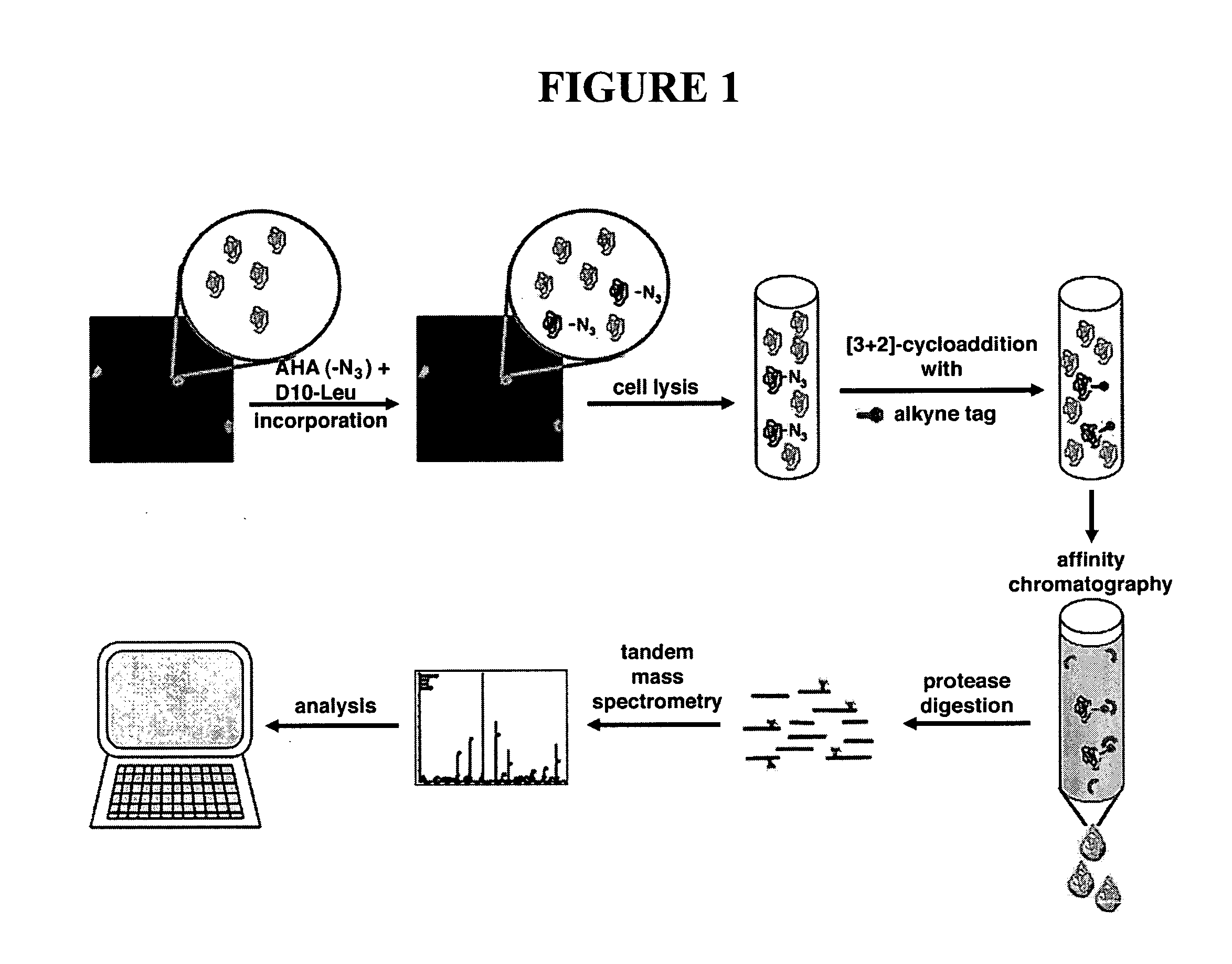
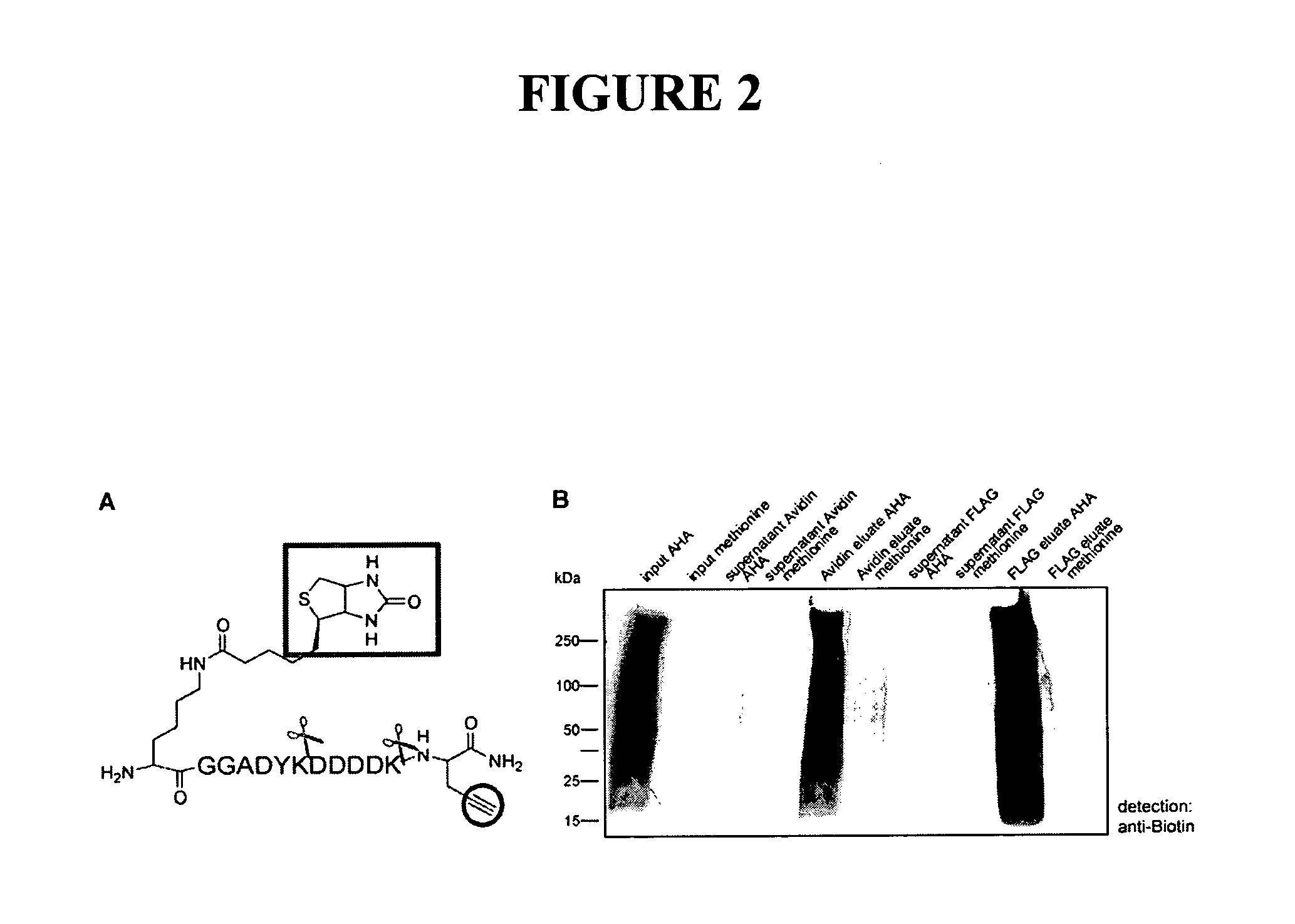
Use of non-canonical amino acids as metabolic markers for rapidly-dividing cells
InactiveUS20100247433A1Inhibit progressFacilitates killingBiocidePeptide/protein ingredientsHigh-Throughput Screening MethodsCancer cell
The invention provides methods, reagents and systems to preferentially mark fast-proliferating cells / tissues (such as cancer), by incorporating non-natural amino acids into proteins, preferably in vivo, using the endogenous protein synthesis machinery of an organism. The incorporated non-natural amino acids contain reactive groups for further chemical reagents, which may serve as a “handle” to for a number of uses, such as imaging of cancer cells, targeting drugs to preferentially kill cancer cells, and proteomic analysis in the context of large scale or high throughput screening for candidate drug leads that affects the proliferation of a target cell, etc.
Owner:CALIFORNIA INST OF TECH
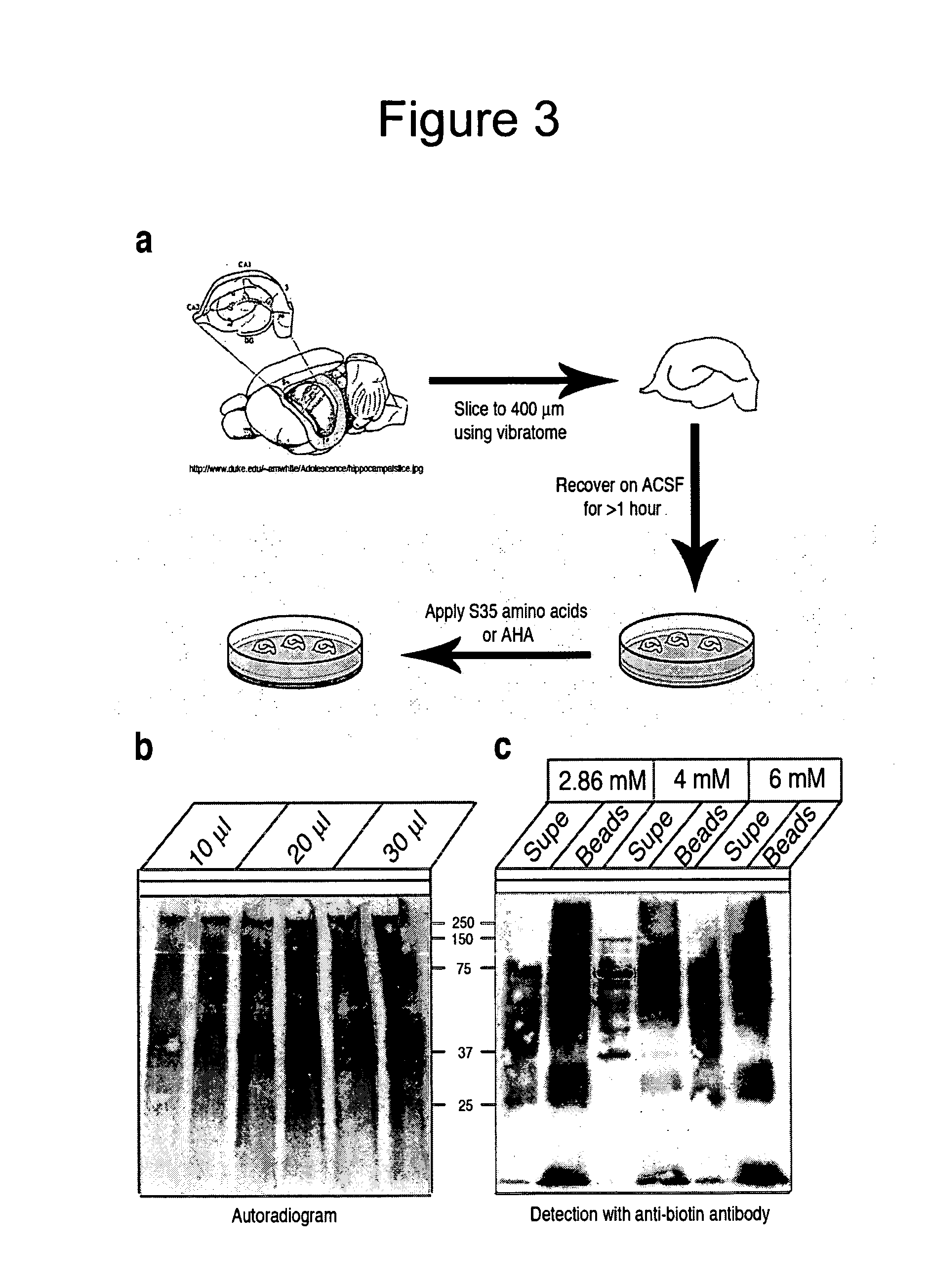
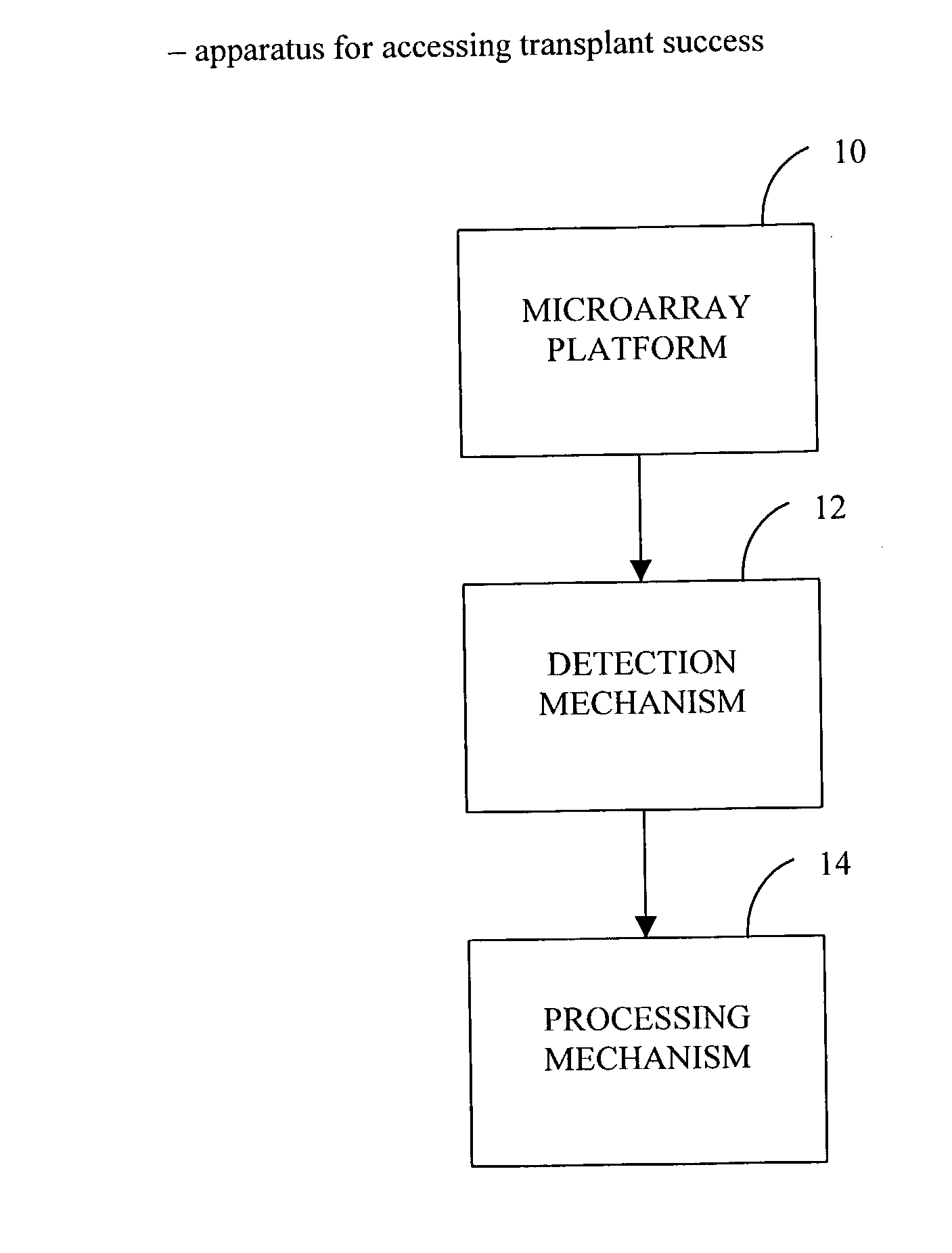
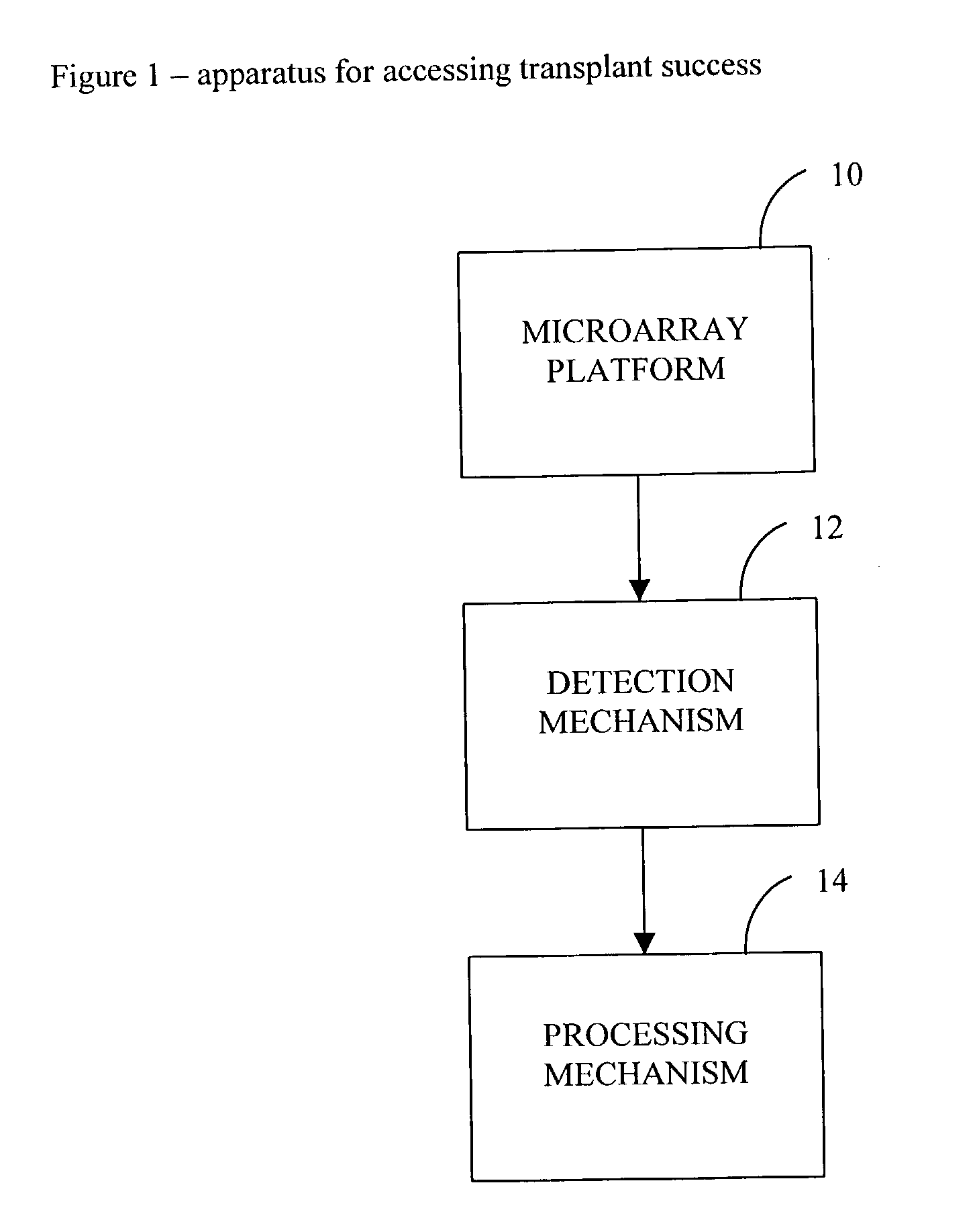
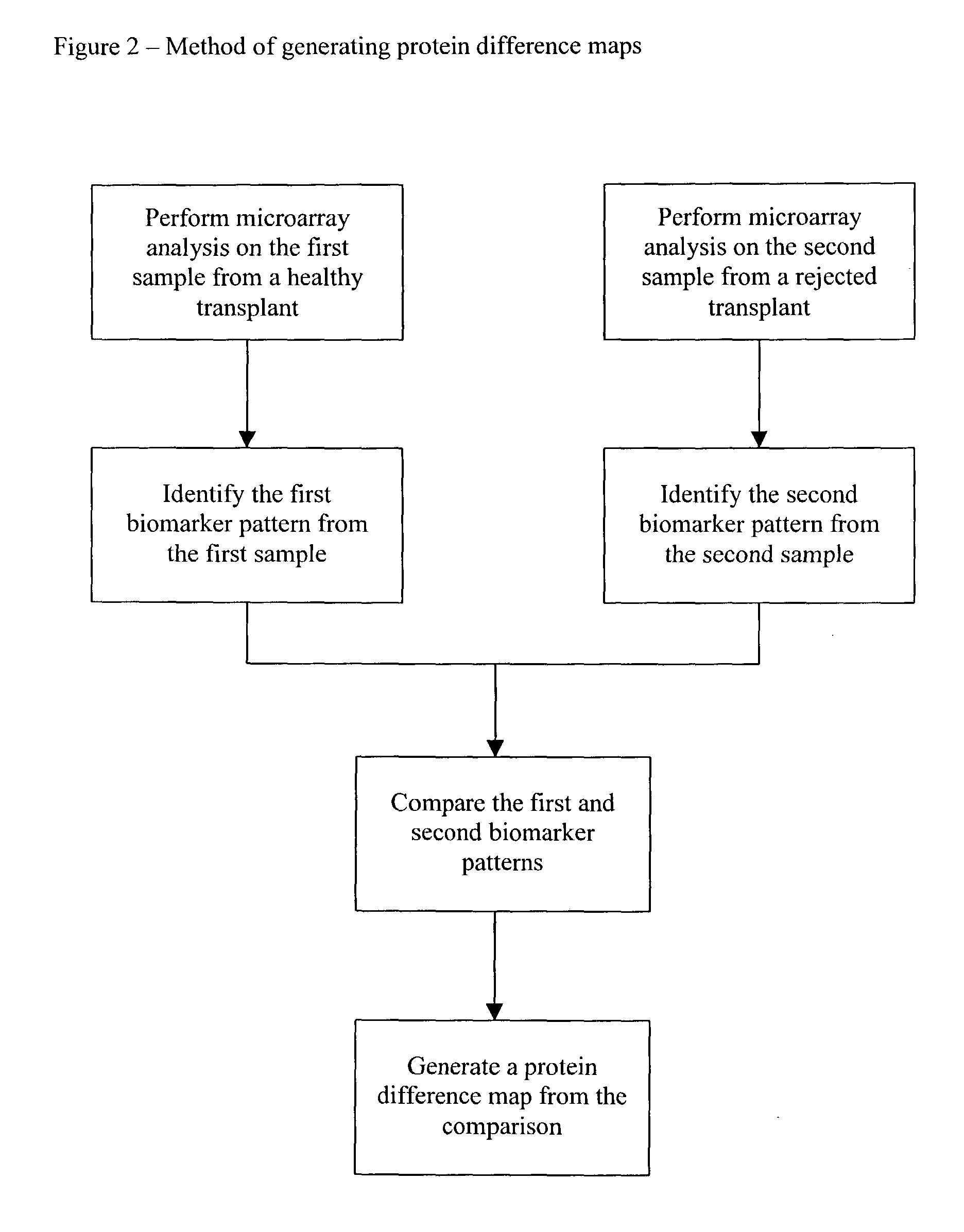
Method and use of protein microarray technology and proteomic analysis to determine efficacy of human and xenographic cell, tissue and organ transplant
InactiveUS20030232396A1Bioreactor/fermenter combinationsBiological substance pretreatmentsOrgan transplantationComputerized system
The present invention is directed to systems and methods for assessing the success of the transplant of a cell, tissue, or organ before and after transplant. Protein array technology is used to obtain a biomarker pattern for the cell, tissue, or organ that is being considered for transplant or that has been transplanted. Samples for the identification of biomarkers and biomarker patterns are obtained from the cell, tissue or organ itself, or from a body fluid of the donor or recipient. Sample biomarker data are compared to reference biomarker data obtained from donors, recipients or cells, tissues or organs that have been transplanted. Correlation of a sample biomarker pattern with the reference biomarker pattern, where transplant outcome for the samples used for the reference biomarkers is known, permits a suggested treatment determination. A computerized system to identify the condition of transplant before or after implantation is also provided.
Owner:BIOLIFE SOLUTIONS INC
Highly sensitive proteomic analysis methods, and kits and systems for practicing the same
InactiveUS20030036095A1Preparing sample for investigationBiological material analysisProteomic ProfileIon
Methods of determining whether a sample includes one or more analytes, particularly proteinaceous analytes, of interest are provided. In the subject methods, an array of binding agents, where each binding agent includes an epitope binding domain of an antibody, is contacted with the sample. In many embodiments, contact occurs in the presence of a metal ion chelating polysaccharide, e.g., a pectin. Following contact, the presence of binding complexes on the array surface are detected and the resultant data is employed to determine whether the sample includes the one or more analytes of interest. Also provided are kits, systems and other compositions of matter for practicing the subject methods. The subject methods and compositions find use in a variety of applications, including proteomic applications such as protein expression analysis, e.g., differential protein expression profiling.
Owner:CLONTECH LAB
Labeling of proteomic samples during proteolysis for quantitation and sample multiplexing
InactiveUS6864089B2Improve throughputBiological testingFermentationMultiplexingMass Spectrometry-Mass Spectrometry
Owner:PROTANA
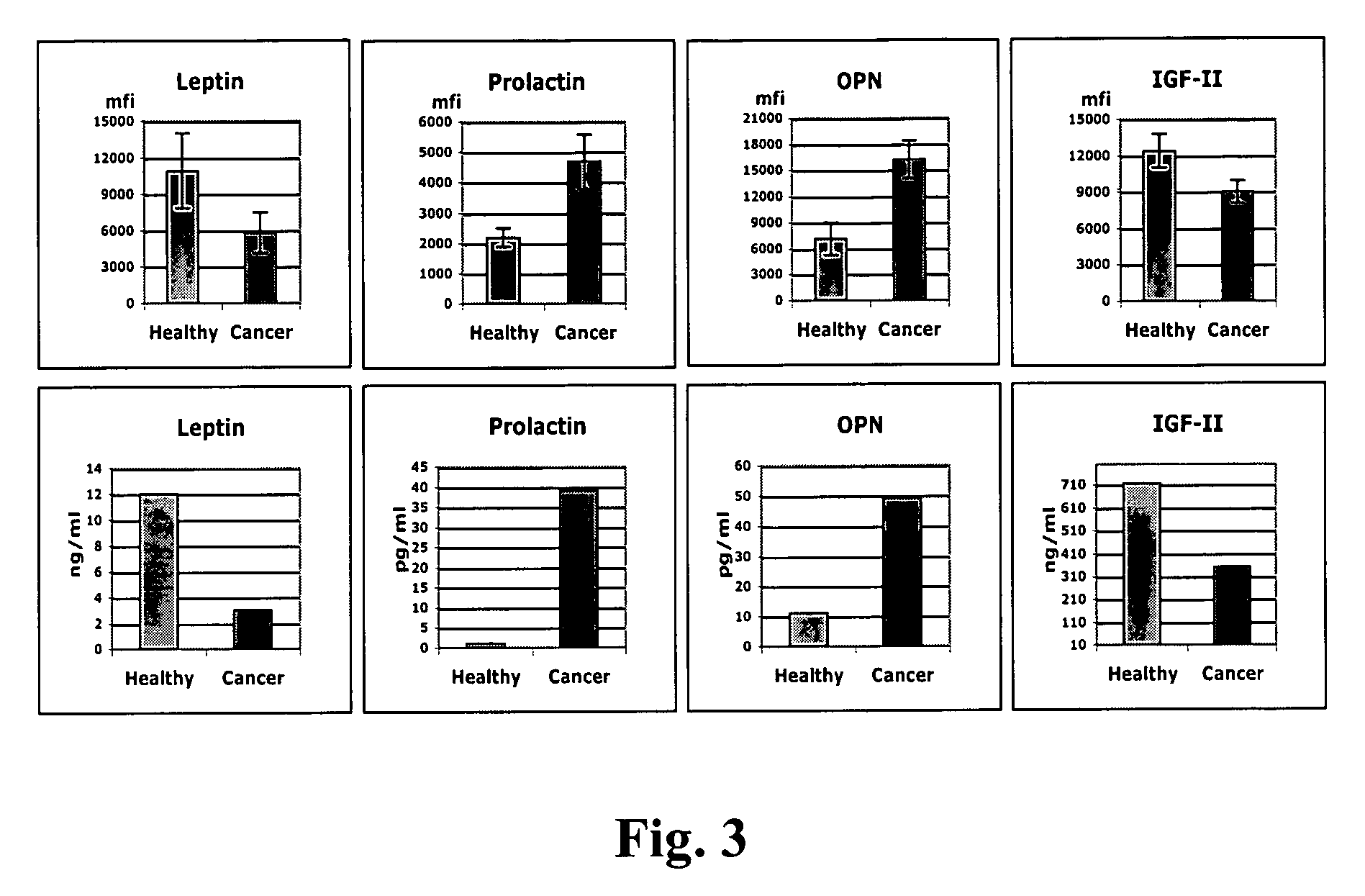
Identification of cancer protein biomarkers using proteomic techniques
The claimed invention describes methods to diagnose or aid in the diagnosis of cancer. The claimed methods are based on the identification of biomarkers which are particularly well suited to discriminate between cancer subjects and healthy subjects. These biomarkers were identified using a unique and novel screening method described herein. The biomarkers identified herein can also be used in the prognosis and monitoring of cancer. The invention comprises the use of leptin, prolactin, OPN and IGF-II for diagnosing, prognosis and monitoring of ovarian cancer.
Owner:YALE UNIV
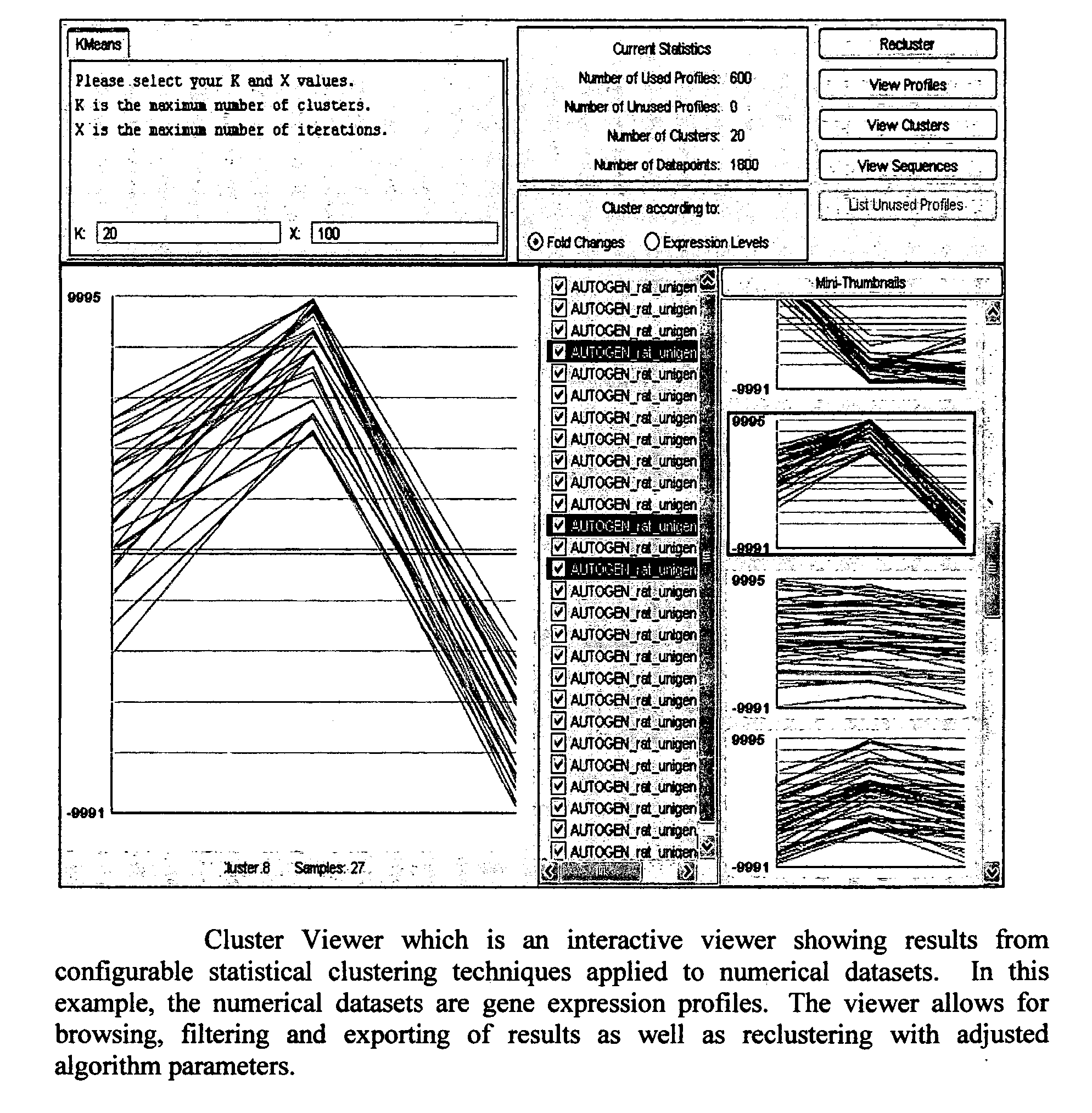
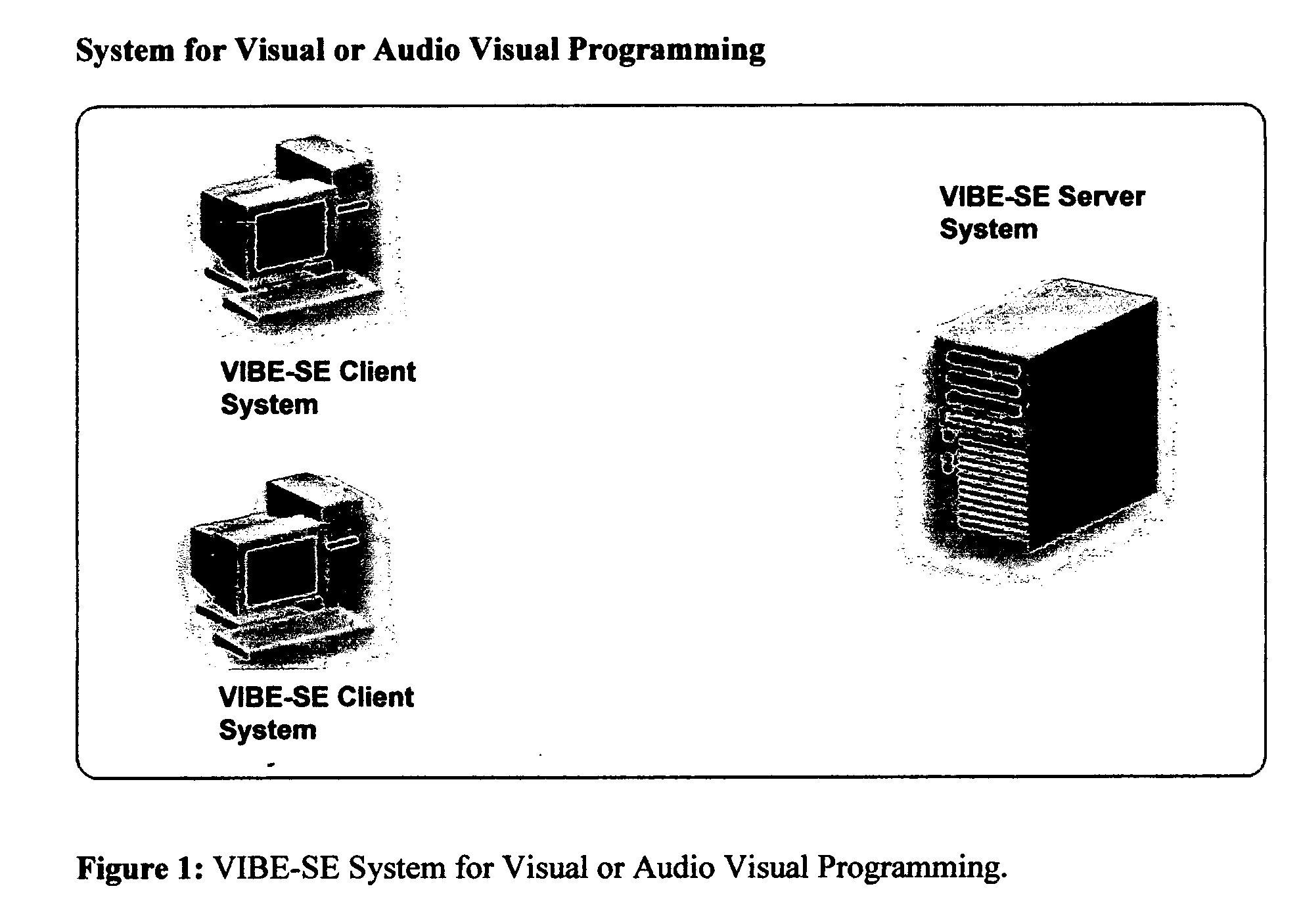
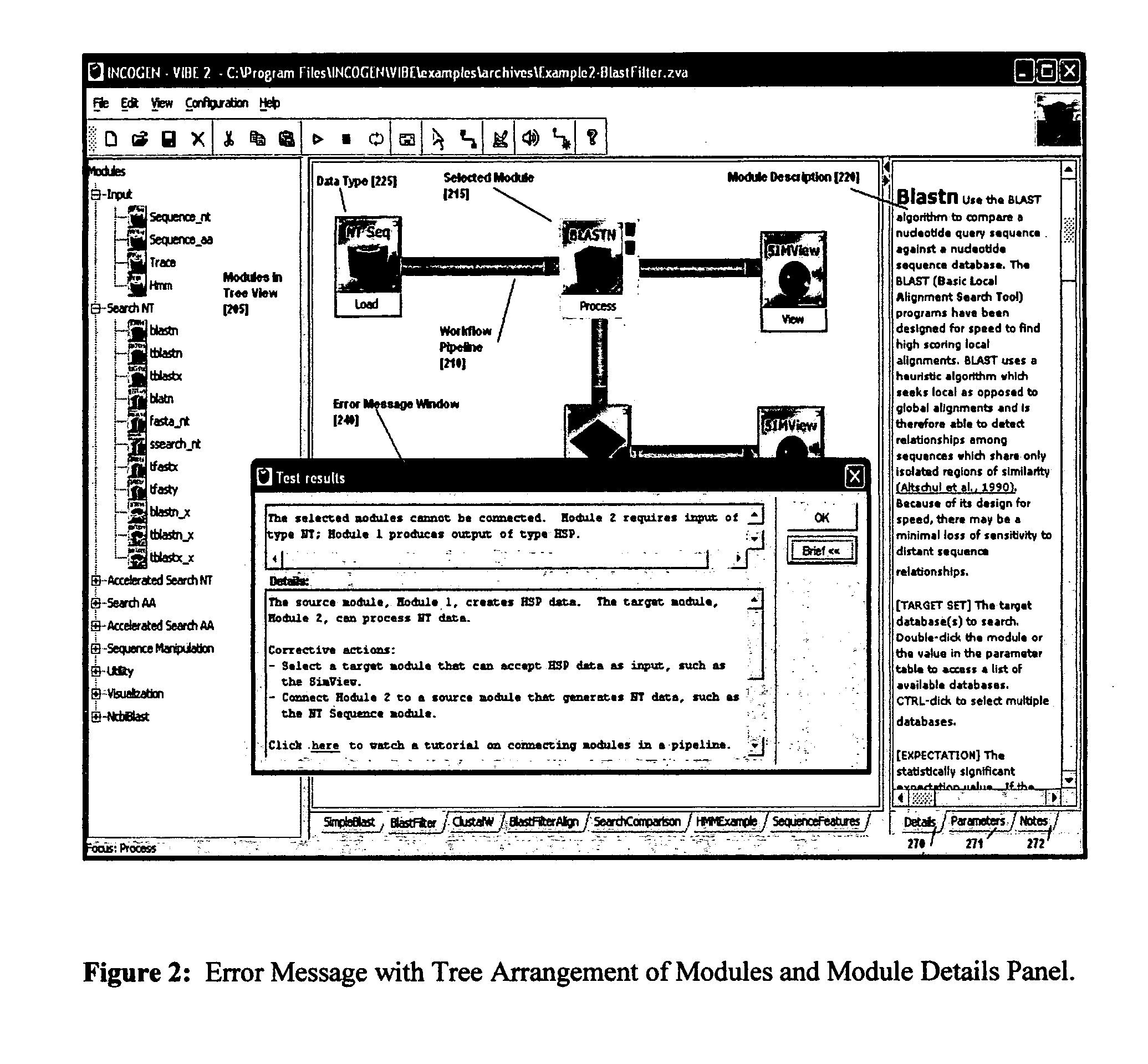
System and method using a visual or audio-visual programming environment to enable and optimize systems-level research in life sciences
InactiveUS20060190184A1Enabling and optimizing systems-level researchEnhances with user-level extensibilityData visualisationBiological testingAnalysis dataProteomic Profile
The current invention provides a visual or audio-visual programming environment for life science and bioinformatics. It is based on the VIBE platform, which is a flexible, extensible, and integrated workflow construction and management platform. The current invention enables researchers to consolidate molecular profiling data from complementary experimental techniques, intelligently reduce the volume of the data, construct disease-specific molecular fingerprints, construct relationship networks among functionally significant genomic, transcriptomic, metabonomic, and proteomic data, integrate information from existing biological databases into those networks, optimize the process through iterative feedback loops, and to generate and validate hypotheses based on the above process. The uses of this integrative, systems-based approach include, but are not limited to, the identification of potential biomarkers, characterization and classification of diseases and pathogens, and discovery of drug targets.
Owner:INCOGEN
Labeling of proteomic samples during proteolysis for quantitation and sample multiplexing
InactiveUS20020076817A1Microbiological testing/measurementBiological testingMultiplexingMass Spectrometry-Mass Spectrometry
This invention related to methods useful in the labeling of multiple polypeptide samples and subsequent analysis of these samples by mass spectrometry, particularly in the high throughput proteomic setting.
Owner:PROTANA
Protein microarrays on mirrored surfaces for performing proteomic analyses
InactiveUS7148058B2Enhanced signalFacilitating drug discoveryCompound screeningMaterial nanotechnologyAntigenStructure analysis
Provided are protein microarrays, their manufacture, use, and application. Protein microarrays in accordance with the present invention are useful in a variety preoteomic analyses. Various protein arrays in accordance with the present invention may immobilize large arrays of proteins that may be useful for studying protein-protein interactions to improve understanding of disease processes, facilitating drug discovery, or for identifying potential antigens for vaccine development. The protein array elements of the invention are native or modified proteins (e.g., antibodies or fusion proteins). The protein array elements may be attached directly to a organic functionalized mirrored substrate by a binding reaction between functional groups on the substrate (e.g., amine) and protein (e.g., activated carboxylic acid). Techniques for chemical blocking of the arrays are also provided. The invention contemplates spotting of array elements onto solid planar substrates, labeling of complex protein mixtures, and the analysis of protein binding to the array. The invention also enables the enrichment or purification, and subsequent sequencing or structural analysis of proteins that are identified as differential by the array screen. Kits including protein-binding microarrays for proteomic analysis in accordance with the present invention are also provided.
Owner:CHIRON CORP
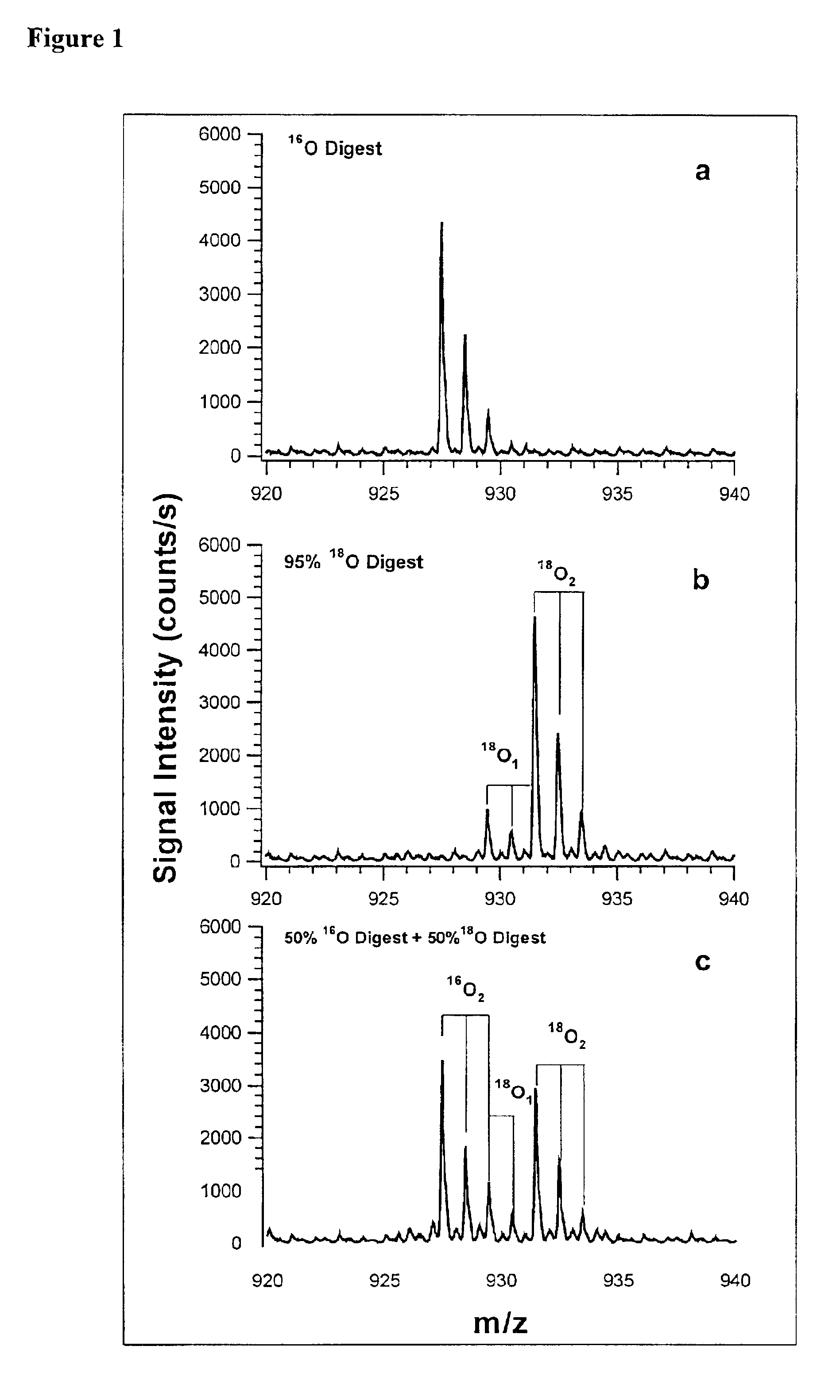
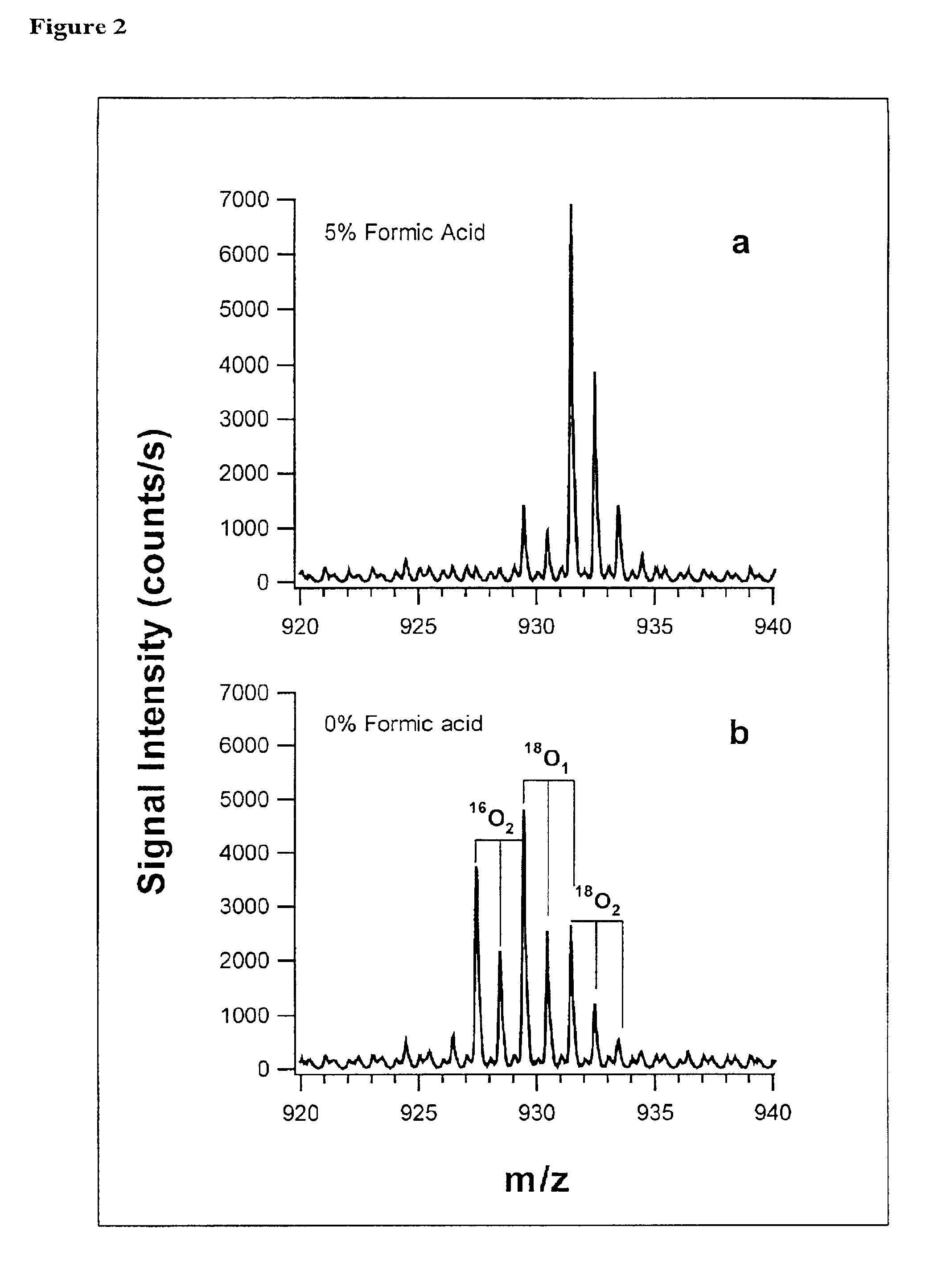
Polypeptide marker for early diagnosis of diabetes mellitus
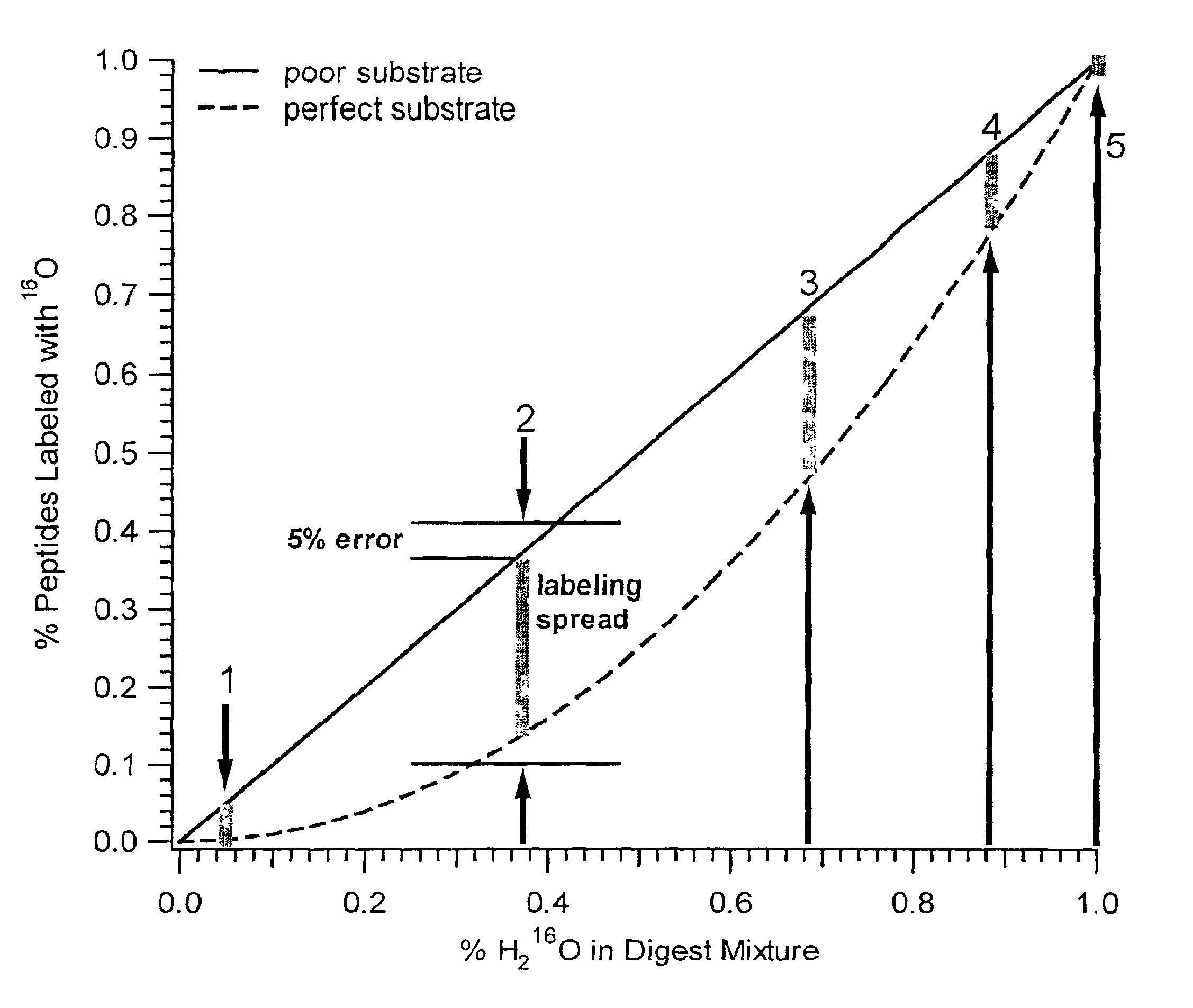
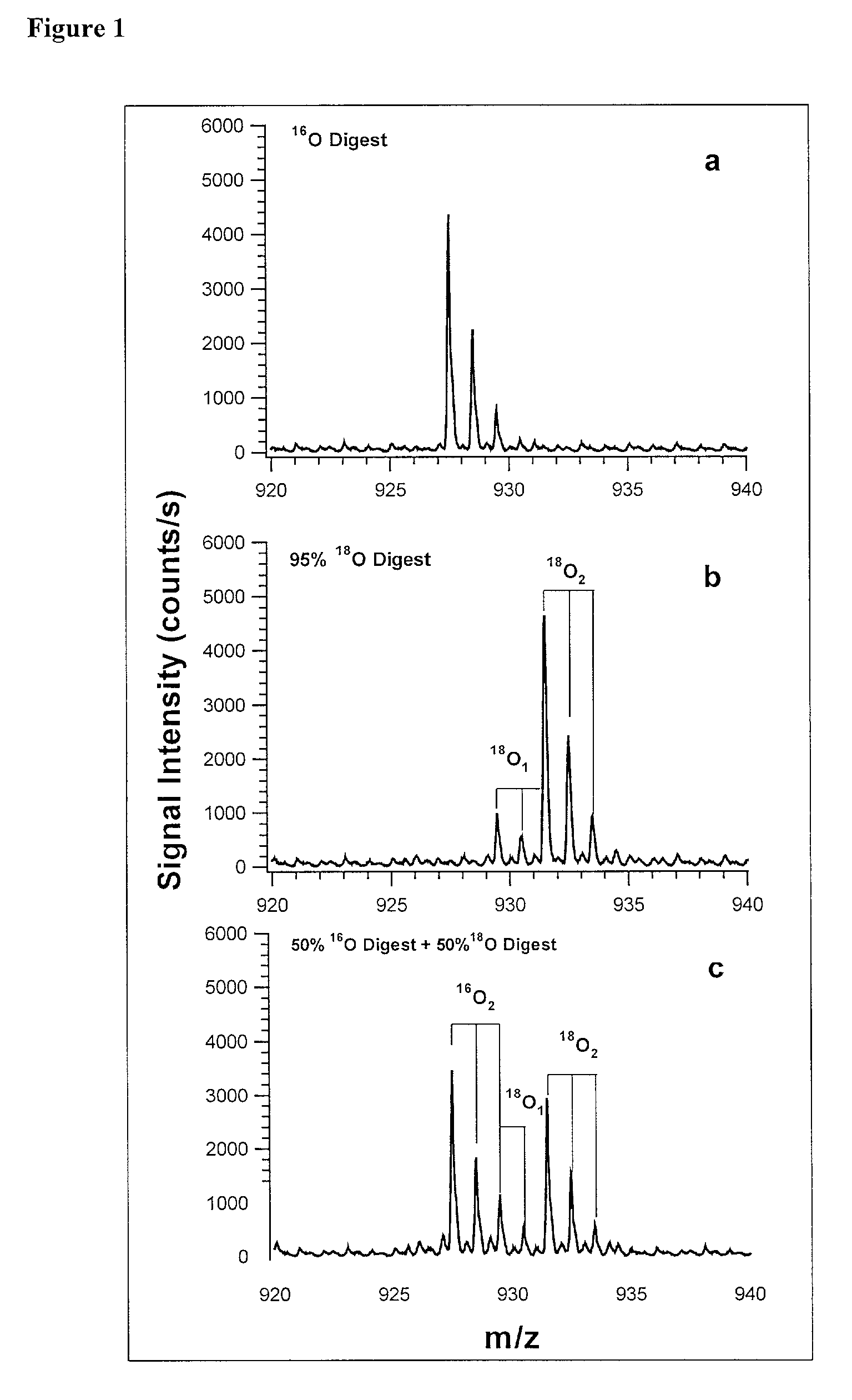
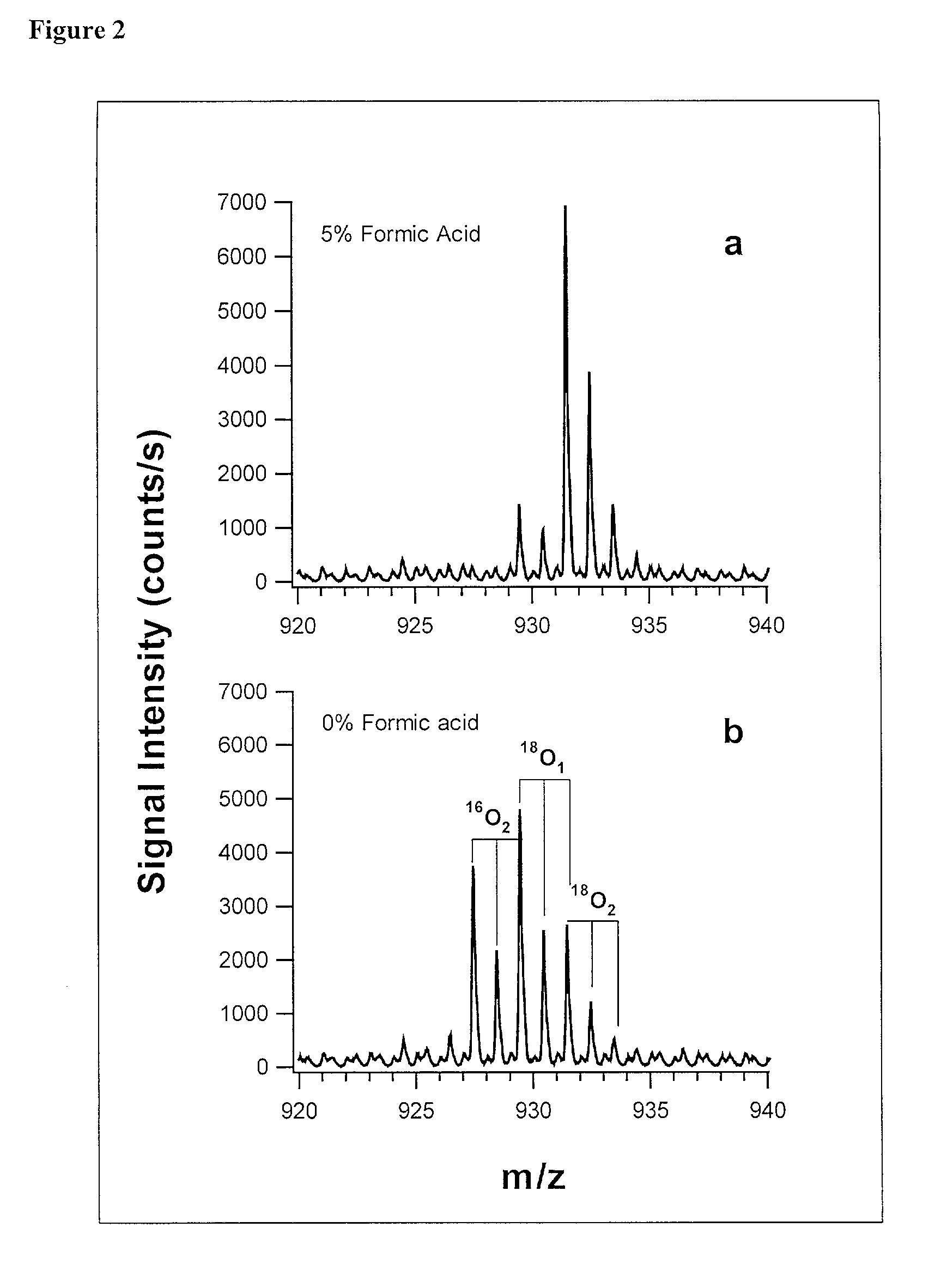
The invention provides a polypeptide marker for early diagnosis of diabetes mellitus. The operating method comprises the following steps: establishing an in-vitro simulated glycosylation model by selecting human peripheral blood high-abundant protein HAS, performing enzyme digestion under optimized conditions, performing 18O marking on a blank control group sample subjected to enzyme digestion, mixing the blank control group sample with a reaction group sample subjected to parallel treatment by using 16O according to a volume percent of 1:1, and performing HPLC / ESI-TOFMS detection. According to quantitative analysis of HSA peptide fragments, a peptide fragment, which is the most difficultly modified by glucose and is shown as SEQ ID NO.1, serves as an internal standard peptide fragment, a ratio of a peptide fragment which is easily subjected to glycosylation modification to the peak area of the internal standard peptide fragment is used for measuring the glycosylation modification degree of a glucose sensitive peptide, and according to a proteomics method, three peptide fragments, namely FKDLGEENFK, LDELRDEGK and KVPQVSTPTLVEVSR are finally discovered to serve as biological markers to be used for early diagnosis of diabetes mellitus.
Owner:BEIJING INSTITUTE OF TECHNOLOGYGY
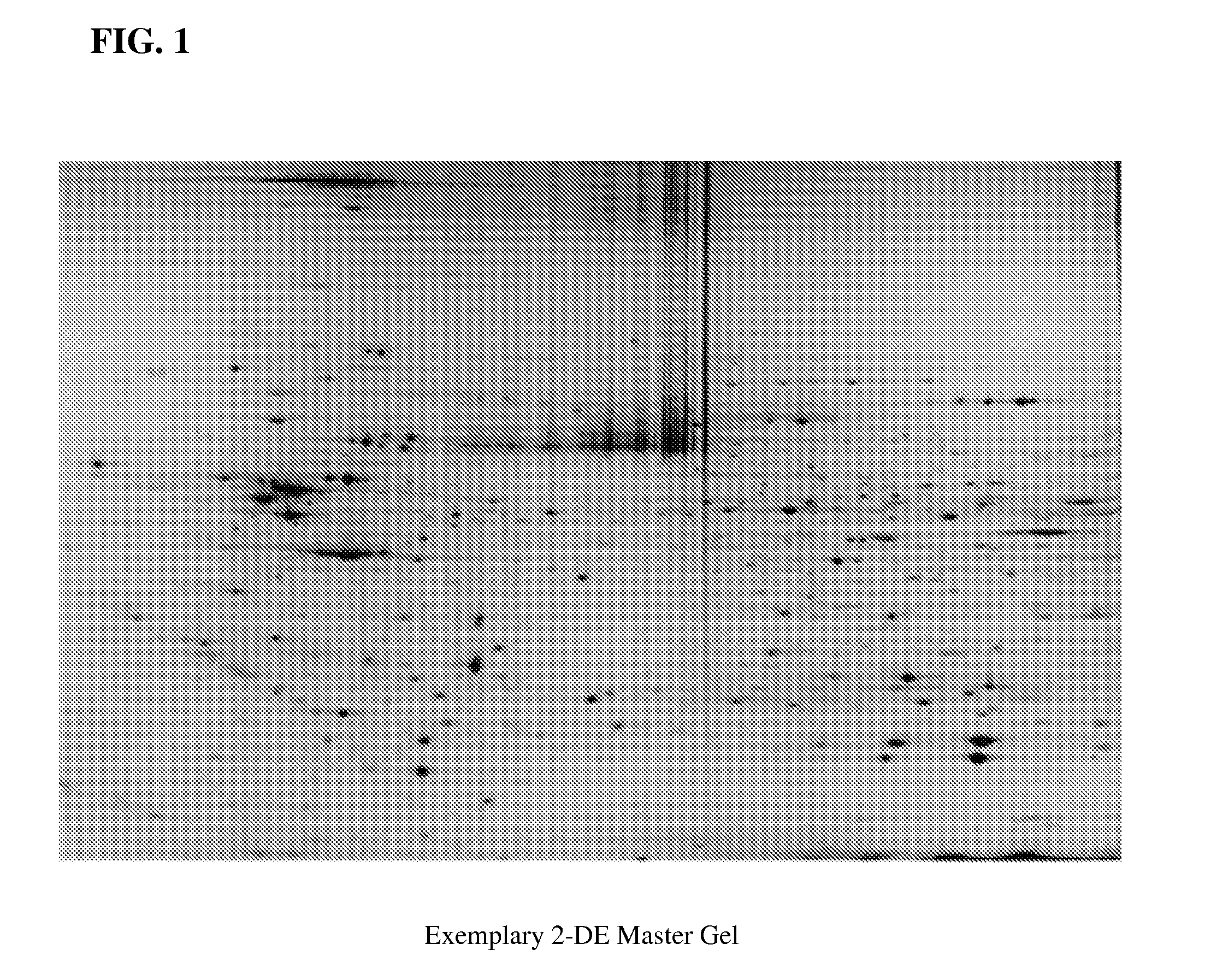
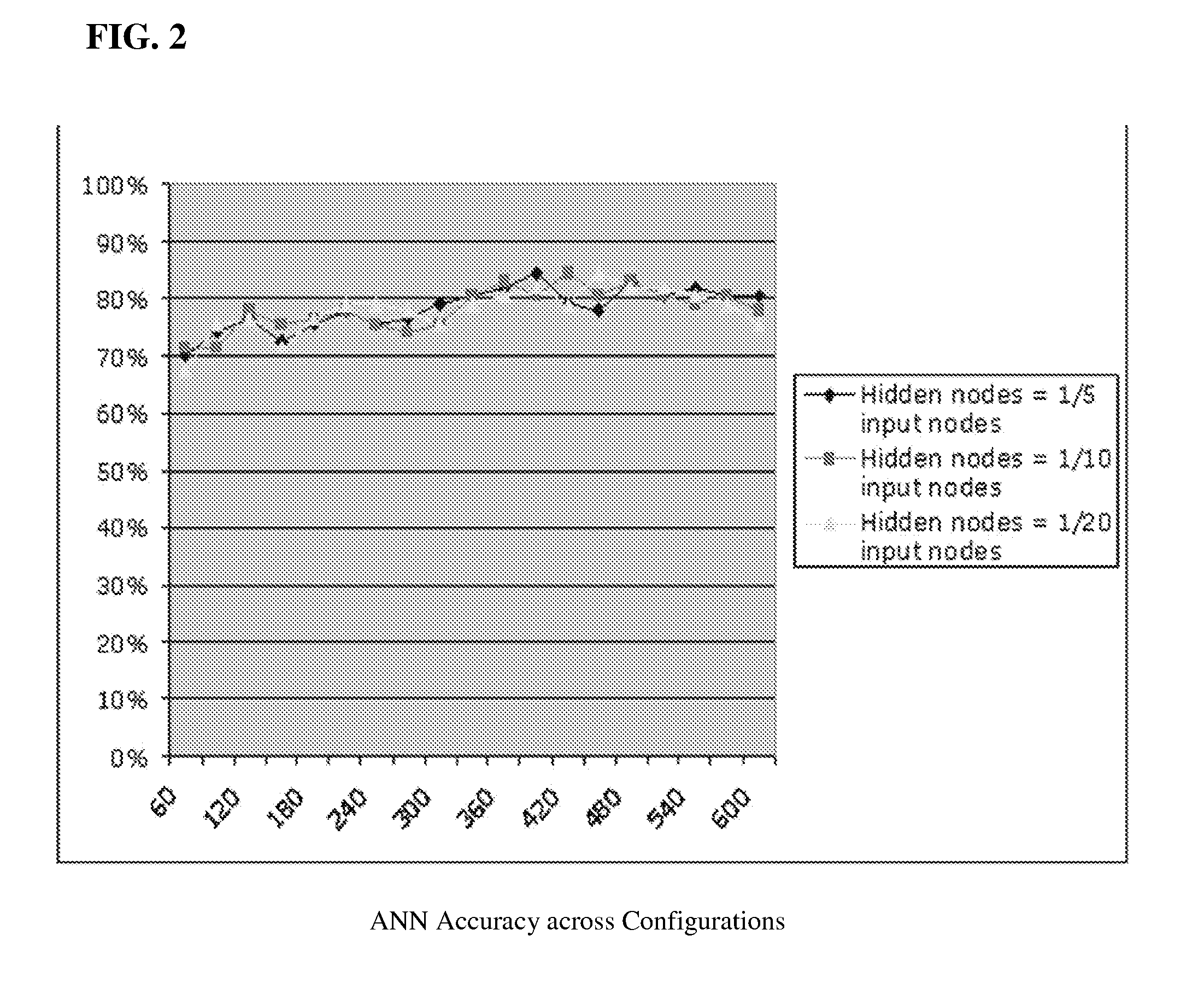
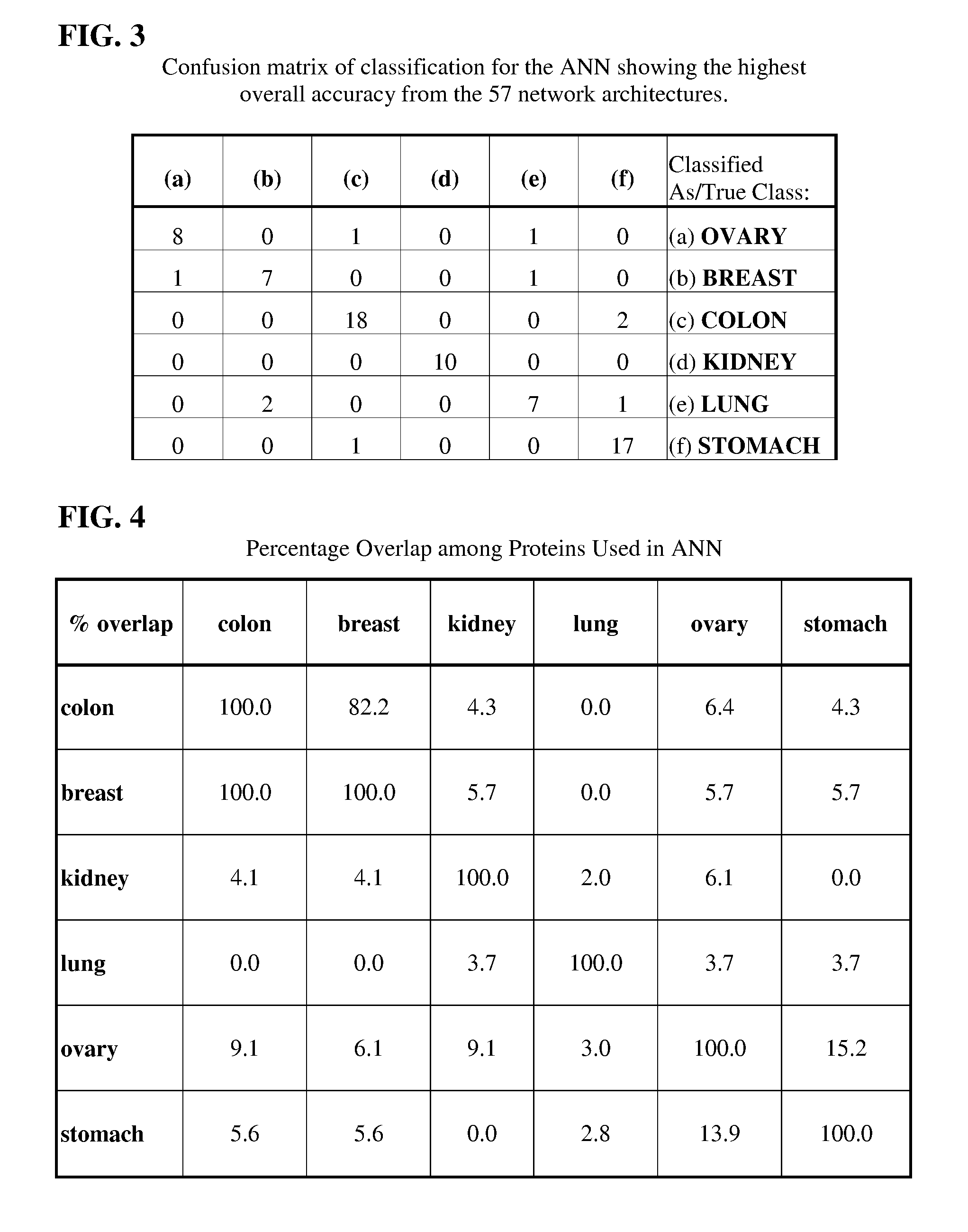
Artificial neural network proteomic tumor classification
Here the inventors describe a tumor classifier based on protein expression. Also disclosed is the use of proteomics to construct a highly accurate artificial neural network (ANN)-based classifier for the detection of an individual tumor type, as well as distinguishing between six common tumor types in an unknown primary diagnosis setting. Discriminating sets of proteins are also identified and are used as biomarkers for six carcinomas. A leave-one-out cross validation (LOOCV) method was used to test the ability of the constructed network to predict the single held out sample from each iteration with a maximum predictive accuracy of 87% and an average predictive accuracy of 82% over the range of proteins chosen for its construction.
Owner:UNIV OF SOUTH FLORIDA +1
Protein micro-arrays and multi-layered affinity interaction detection
InactiveUS20050153298A1Improve throughputHigh analysisMicrobiological testing/measurementBiological testingPost translationalHigh flux
The present invention provides proteomic techniques that extend sensitive and quantitative analysis of proteins to post-translational modifications. Protein micro-arrays and / or multiplex coded-microbeads are used in combination with multilayered affinity interaction detection (MAID) methods that permit high throughput analysis of cellular protein modifications and functional protein interactions.
Owner:GEMBITSKY DMITRY S +1
Molecular markers predicting response to adjuvant therapy, or disease progression, in breast cancer
Predicting response to adjuvant therapy or predicting disease progression in breast cancer is realized by (1) first obtaining a breast cancer test sample from a subject; (2) second obtaining clinicopathological data from said breast cancer test sample; (3) analyzing the obtained breast cancer test sample for presence or amount of (a) one or more molecular markers of hormone receptor status, one or more growth factor receptor markers, (b) one or more tumor suppression / apoptosis molecular markers; and (c) one or more additional molecular markers both proteomic and non-proteomic that are indicative of breast cancer disease processes; and then (4) correlating (a) the presence or amount of said molecular markers and, with (b) clinicopathological data from said tissue sample other than the molecular markers of breast cancer disease processes. A kit of (1) a panel of antibodies; (2) one or more gene amplification assays; (3) first reagents to assist said antibodies with binding to tumor samples; (4) second reagents to assist in determining gene amplification; permits, when applied to a breast cancer patient's tumor tissue sample, (A) permits observation, and determination, of a numerical level of expression of each individual antibody, and gene amplification; whereupon (B) a computer algorithm, residing on a computer can calculate a prediction of treatment outcome for a specific treatment for breast cancer, or future risk of breast cancer progression.
Owner:LINKE STEVEN +2
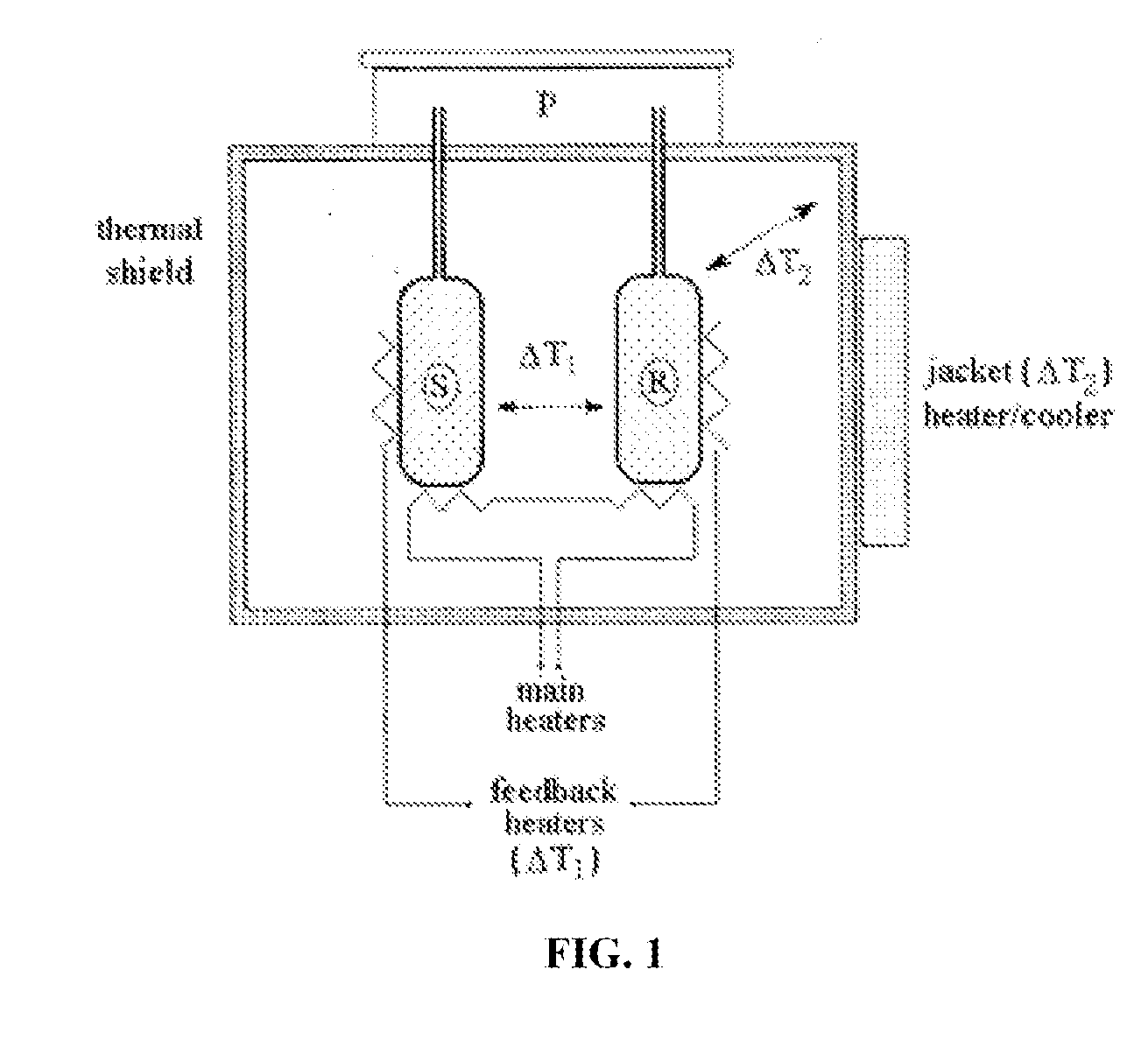
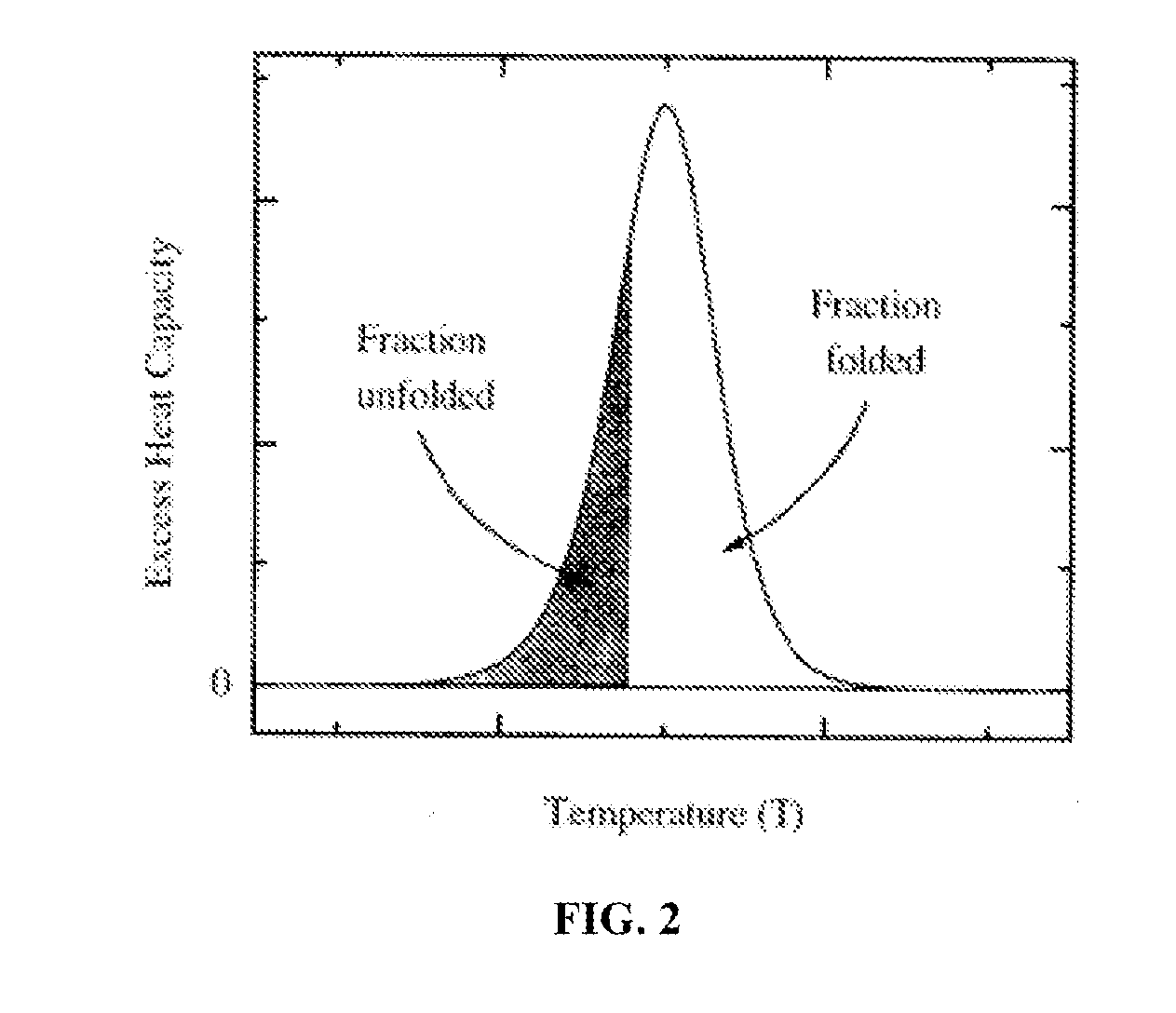
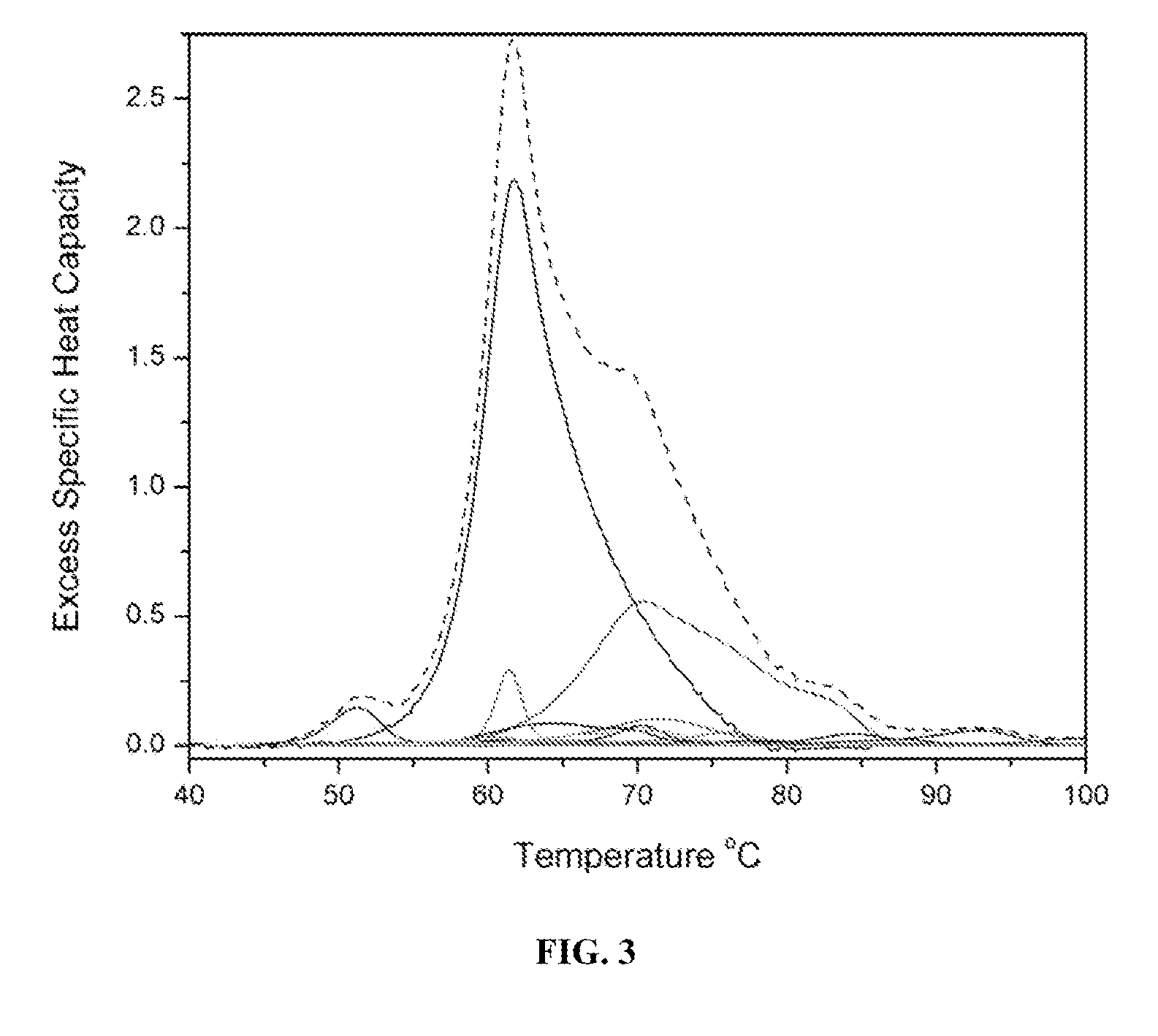
Proteomic profiling method useful for condition diagnosis and monitoring, composition screening, and therapeutic monitoring
InactiveUS20080172184A1Low costLower levelCompound screeningApoptosis detectionTherapy monitoringProtein composition
A method of diagnosing or monitoring a condition of interest in a subject includes comparing thermograms generated using differential scanning calorimetery. A signature thermogram contains a protein composition pattern for a sample obtained from the subject. The signature thermogram is compared to a standard thermogram. Standard thermograms can include a negative standard thermogram containing a protein composition pattern associated with an absence of the condition of interest, and a positive standard thermogram containing a protein composition pattern associated with a presence of the condition of interest.
Owner:UNIV OF LOUISVILLE RES FOUND INC
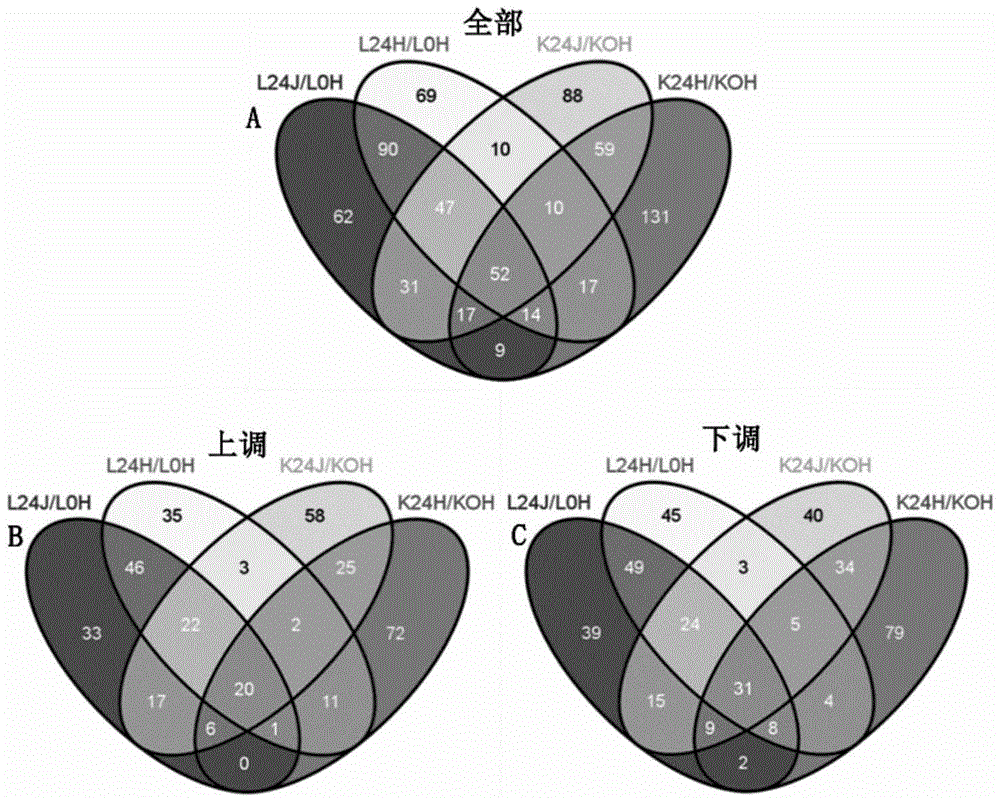
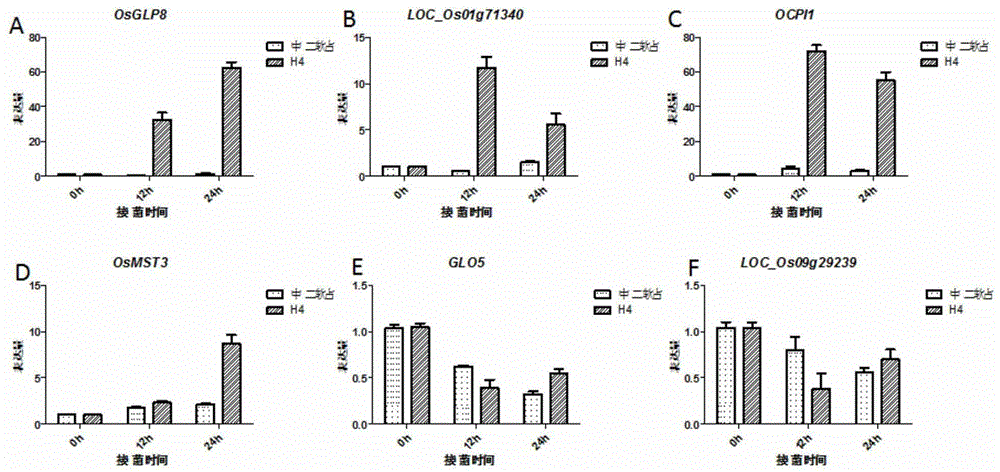
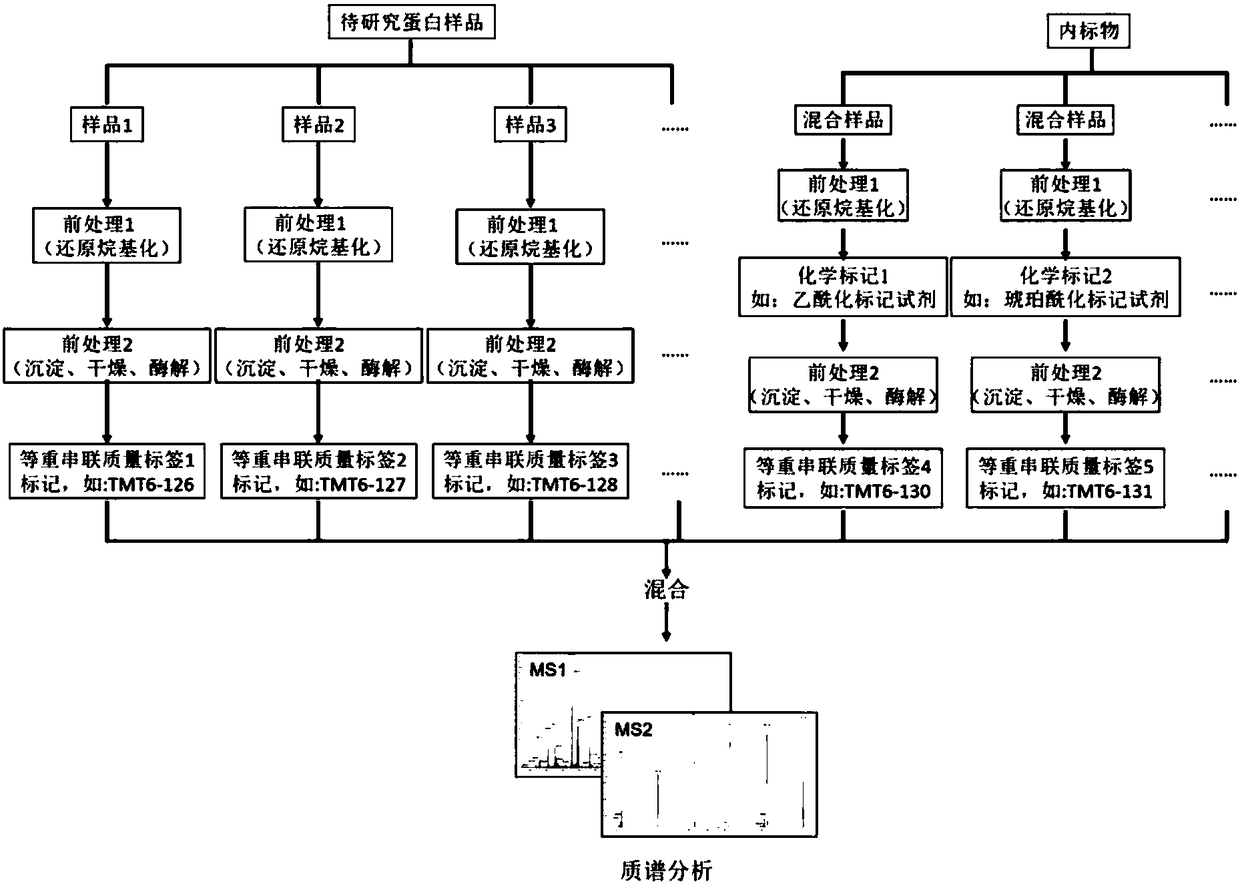
Method of researching change of proteome of rice responding rice blast bacterial infection through iTRAQ technology
InactiveCN104820103AQuick and effective screeningStrong disease resistanceBiological testingDiseaseAgricultural science
The invention discloses a method of researching the change of a proteome of rice responding rice blast bacterial infection through an iTRAQ technology and belongs to the field of plant biotechnology. The method includes following steps: A. extracting a plant materials and leaf protein; B. performing further FASP enzymolysis, peptide fragment marking, SCX classification and LC-MS analysis to the protein sample obtained in the step A; C. performing data query and quantitative analysis to the result after the LC-MS analysis to obtain the change situation of the whole proteome during an interaction process between rice and rice blast bacteria. The method can quickly and effectively screening a candidate gene relative to disease resistance of rice, can be used for researching the candidate gene relative to the disease resistance of the rice well with the proteomics researching method, iTRAQ, which is high-throughput and accurate, can be widely used for researching and developing the characters of plant disease resistance and breeding new plant varieties being strong in disease resistance with combination of the transgenic technology.
Owner:SOUTH CHINA AGRI UNIV
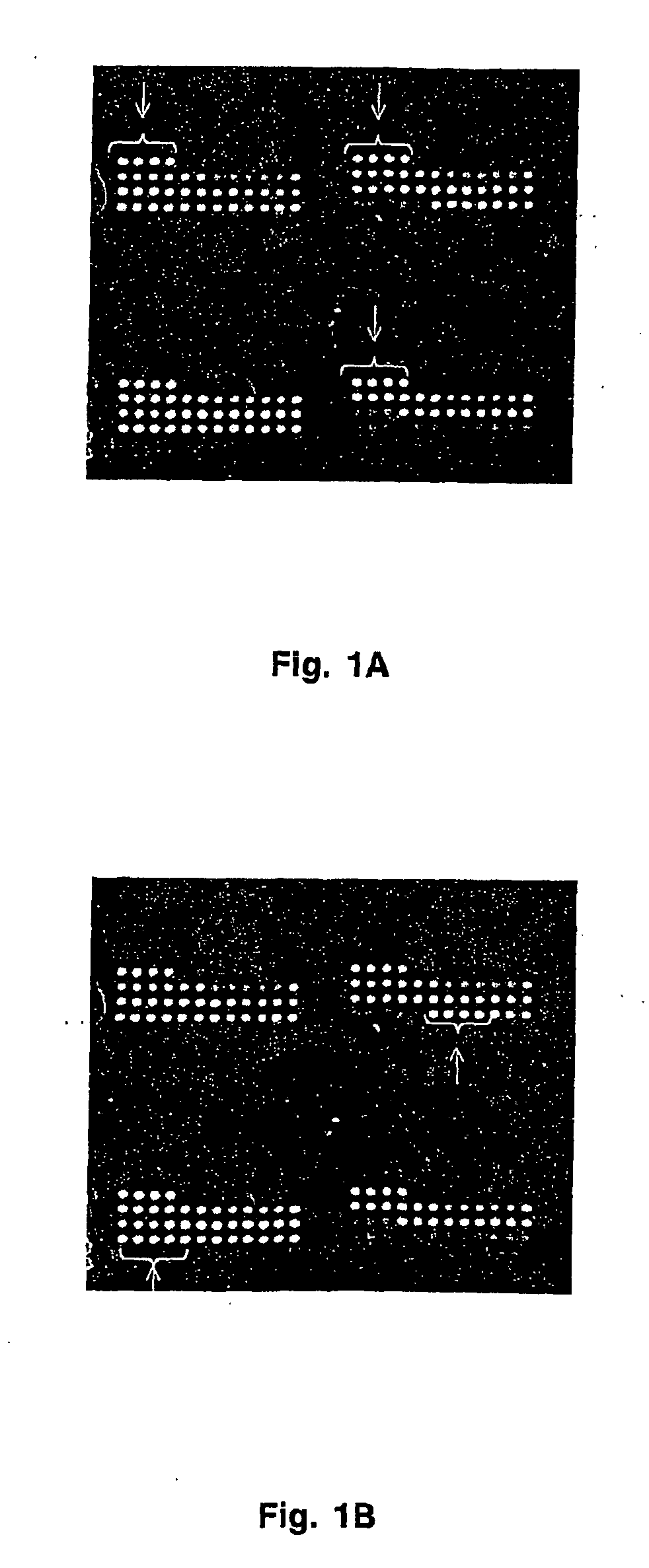
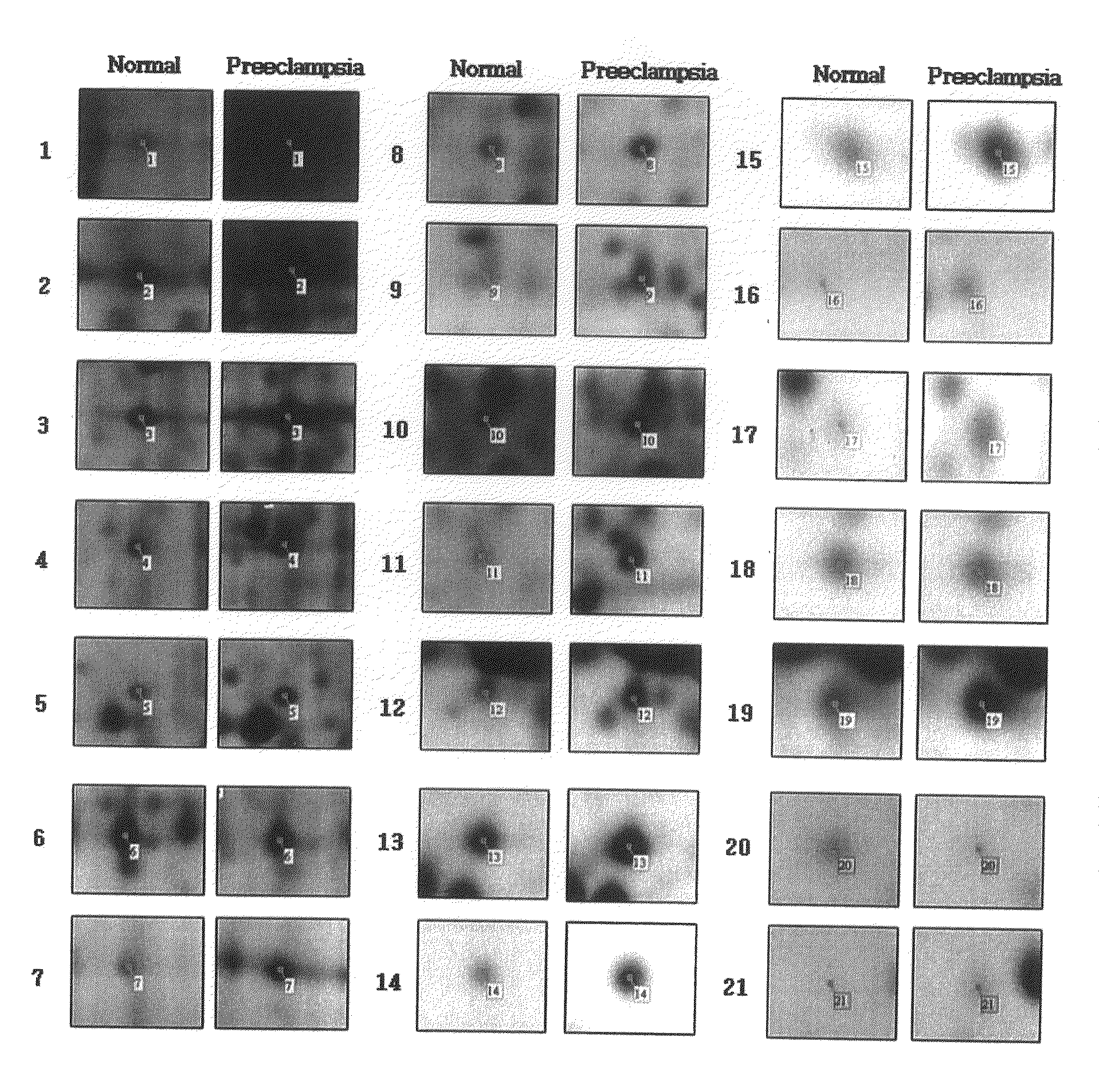
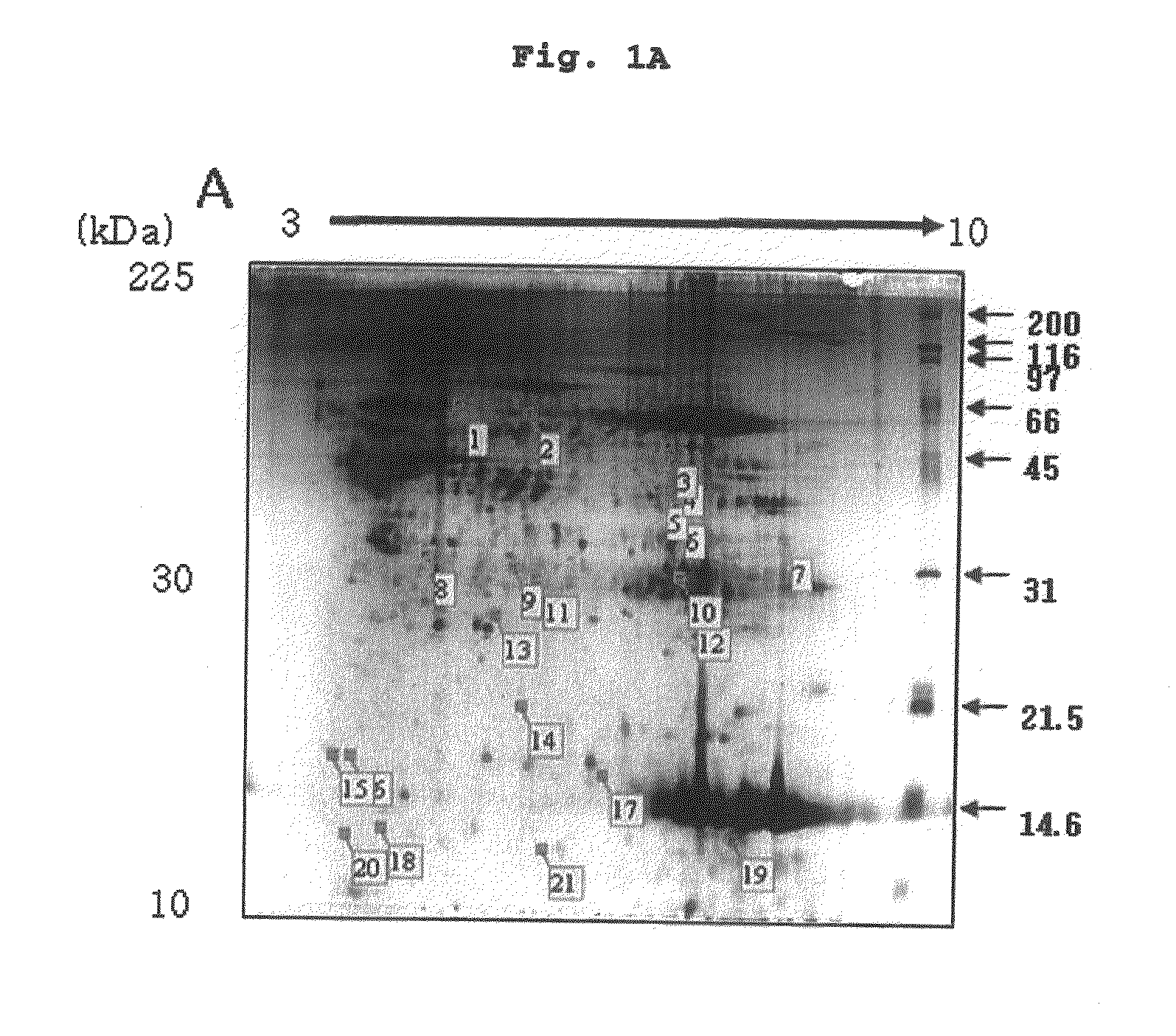
Method of screening placental proteins responsible for pathophysiology of preeclampsia, and marker for early diagnosis and prediction of preeclampsia
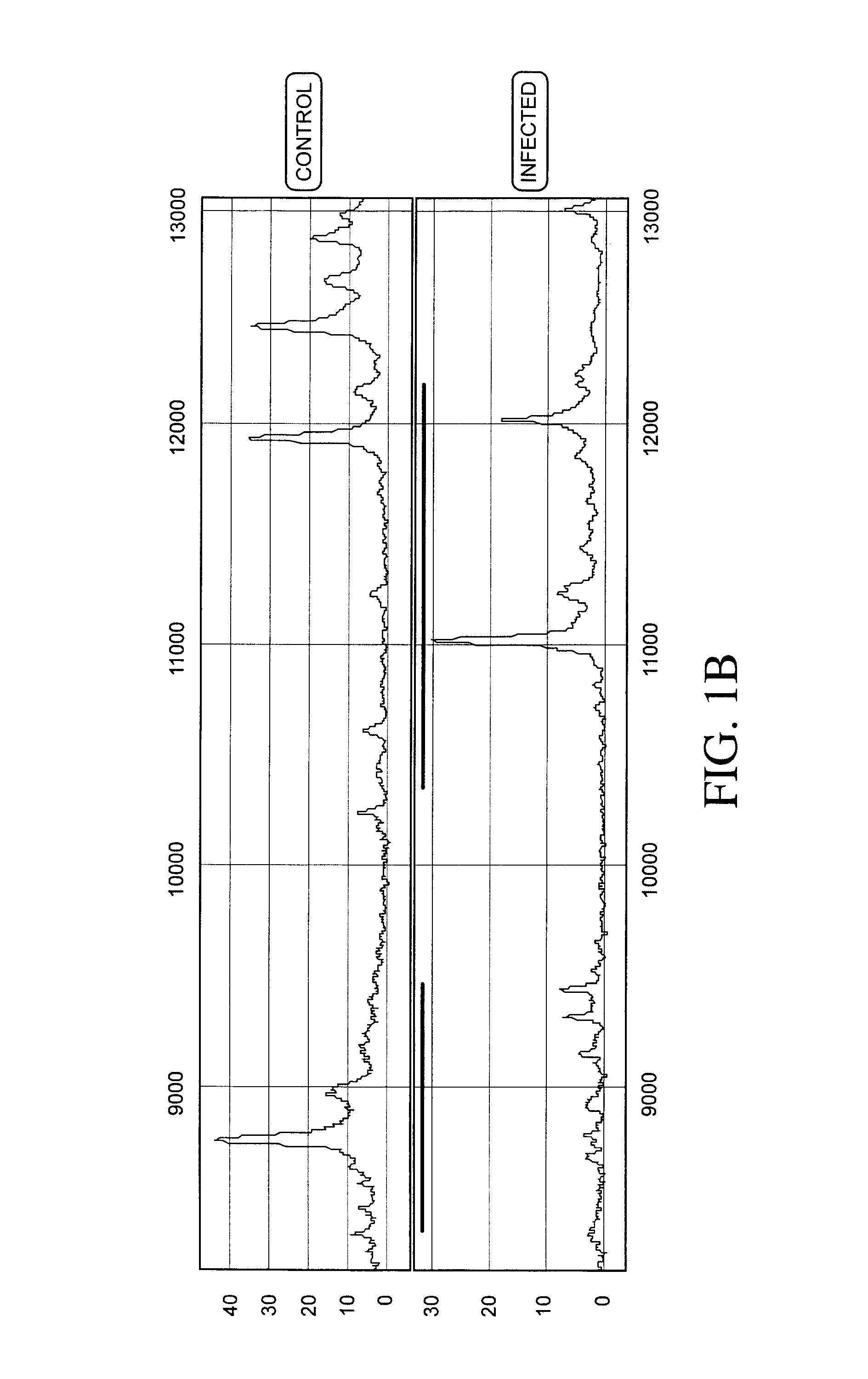
The present invention relates to a method of screening placental proteins responsible for pathophysiology of preeclampsia, and a marker for early diagnosis and prediction of preeclampsia. In accordance with one aspect of the present invention, there is provided a method of screening placental proteins responsible for pathophysiology of preeclampsia by 2D E-proteomics analysis, comprising: isolating placental proteins from a placental tissue; separating the isolated proteins two-dimensionally through 2D electrophoresis; and comparing and analyzing the separated proteins based on scanned gel images and differences in the images between normal placental proteins and preeclamptic placental proteins, wherein the comparison and analysis of the placental proteins based on the scanned gel images and differences in the images are accomplished by selecting proteins with differences of 140% or more between two placentas.
Owner:HAN OU JIN
System for detecting infection in synovial fluid
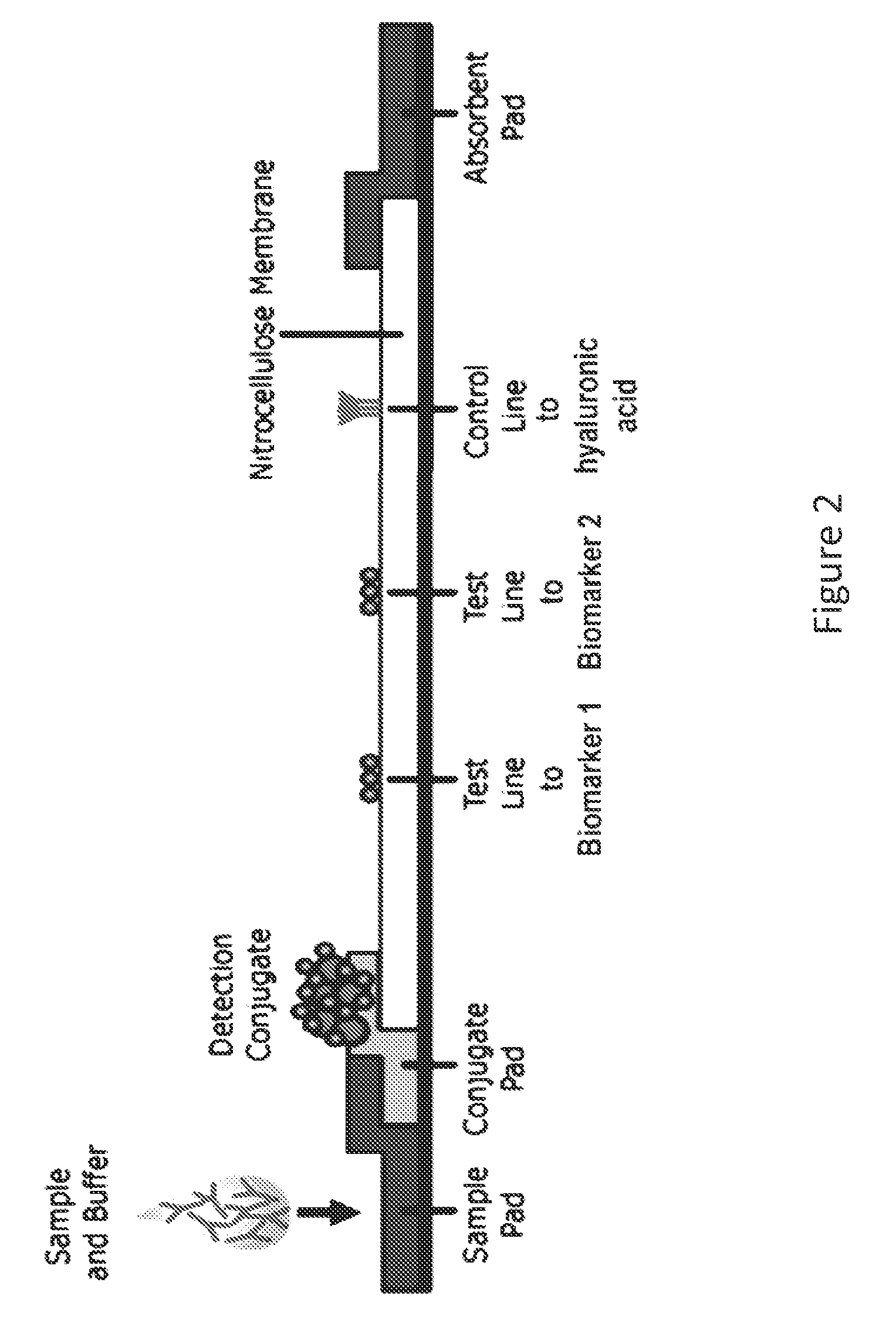
ActiveUS20150011412A1Improve abilitiesBioreactor/fermenter combinationsPeptide librariesProteomics methodsRadioimmunoassay
The invention provides methods and systems for detecting a biomarker in a synovial fluid wherein the system also includes a control to ensure that the test sample is indeed synovial fluid. The biomarkers and the control for synovial fluid can be identified using proteomic methods, including but not limited to antibody based methods, such as an enzyme-linked immunosorbant assay (ELISA), a radioimmunoassay (RIA), or a lateral flow immunoassay.
Owner:CD DIAGNOSTICS
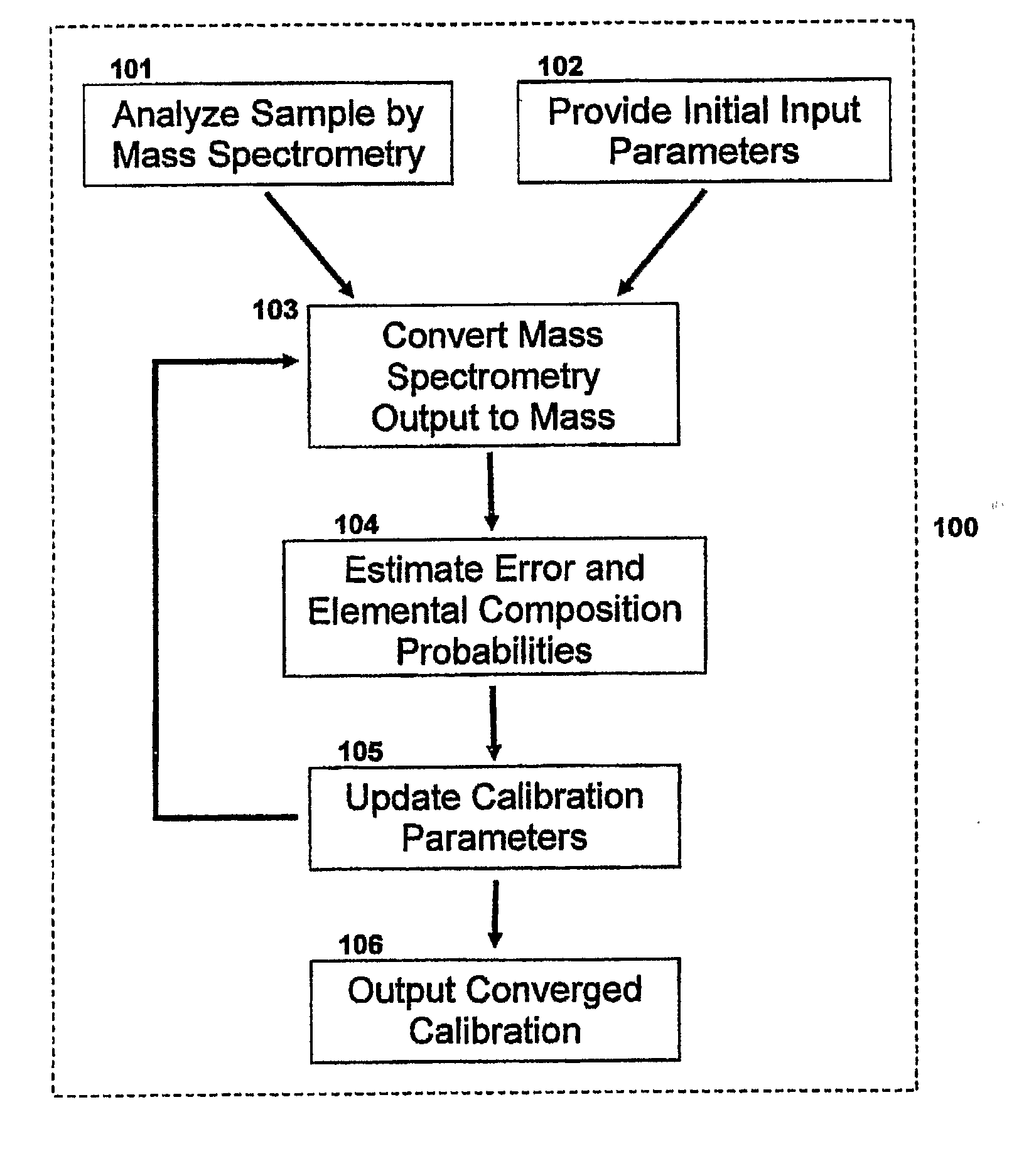
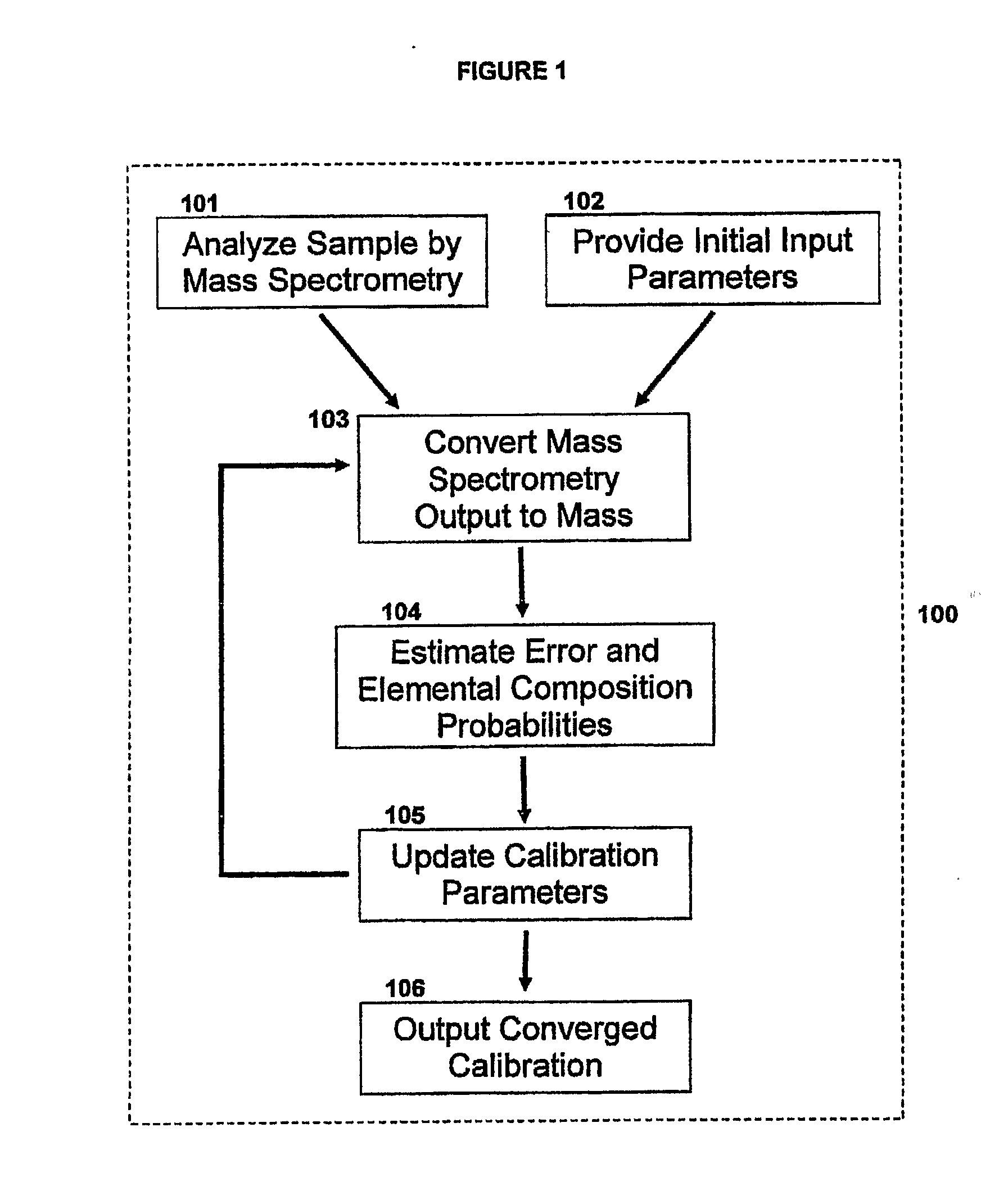
Method For Simultaneous Calibration of Mass Spectra and Identification of Peptides in Proteomic Analysis
InactiveUS20080203284A1Calibration apparatusIsotope separationMass Spectrometry-Mass SpectrometryProteomic Profile
The invention relates to a mass spectrometry calibration system that may be performed in real-time using the information contained within a sample without the addition of specific calibrants. When applied to a sample, such as a proteomic sample, the calibration system may identify the exact masses of peptides in the sample. The system involves the use of mathematical algorithms that iteratively estimate the error in the measurement and update the calibration parameters accordingly; thereby resulting in peptide mass identification.
Owner:CEDARS SINAI MEDICAL CENT
Detection and quantification method for post-translational modification proteomics
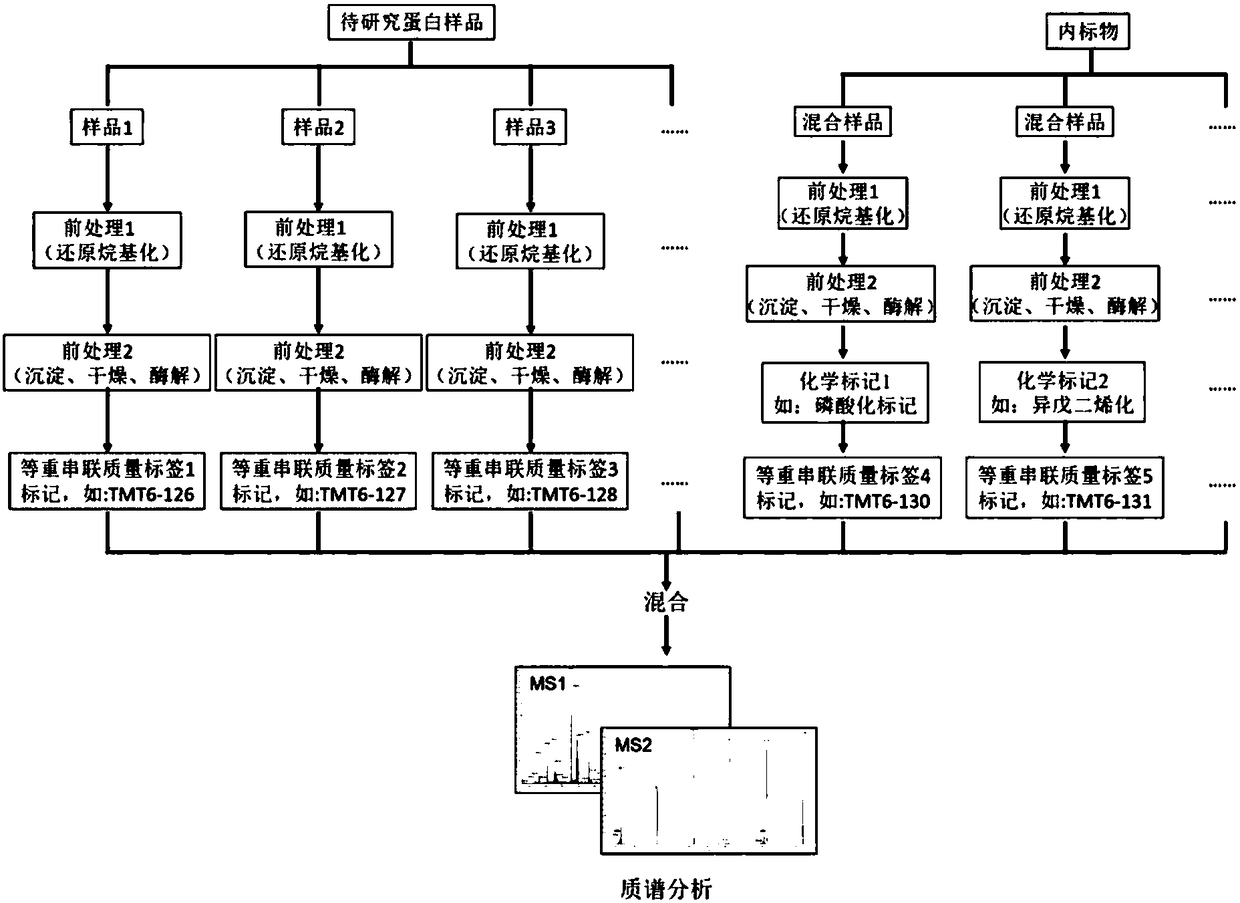
ActiveCN108627647AIncrease the total abundanceImprove the odds of analysisPreparing sample for investigationMass spectrometric analysisInternal standardProteomic Profile
Owner:SICHUAN UNIV
Method and system for identification of protein-protein interaction
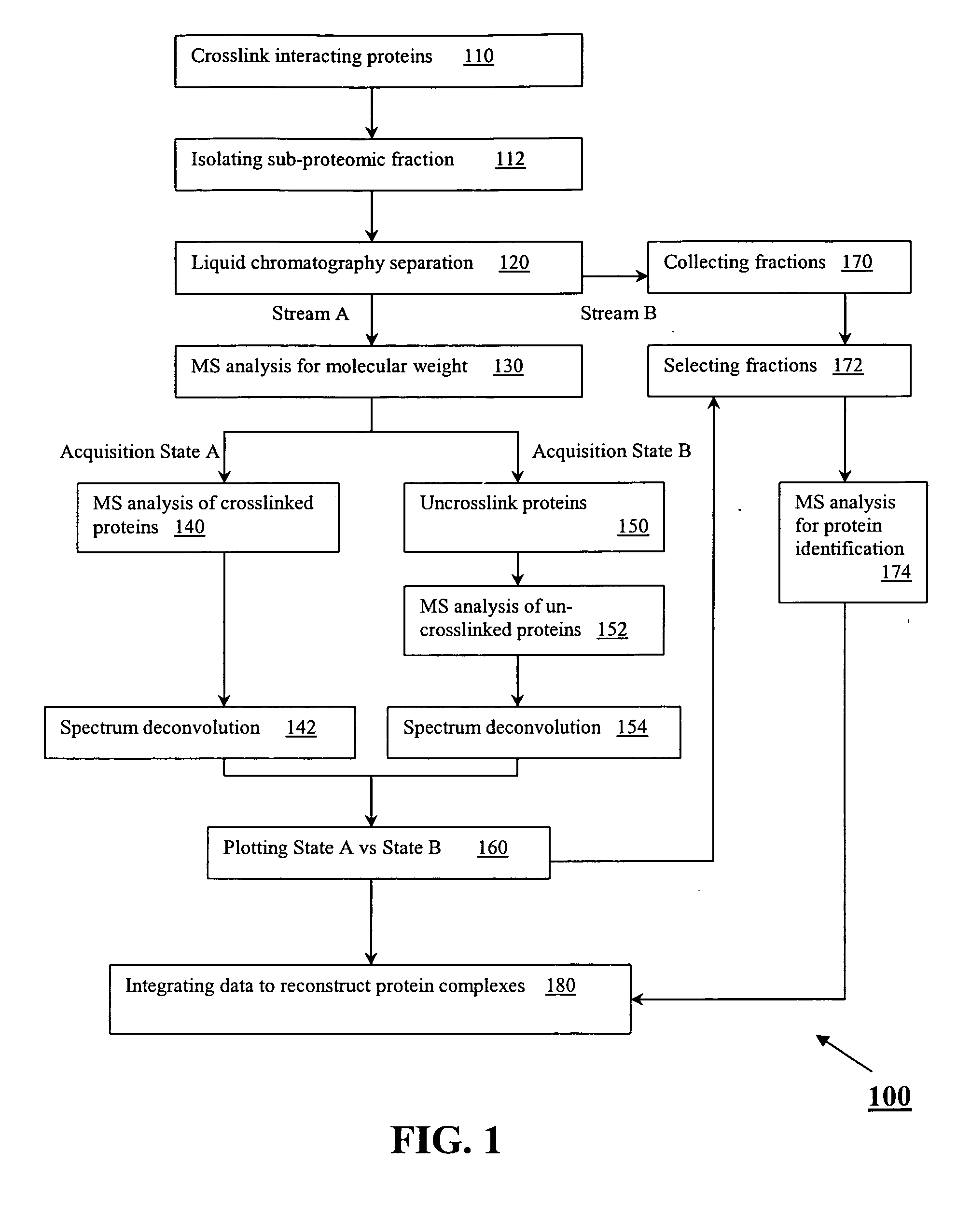
InactiveUS20080090298A1Component separationPeptide preparation methodsChromatographic separationProtein insertion
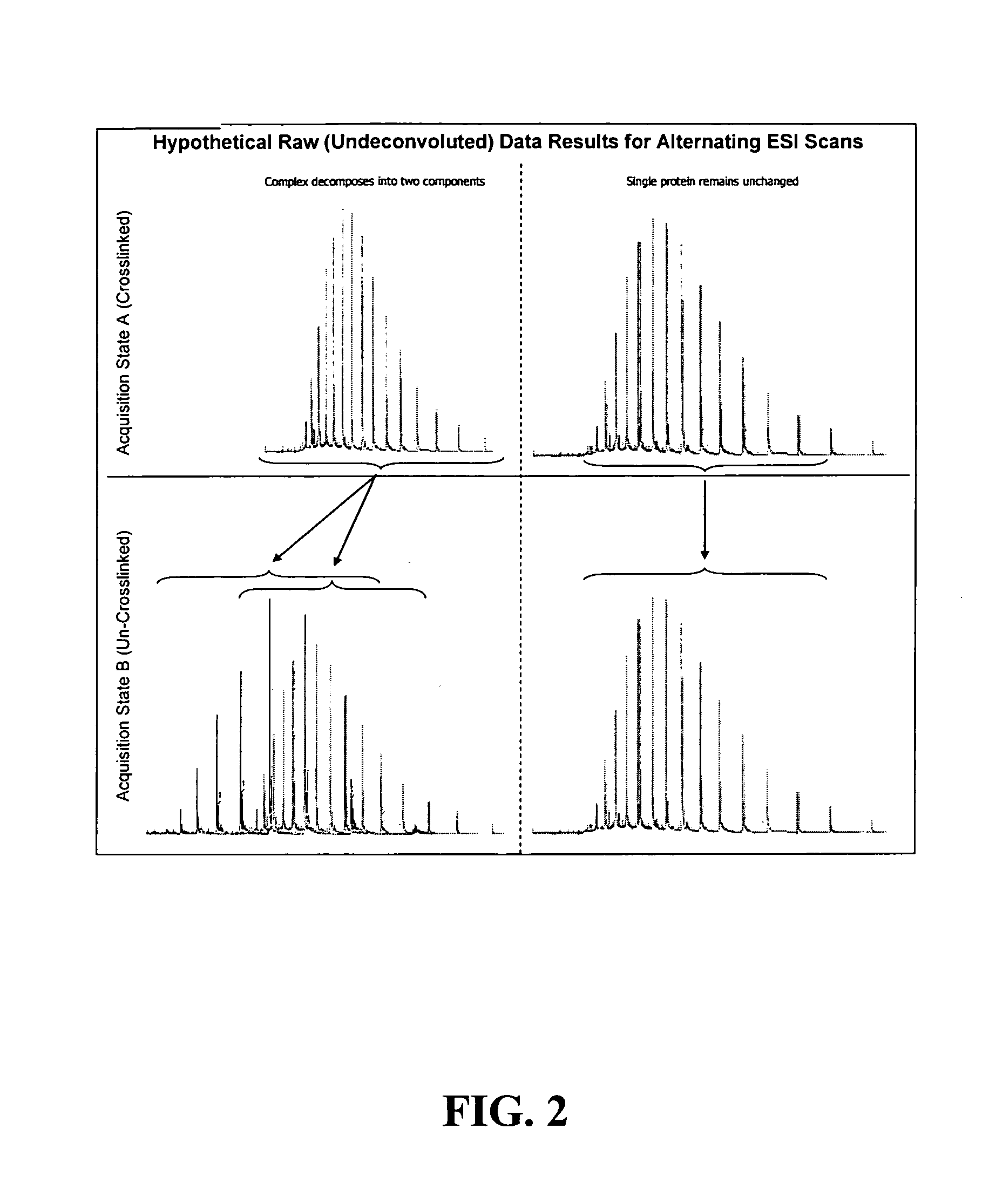
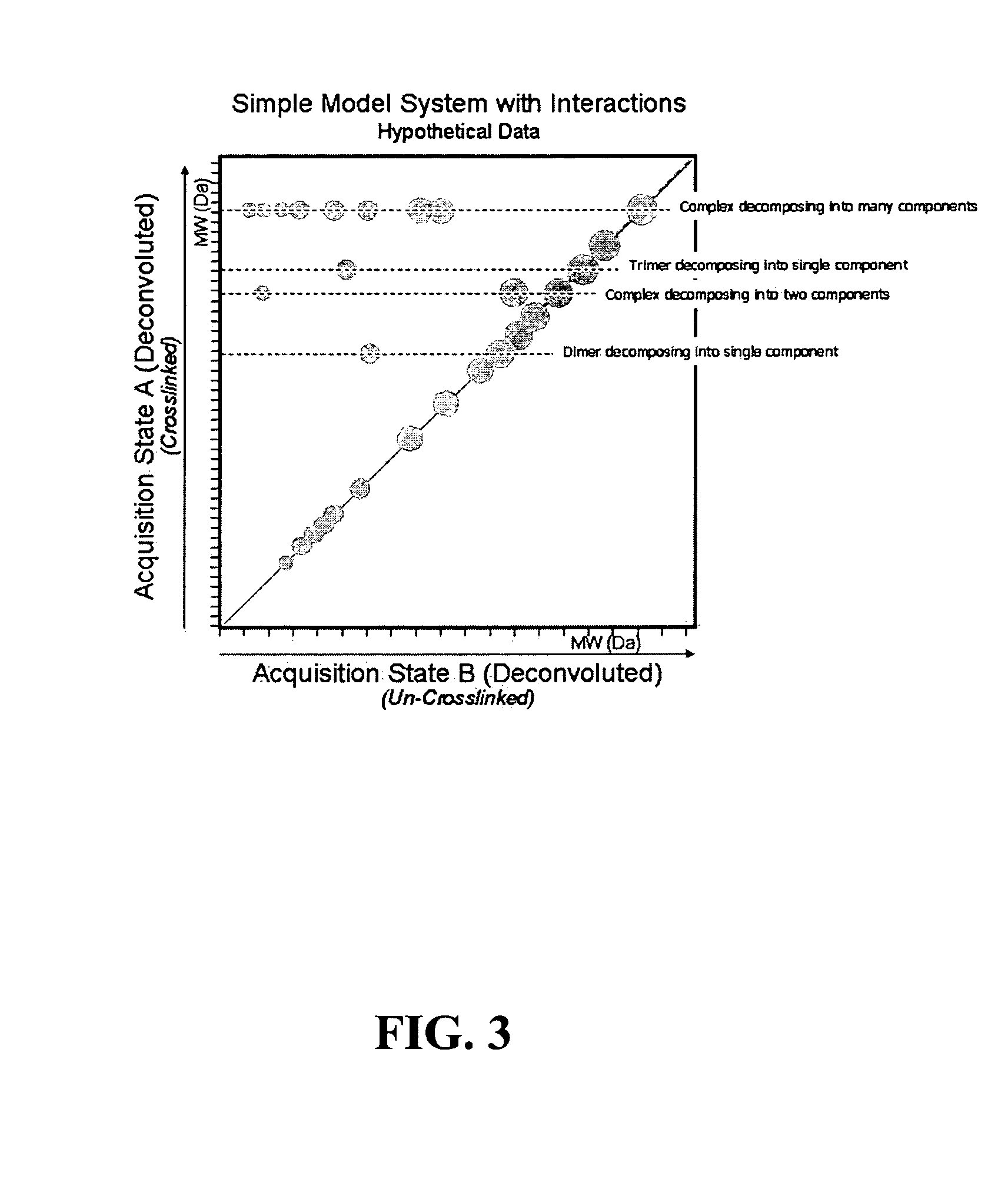
A method for the characterization of protein-protein interactions based on diagonal mass spectrometry is provided. Proteomic samples containing interacting proteins are chemically crosslinked either in vivo or in vitro. After a high resolution chromatographic separation, crosslinked interacting proteins are introduced directly into a mass spectrometer. During the data acquisition, the mass spectrometer alternates between two discrete acquisition states. In the first acquisition state, the crosslinked complexes are analyzed. In the second acquisition state, the crosslinking is cleaved and the mass spectra of the dissociated proteins are collected. Following the data acquisition, the raw mass spectral data is deconvoluted and reconstructed into a diagonal MS plot of crosslinked proteins vs. component proteins to explore protein-protein interactions.
Owner:AGILENT TECH INC
Proteomic Methods For The Identification And Use Of Putative Biomarkers Associated With The Dysplastic State In Cervical Cells Or Other Cell Types
InactiveUS20090221430A1Quality improvementHigh sensitivityMicrobiological testing/measurementLibrary screeningProteomics methodsCervical cells
The invention relates to methods for detecting and identifying potential biomarkers of high-grade cervical dysplasia in an individual human subject. The invention also relates to newly discovered biomarkers, as set forth in Tables 1-4 herein, which are associated with the dysplastic state of cervical cells. It has been discovered that a differential level of expression of any of these markers or combination of these markers correlates with a dysplastic condition in a human subject, e.g., a patient.
Owner:NORTHEASTERN UNIV +1
Molecular markers of hepatocellular carcinoma and their applications
InactiveCN101313220AAccurate Prognosis SystemCompound screeningApoptosis detectionEtiologyProteomics methods
Owner:PROYECTO DE BIOMEDICINA CIMA
Simulating the metabolic pathway dynamics of an organism
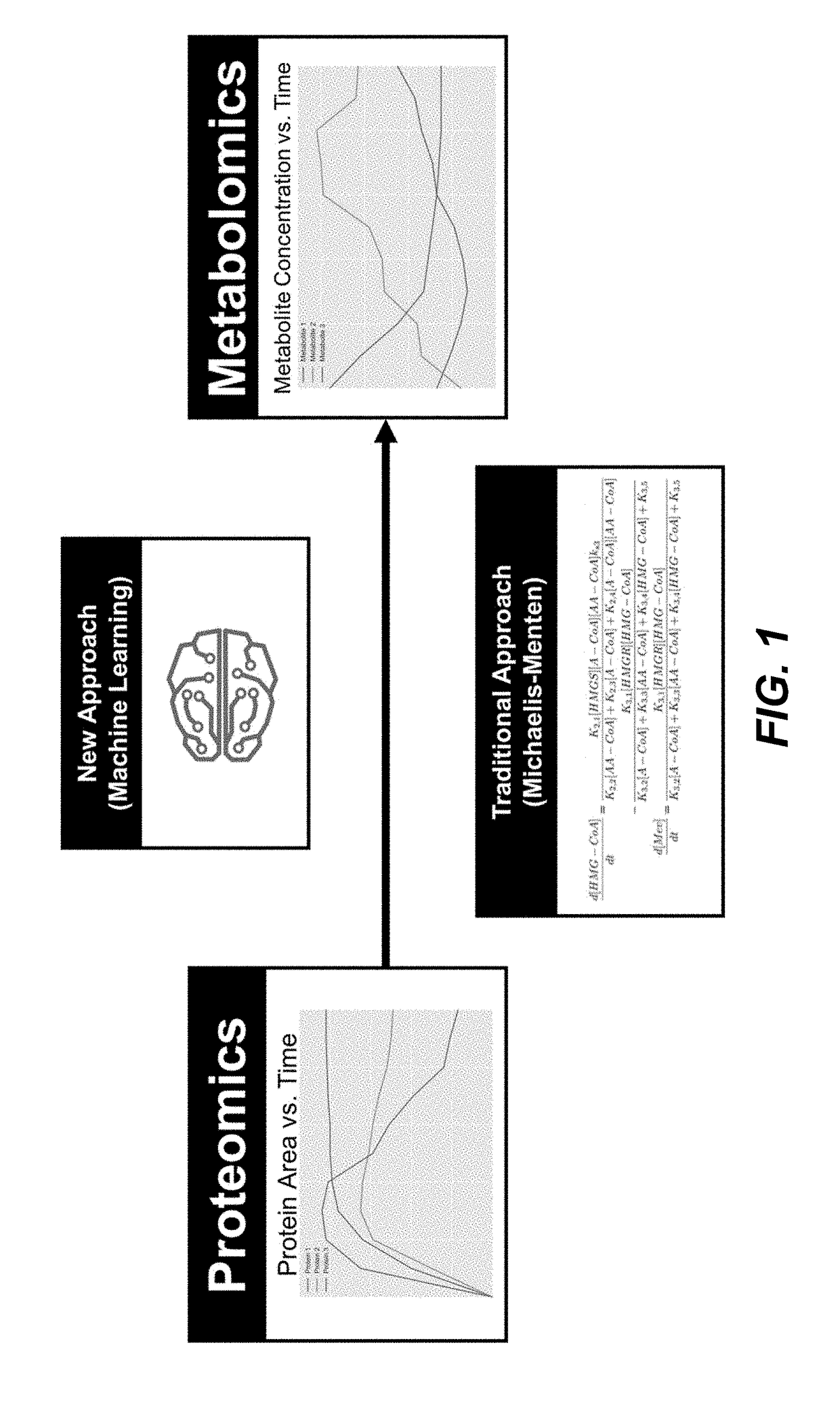
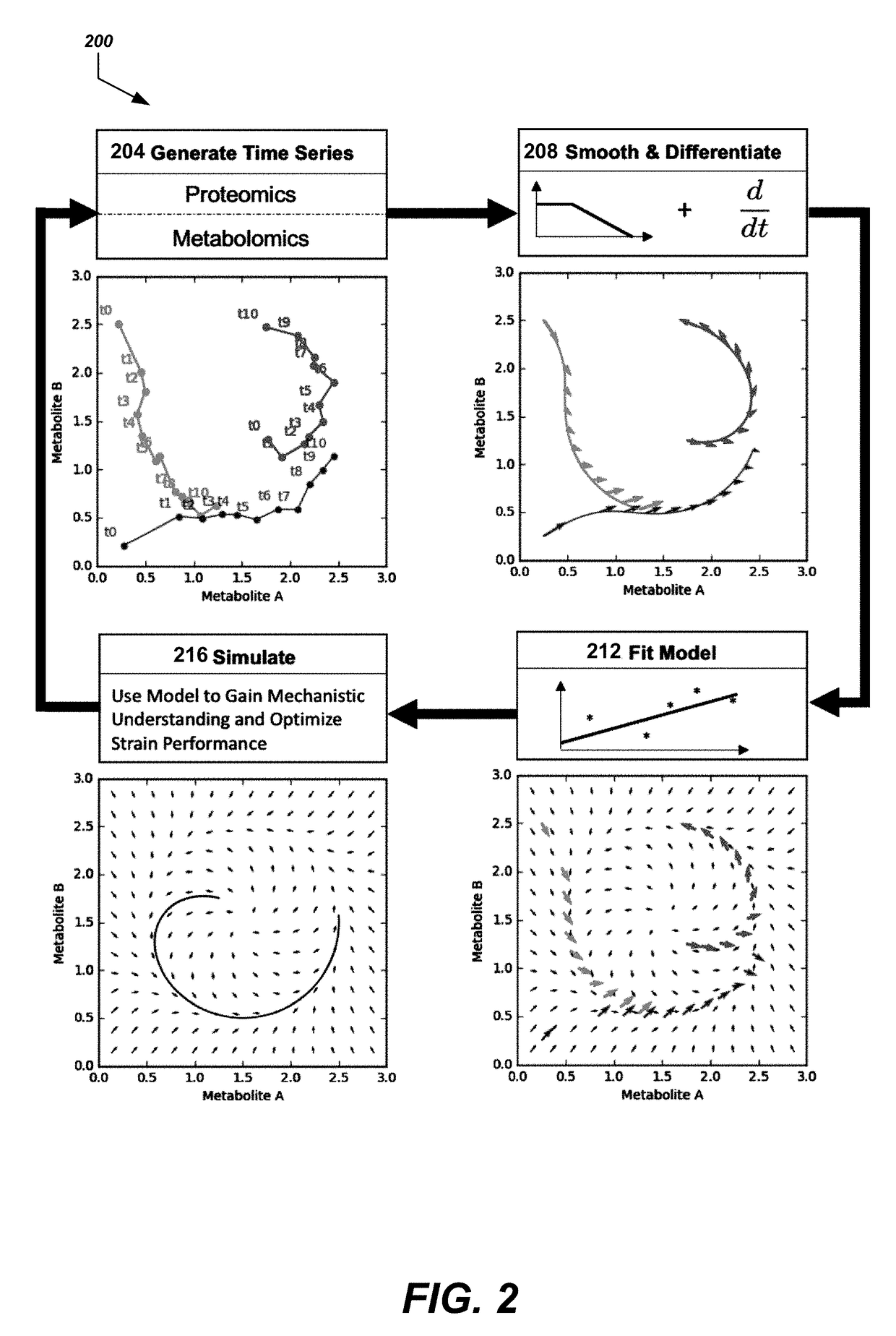
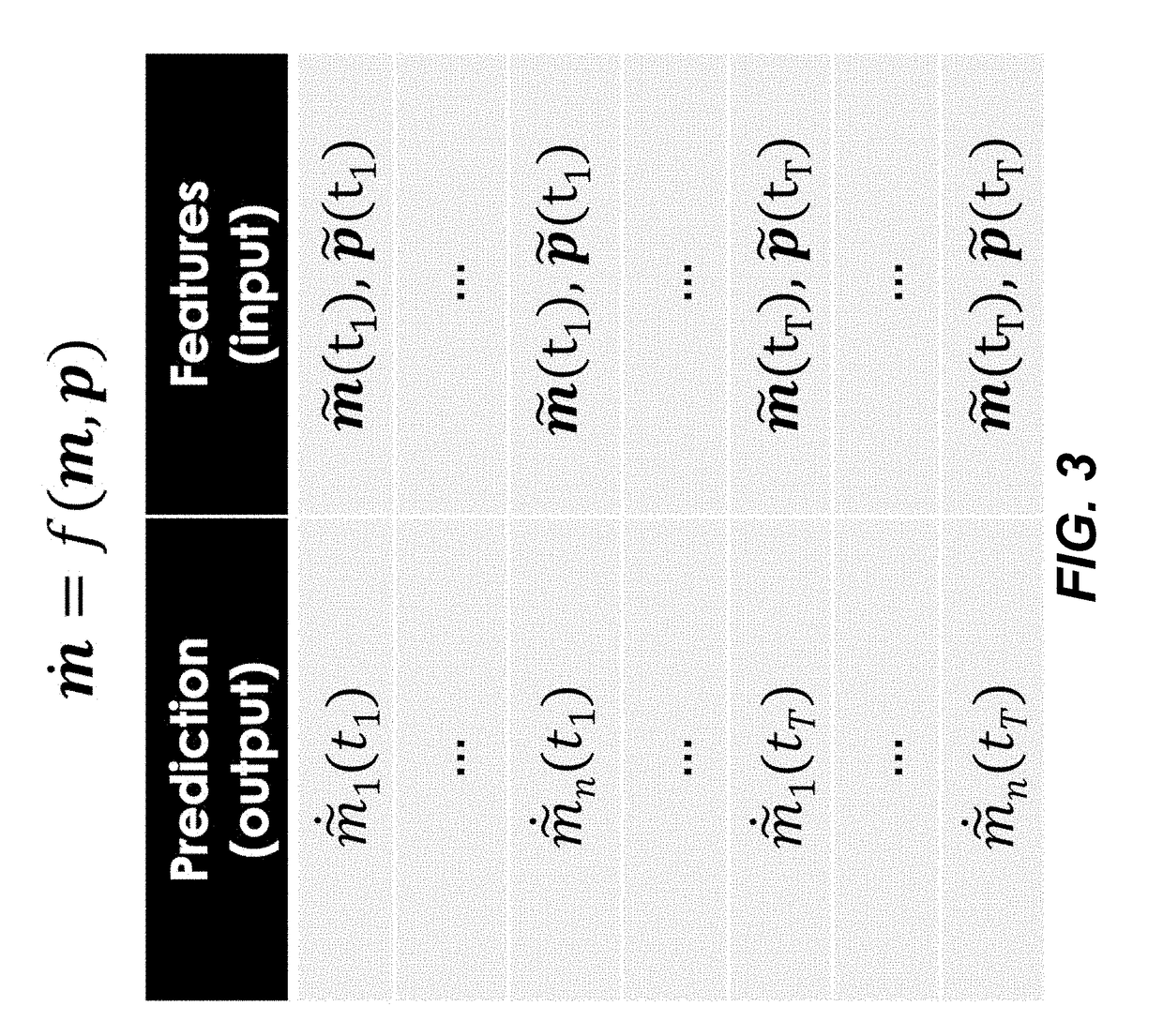
Disclosed herein are systems and methods for determining metabolic pathway dynamics using time series multiomics data. In one example, after receiving time series multiomics data comprising time-series metabolomics data associated a metabolic pathway and time-series proteomics data associated with the metabolic pathway, derivatives of the time series multiomics data can be determined. A machine learning model, representing a metabolic pathway dynamics model, can be trained using the time series multiomics data and the derivatives of the time series multiomics data, wherein the metabolic pathway dynamics model relates the time-series metabolomics data and time-series proteomics data to the derivatives of the time series multiomics data. The method can include simulating a virtual strain of the organism using the metabolic pathway dynamics model.
Owner:RGT UNIV OF CALIFORNIA
Proteomic analysis of biological fluids
InactiveUS20080299594A1Eliminate needMicrobiological testing/measurementBiological testingDiseaseFetal growth
The invention concerns the identification of proteomes of biological fluids and their use in determining the state of maternal / fetal conditions, including maternal conditions of fetal origin, chromosomal aneuploidies, and fetal diseases associated with fetal growth and maturation. In particular, the invention concerns the identification of the proteome of amniotic fluid (multiple proteins representing the composition of amniotic fluid) and the correlation of characteristic changes in the normal proteome with various pathologic maternal / fetal conditions, such as intra-amniotic infection, or chromosomal defects.
Owner:HOLOGIC INC
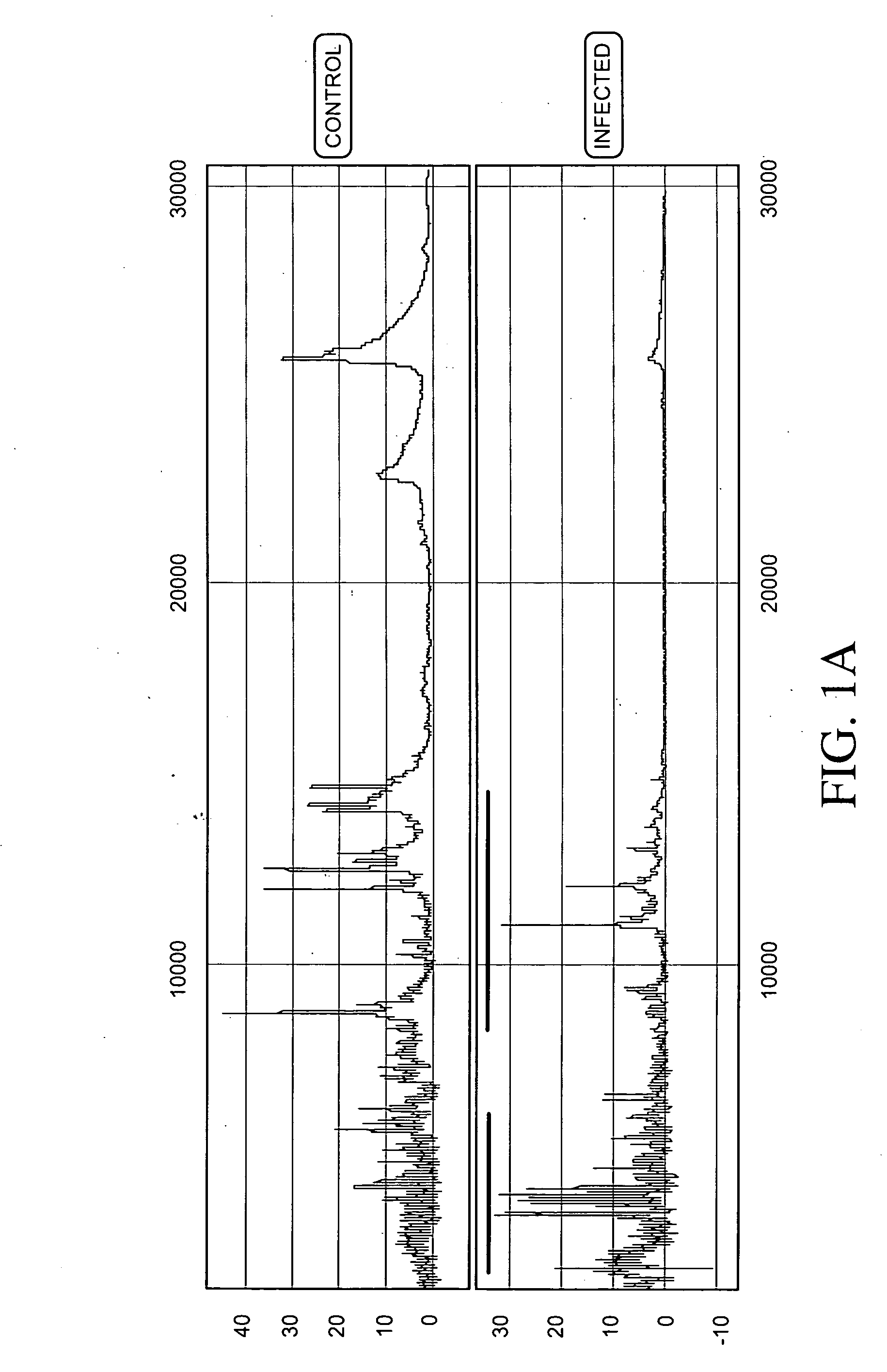
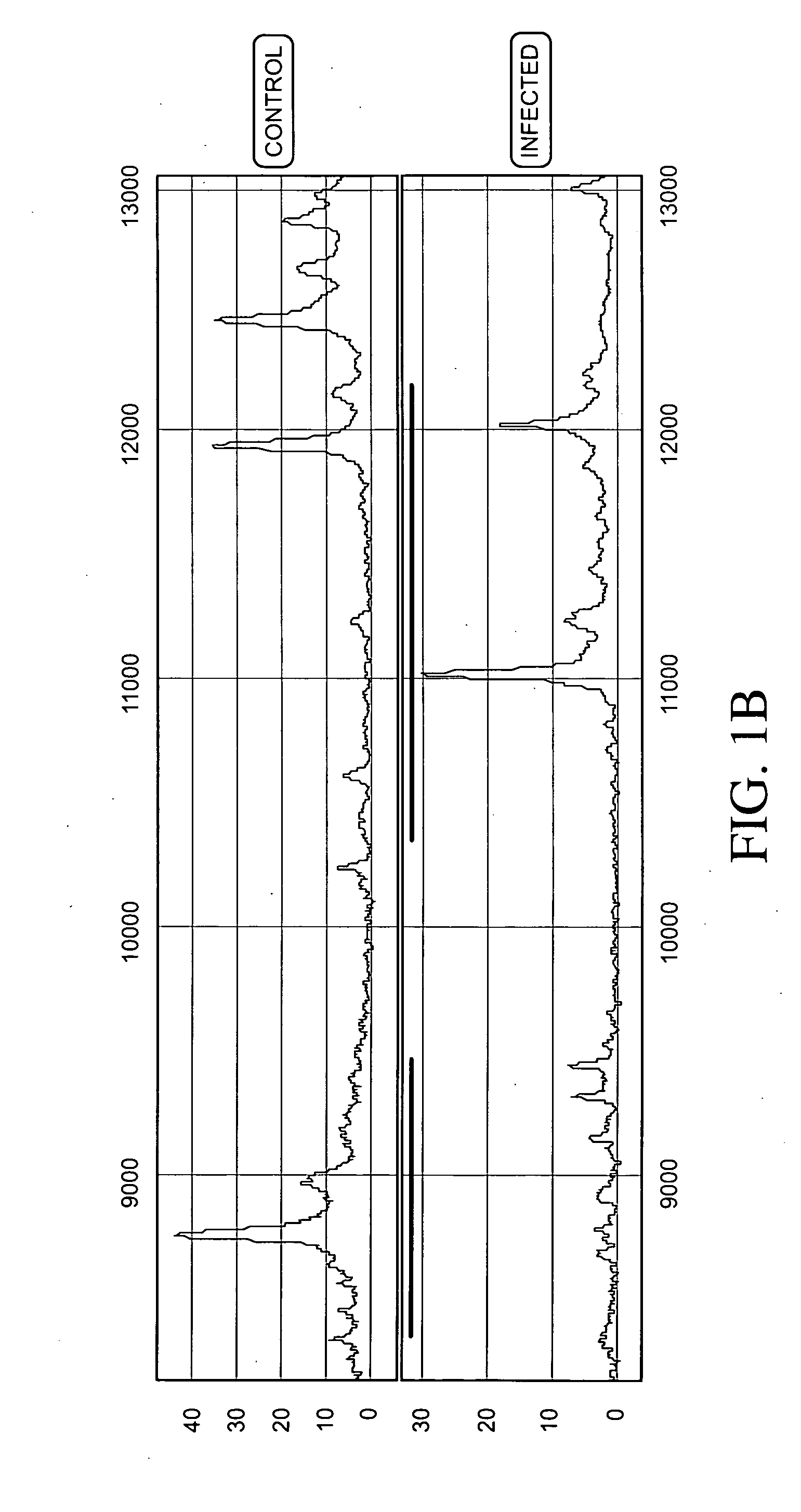
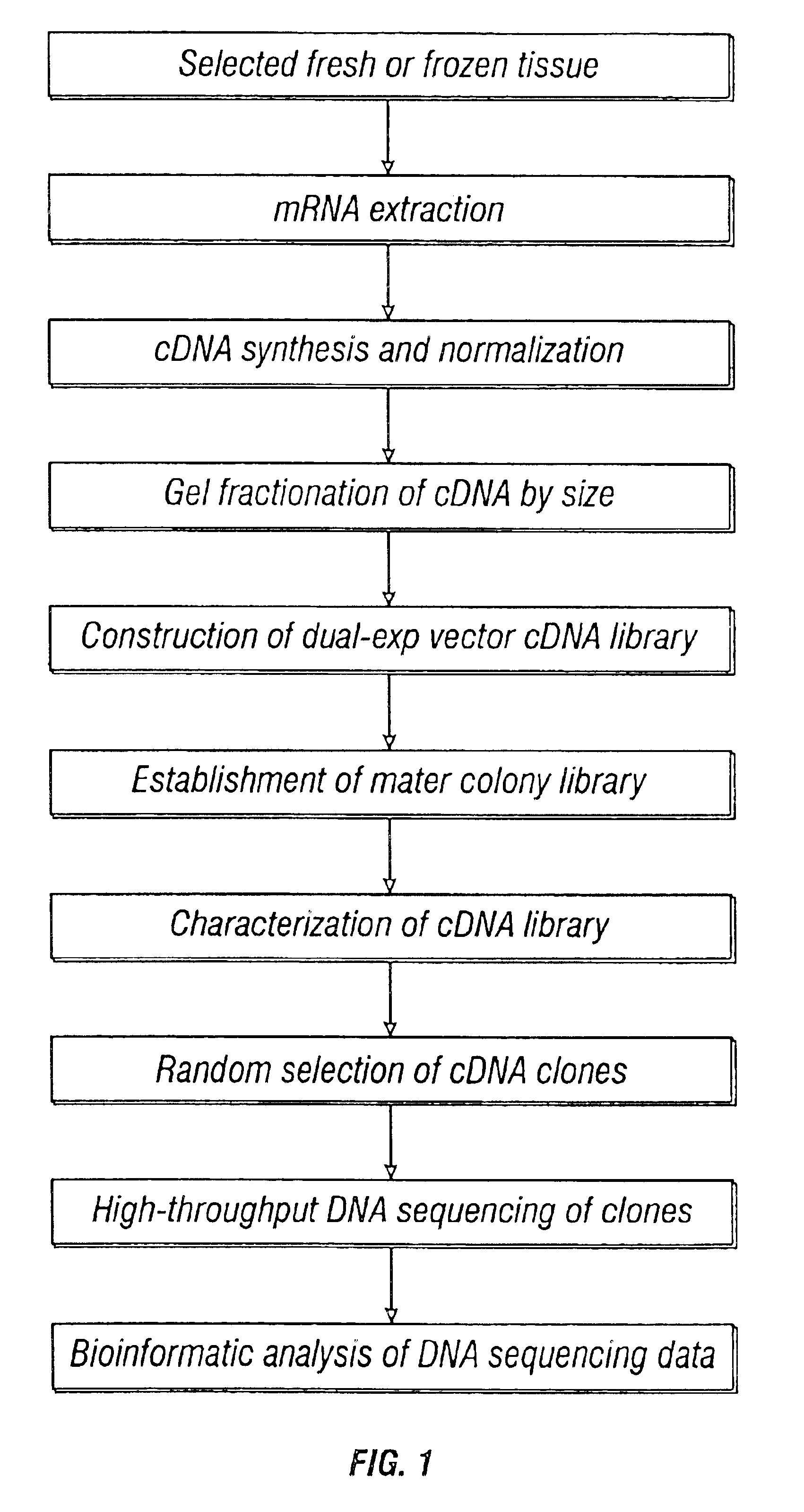
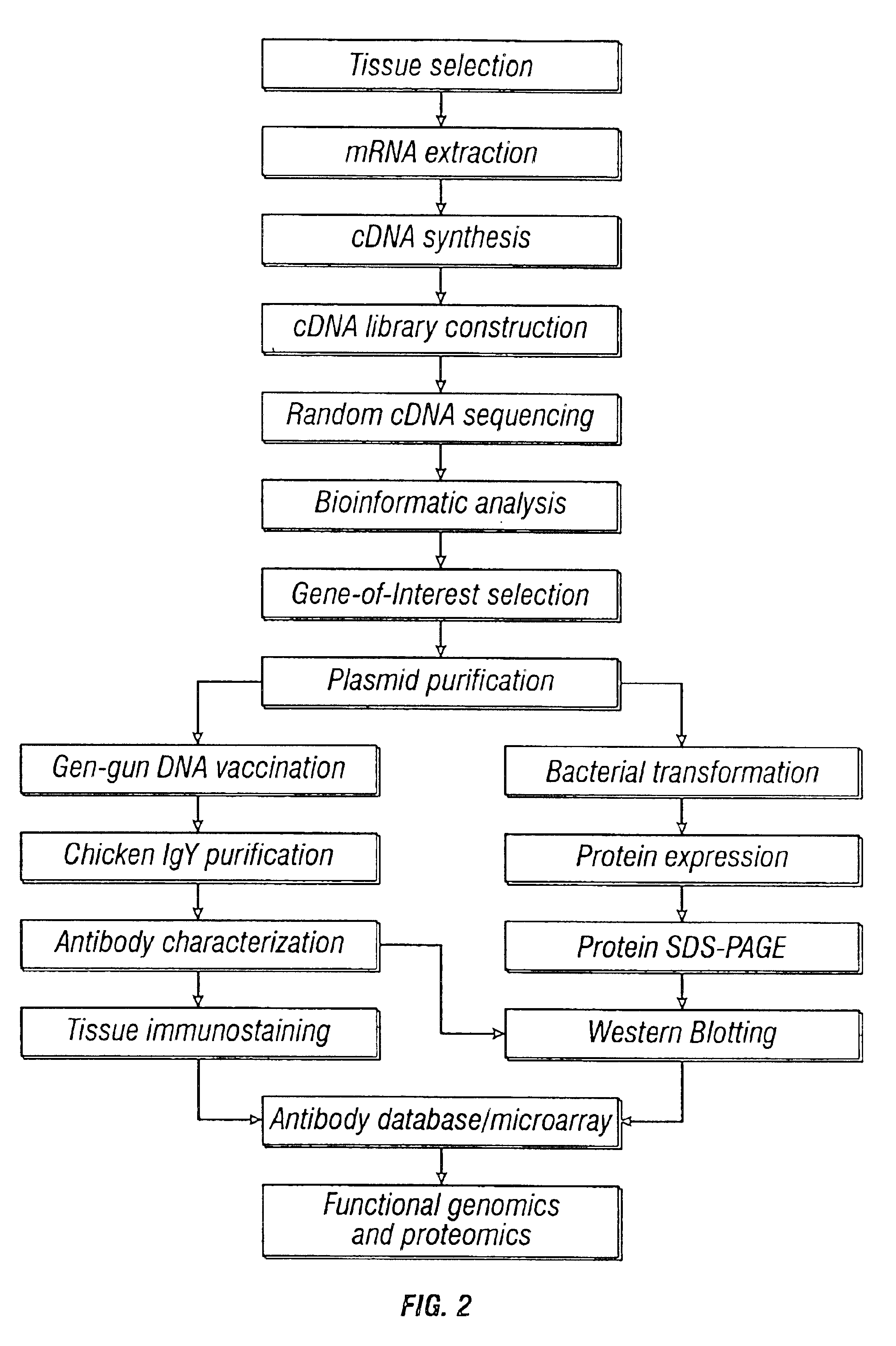
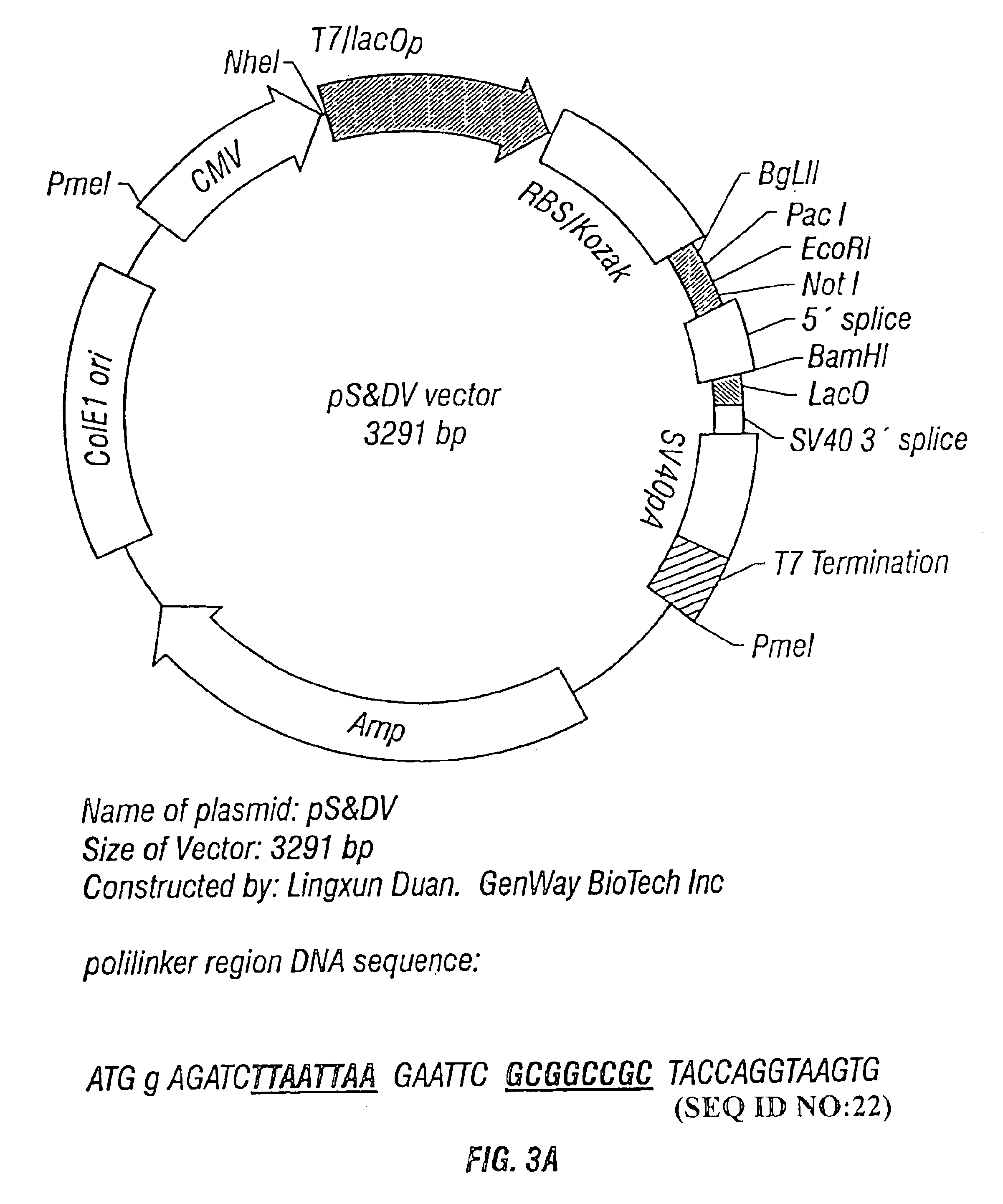
Methods and vectors for generating antibodies in avian species and uses therefor
The present invention relates to processes for producing polyclonal and monoclonal antibodies to an antigen in an avian species, preferably in a chicken, using polynucleotide vaccination. The present invention also relates to processes for determining the proteomics profile of a set of pre-selected DNA sequences isolated from a bio-sample, preferably the proteomics profile of a human cDNA library. The present invention additionally relates to processes for identifying physiologically distinguishable markers associated with a physiologically abnormal bio-sample. The present invention further relates to antibody arrays, integrated databases for identification of genes and proteins, multi-functional gene expression vectors, and methods of producing and using such antibody arrays, integrated databases and multi-functional gene expression vectors.
Owner:GENWAY BIOTECH
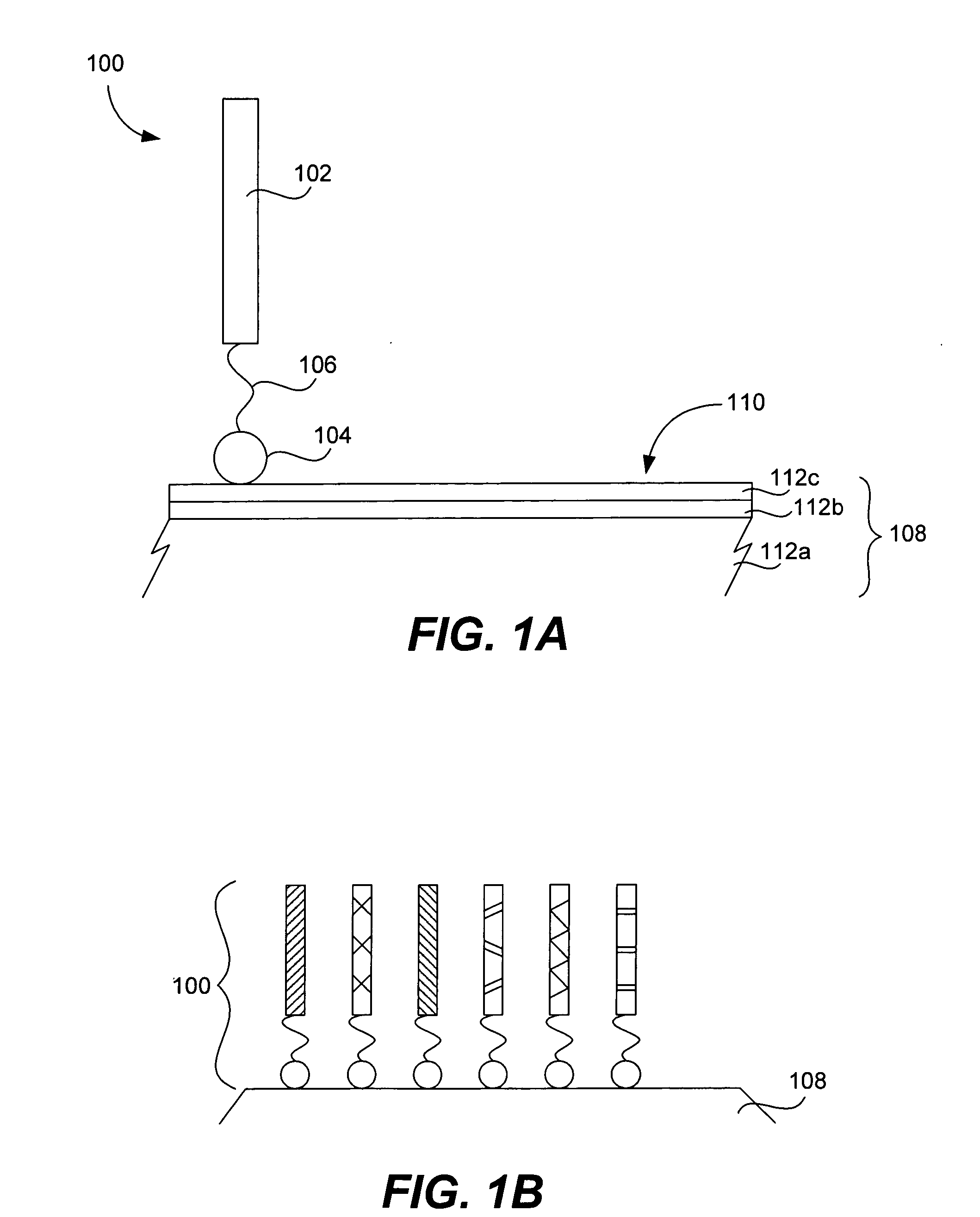
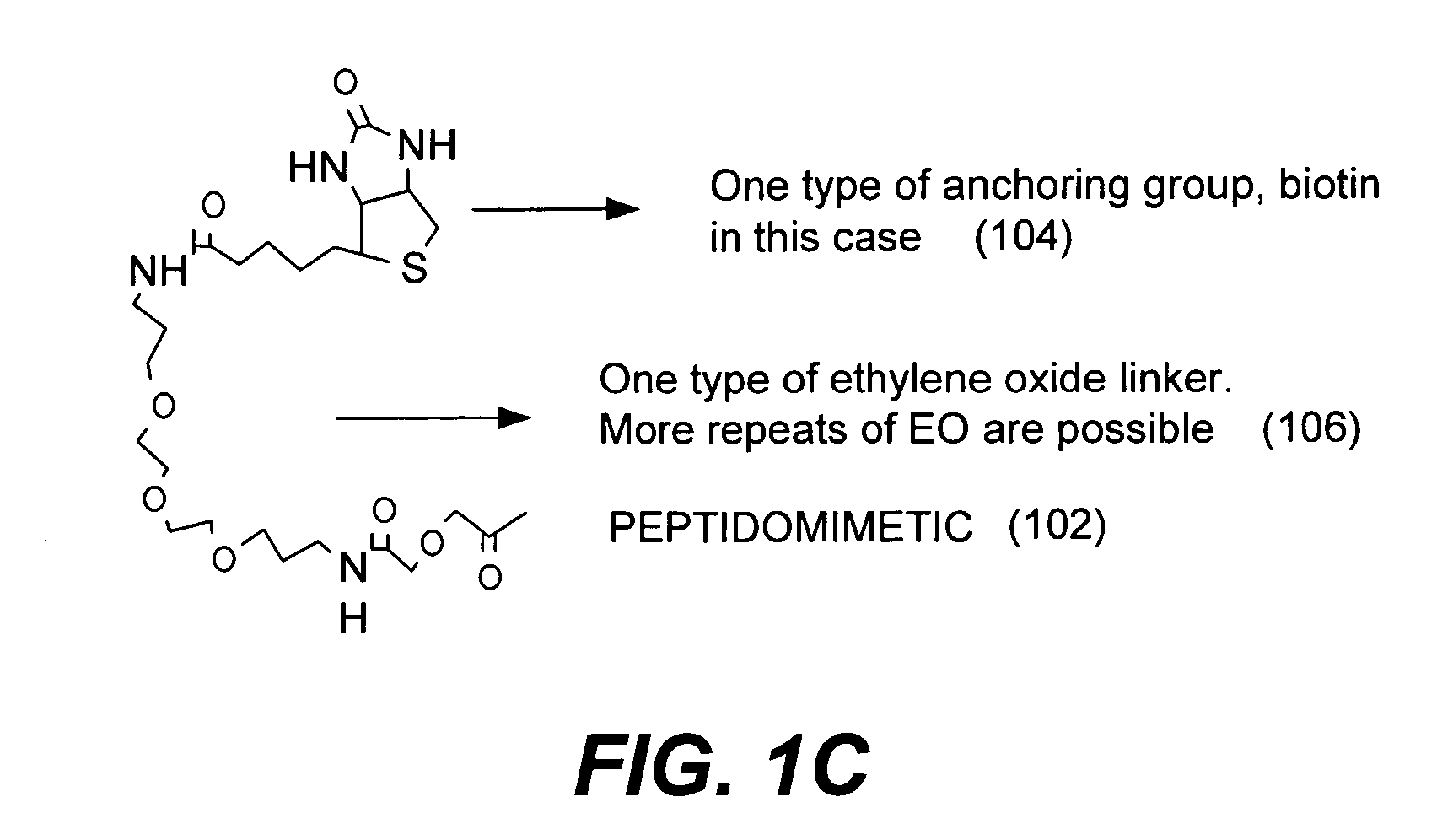
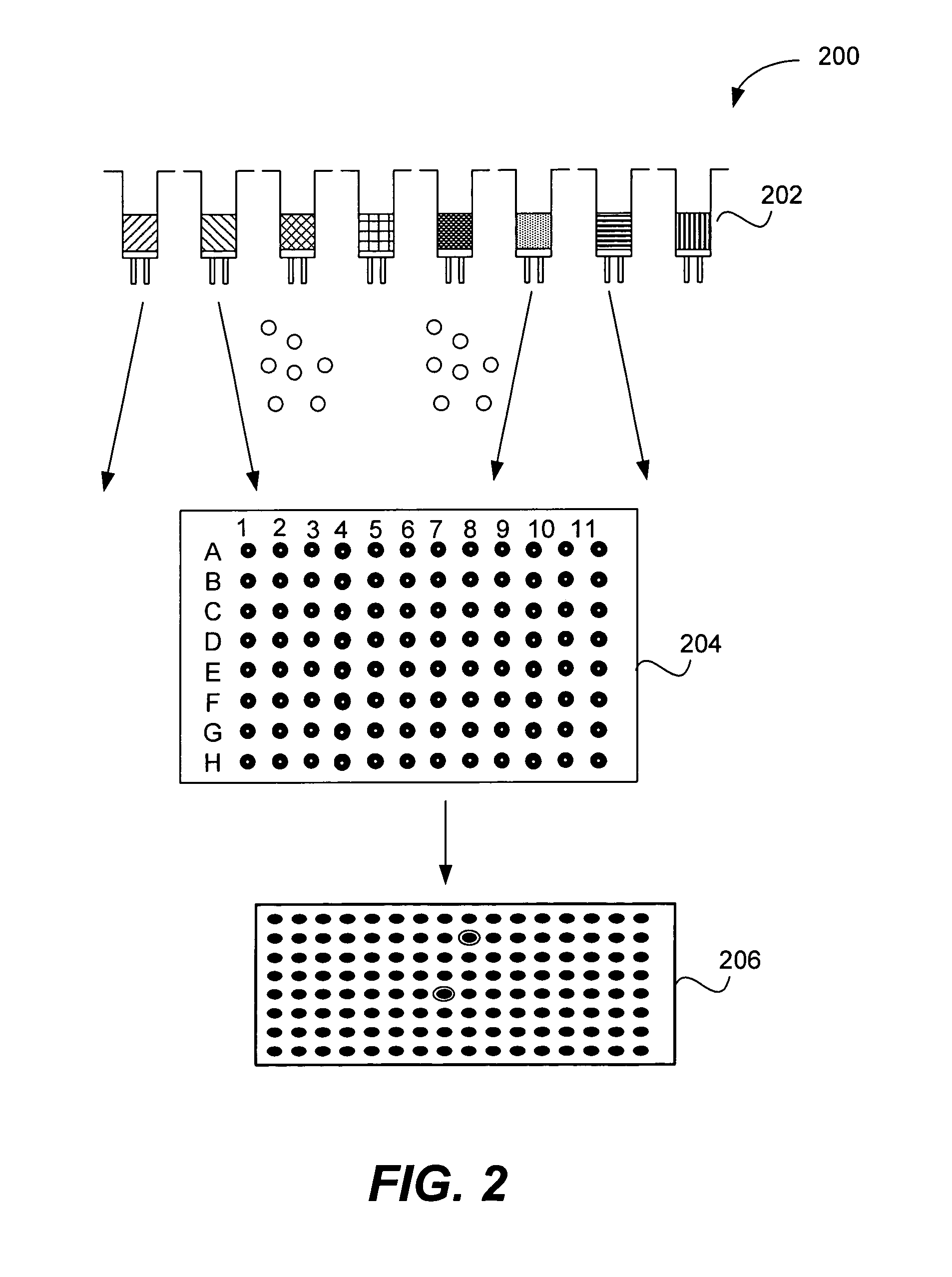
Microarrays on mirrored substrates for performing proteomic analyses
Provided are peptidomimetic protein-binding arrays, their manufacture, use, and application. The protein-binding array elements of the invention include a peptidomimetic segment linked to a solid support via a stable anchor. The invention contemplates peptidomimetic array element library synthesis, distribution, and spotting of array elements onto solid planar substrates, labeling of complex protein mixtures, and the analysis of differential protein binding to the array. The invention also enables the enrichment or purification, and subsequent sequencing or structural analysis of proteins that are identified as differential by the array screen. Kits including proteomic microarrays in accordance with the present invention are also provided.
Owner:NOVARTIS VACCINES & DIAGNOSTICS INC
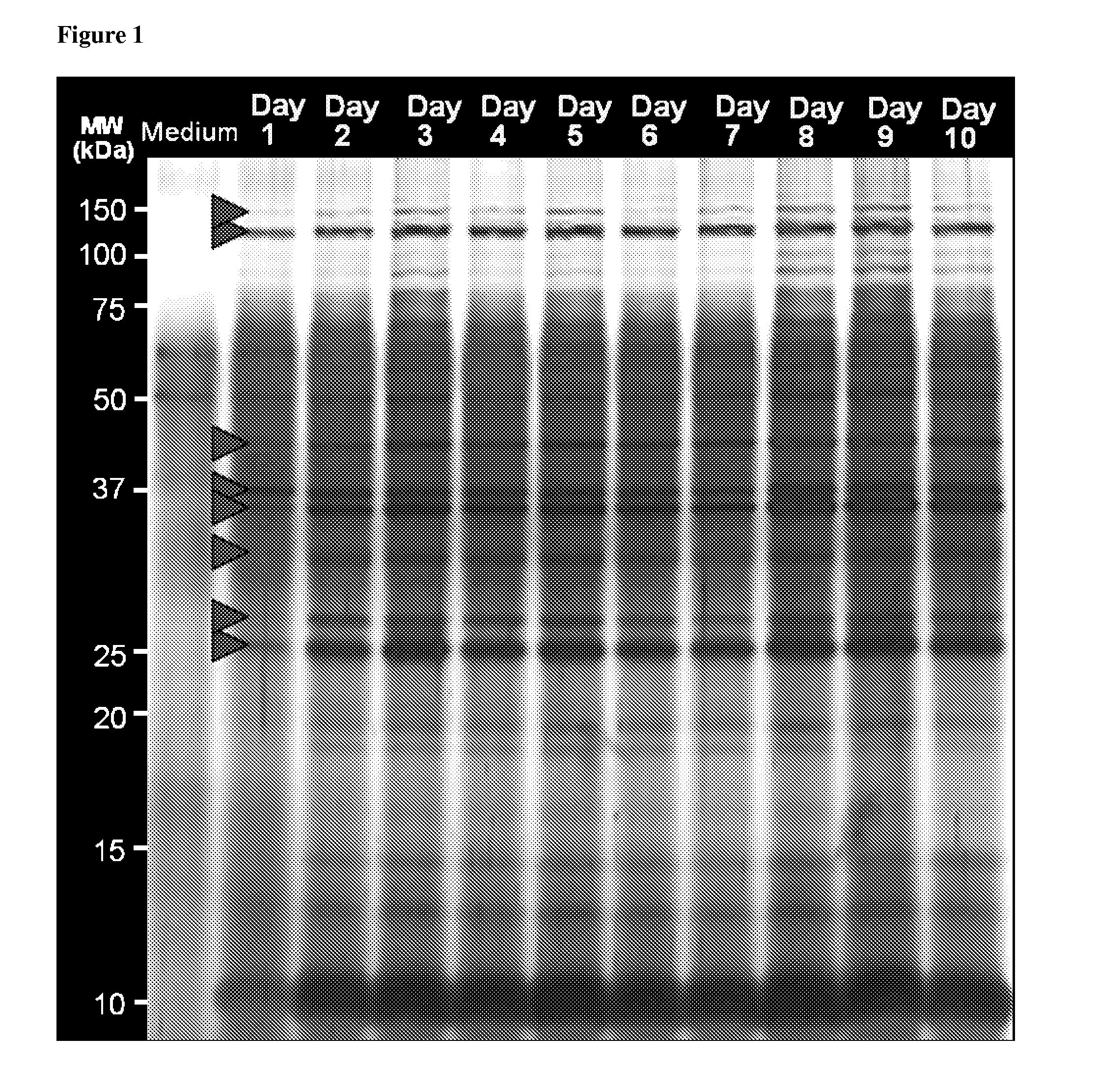
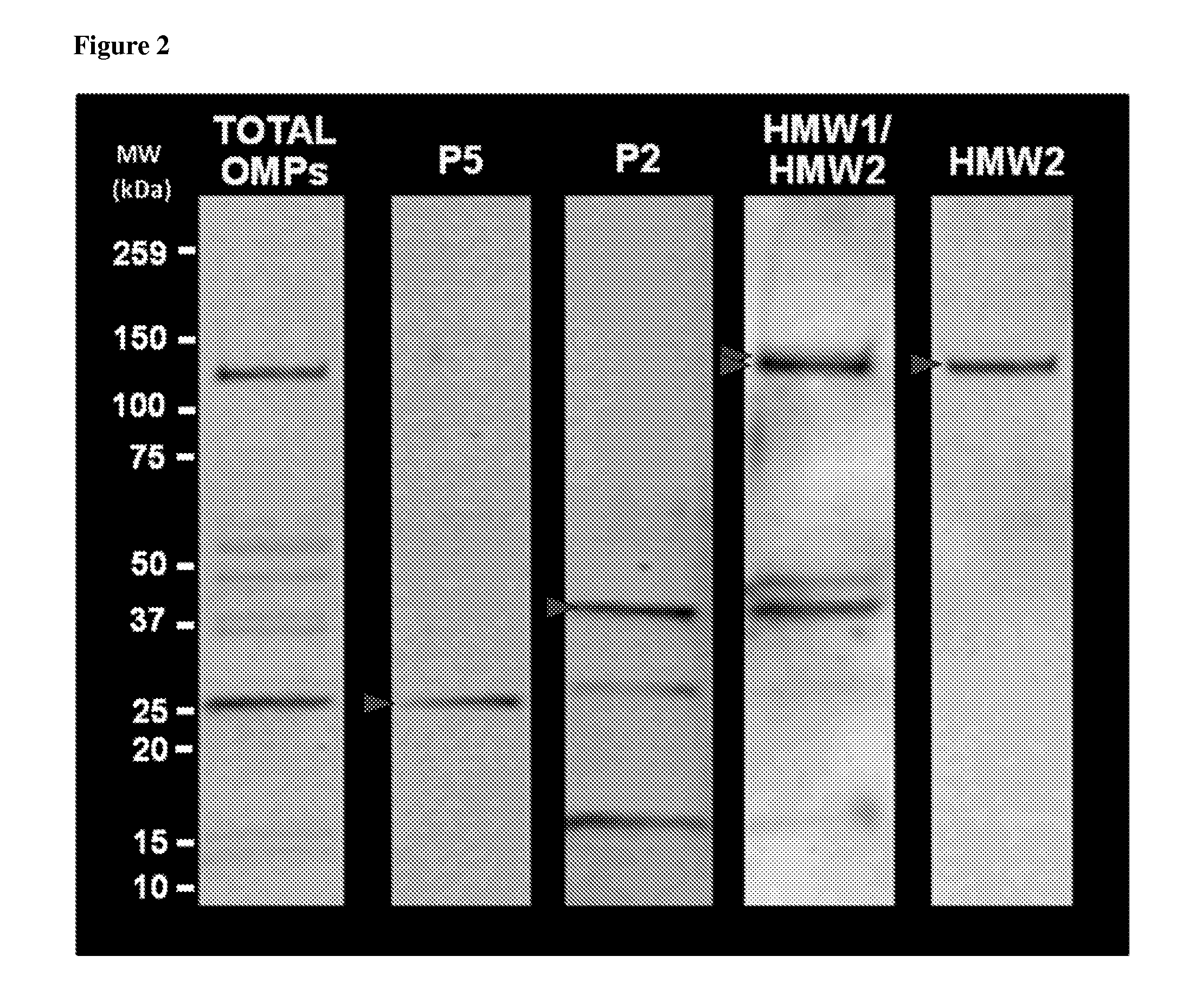
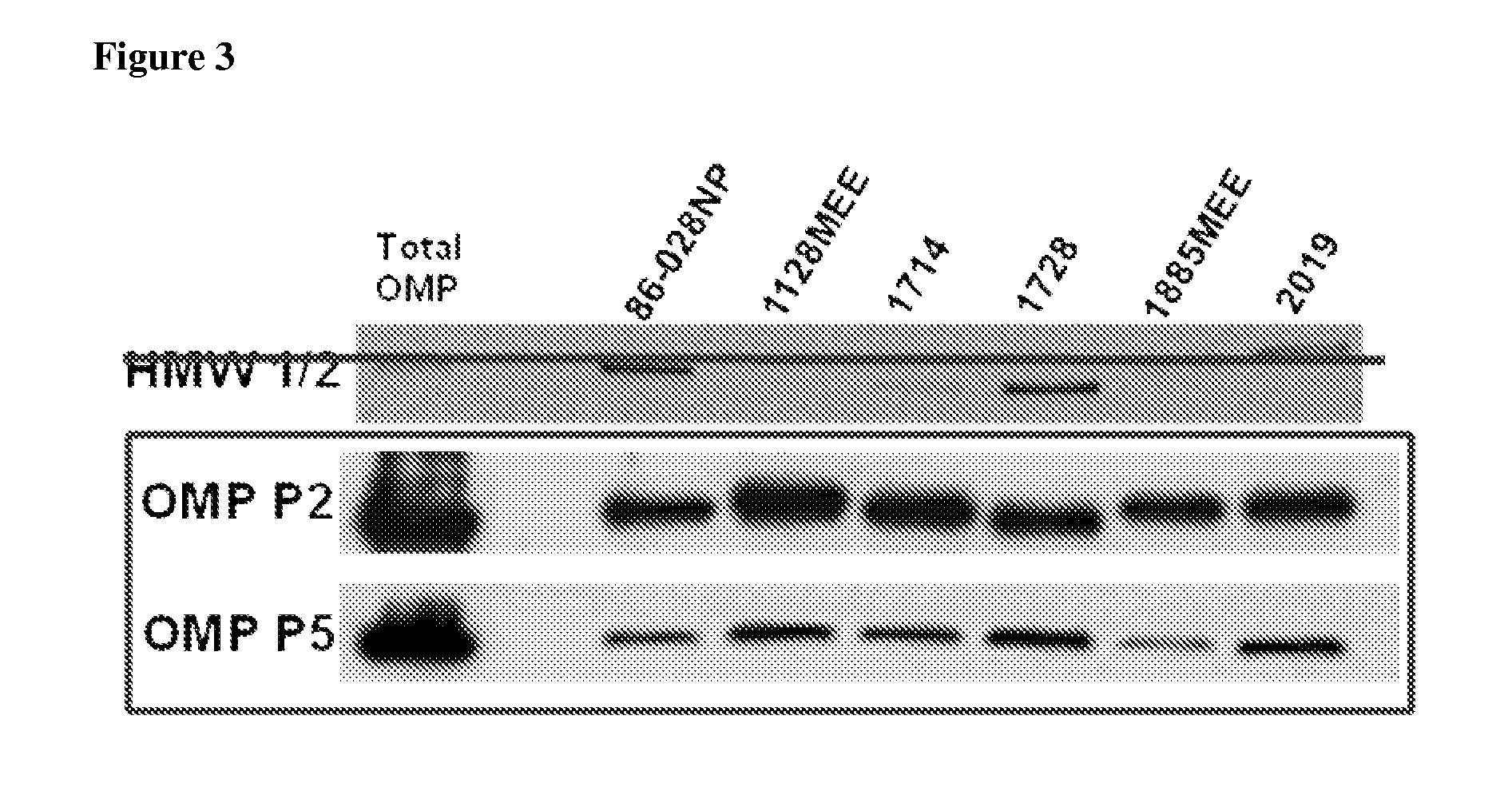
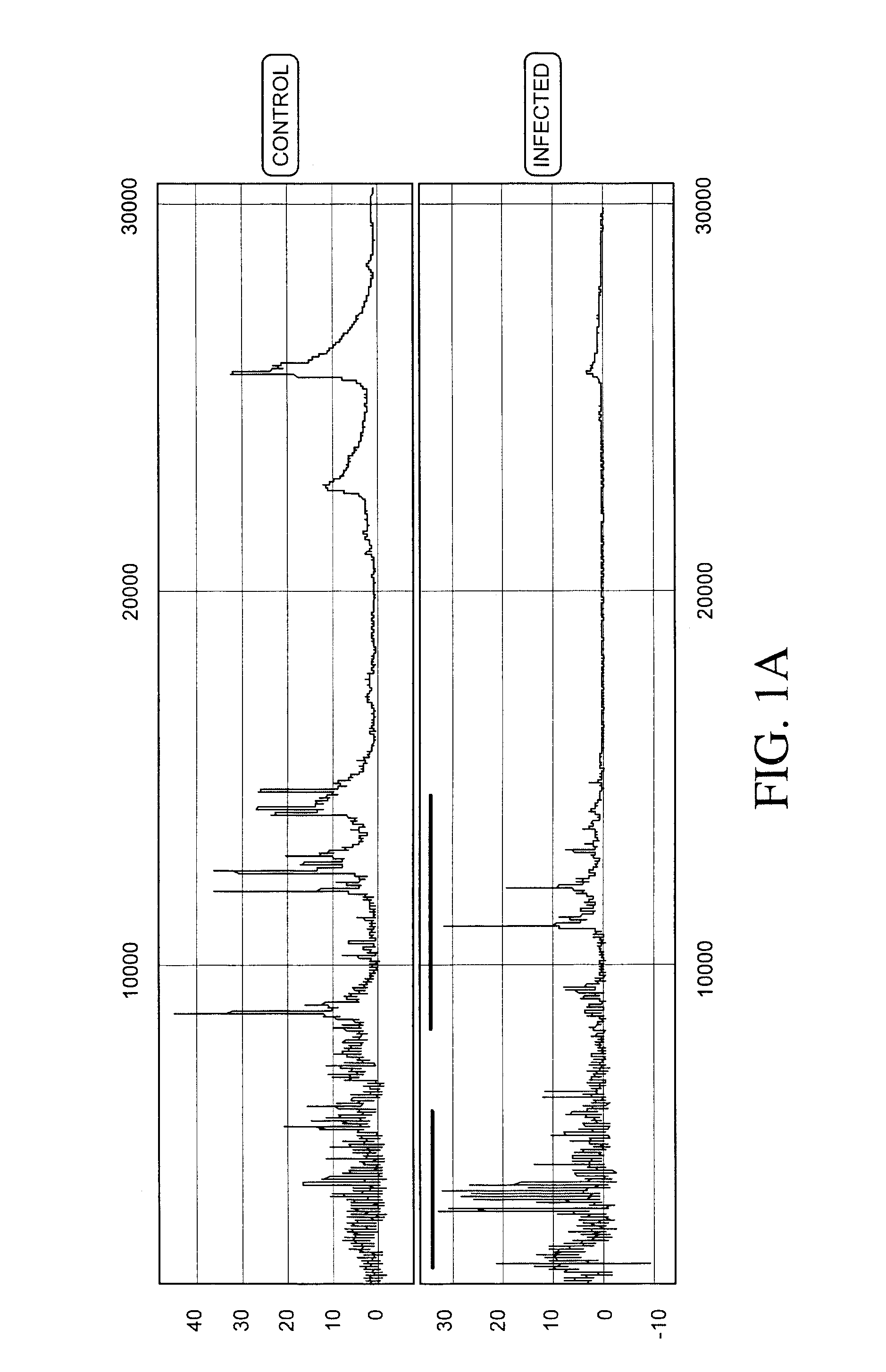
Proteomics based diagnostic detection method for chronic sinusitis
ActiveUS20140314876A1Effective choiceEarlyAntibacterial agentsBioreactor/fermenter combinationsProteomics methodsBacteroides
The invention provides for a proteomic approach for identification of specific bacterial protein profiles that may be used in the development of methods for the diagnosis of bacterial chronic sinusitis. The invention provides for methods for determining the presence of pathogenic bacteria in the upper respiratory tract of a subject using protein profiles of the pathogenic bacteria. The invention also provides for methods of diagnosing a bacterial infection of the upper respiratory tract of a subject using protein profiles of a pathogenic bacteria. In addition, the invention provides for devices, immunoassays and kits for identifying pathogenic bacteria in the upper respiratory tract.
Owner:NATIONWIDE CHILDRENS HOSPITAL +1
Diagnosis of intra-uterine infection by proteomic analysis of cervical-vaginal fluids
ActiveUS8068990B2Eliminate needMicrobiological testing/measurementAnalogue computers for chemical processesDiseaseObstetrics
The invention concerns the identification of proteomes of biological fluids and their use in determining the state of maternal / fetal conditions, including maternal conditions of fetal origin, chromosomal aneuploidies, and fetal diseases associated with fetal growth and maturation. In particular, the invention concerns a comprehensive proteomic analysis of human amniotic fluid (AF) and cervical vaginal fluid (CVF), and the correlation of characteristic changes in the normal proteome with various pathologic maternal / fetal conditions, such as intra-amniotic infection, pre-term labor, and / or chromosomal defects. The invention further concerns the identification of biomarkers and groups of biomarkers that can be used for non-invasive diagnosis of various pregnancy-related disorders, and diagnostic assays using such biomarkers.
Owner:HOLOGIC INC
Proteomics based diagnostic detection method for chronic sinusitis
ActiveUS9568472B2Effective choiceEarlyAntibacterial agentsMicrobiological testing/measurementBacteroidesProtein profiling
Owner:NATIONWIDE CHILDRENS HOSPITAL +1
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com