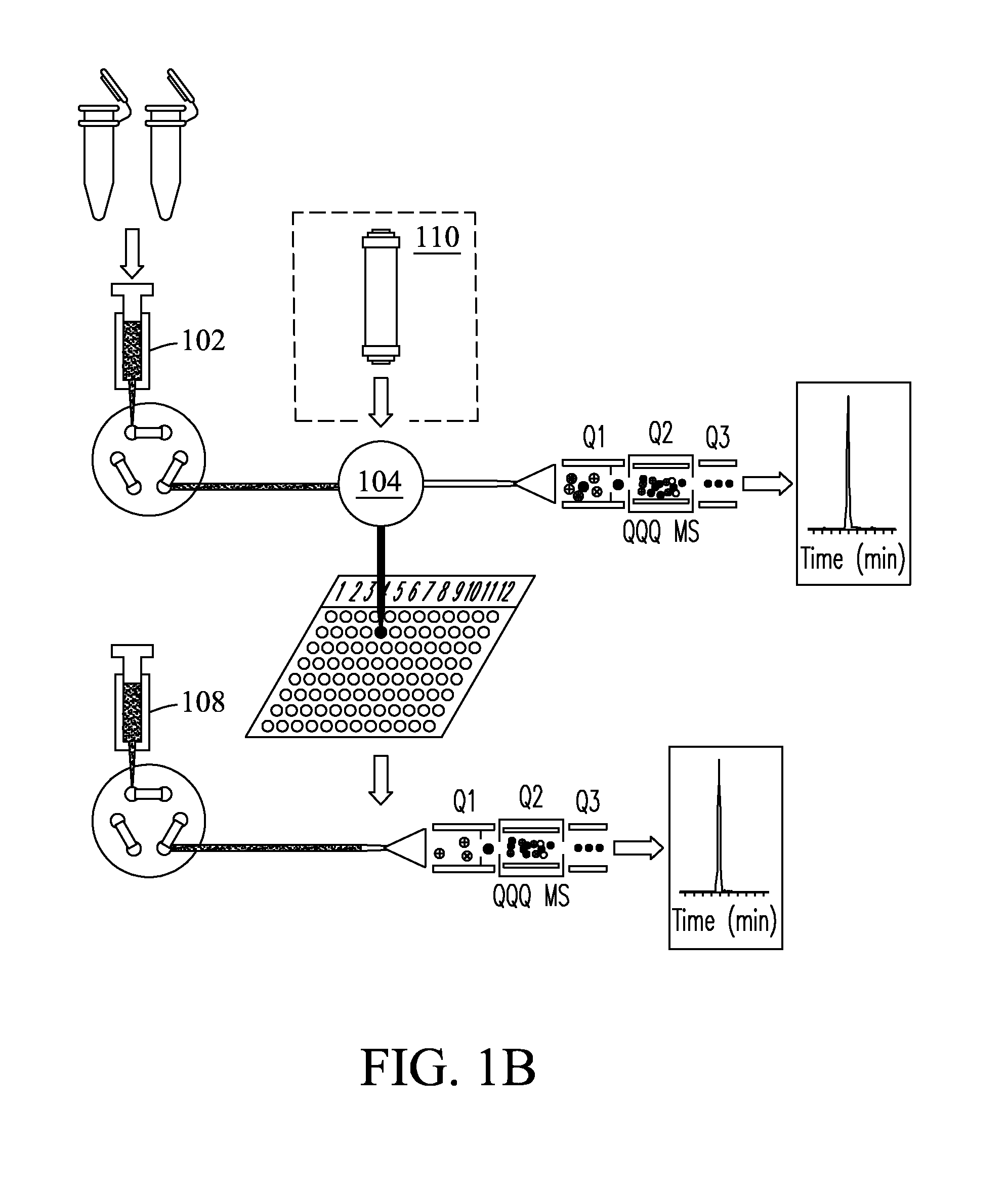
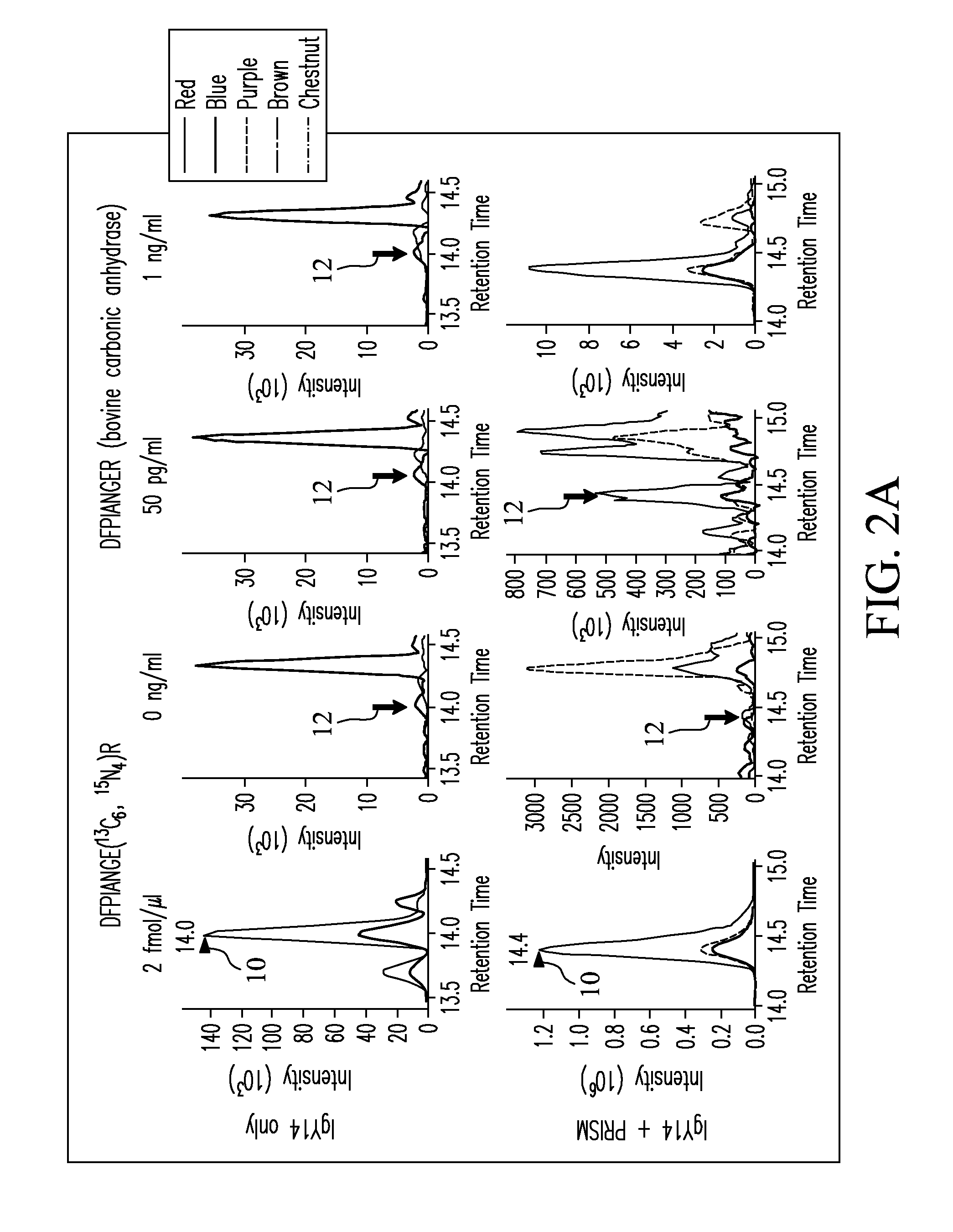
Patents
Literature
30 results about "Selected reaction monitoring" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
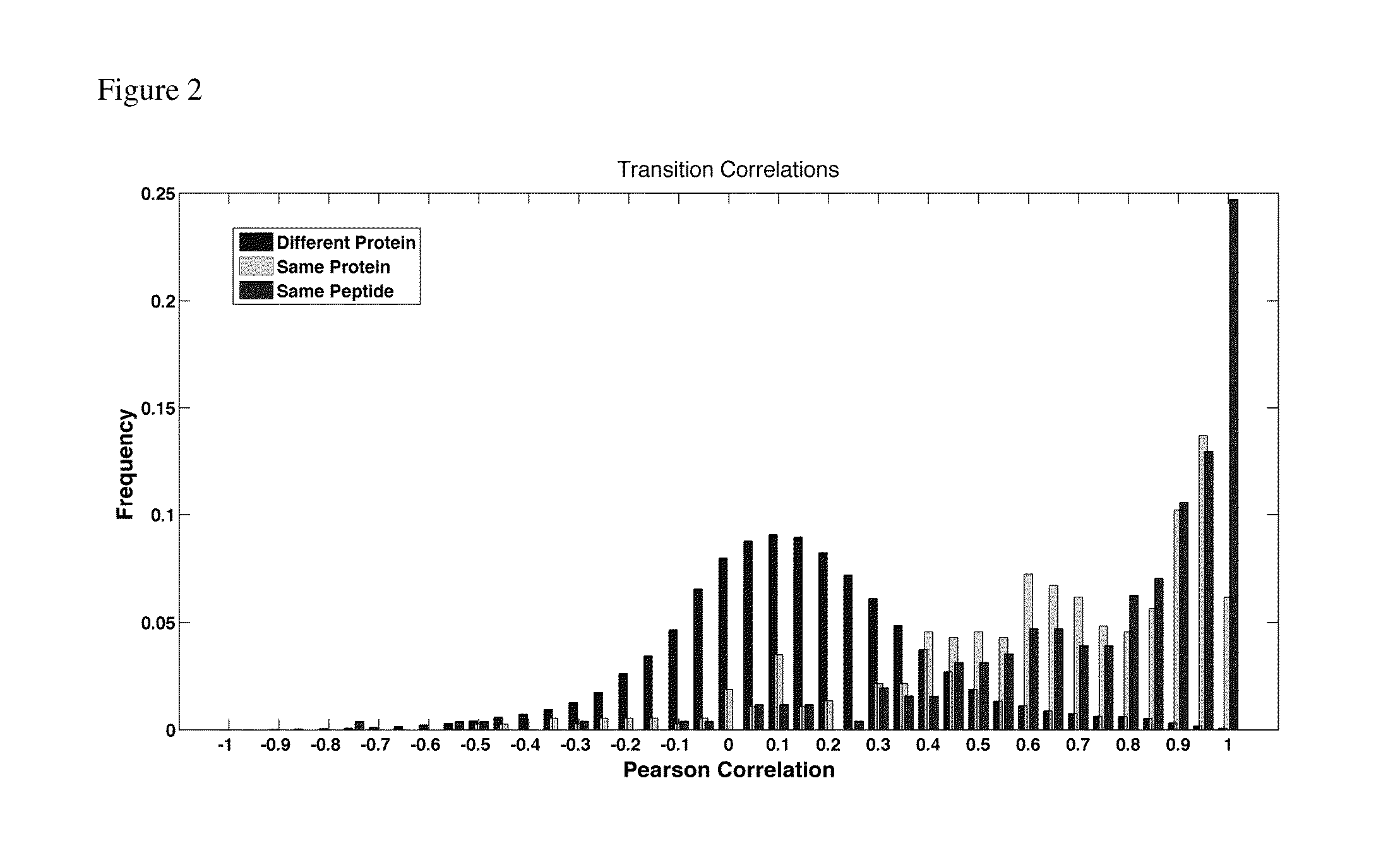
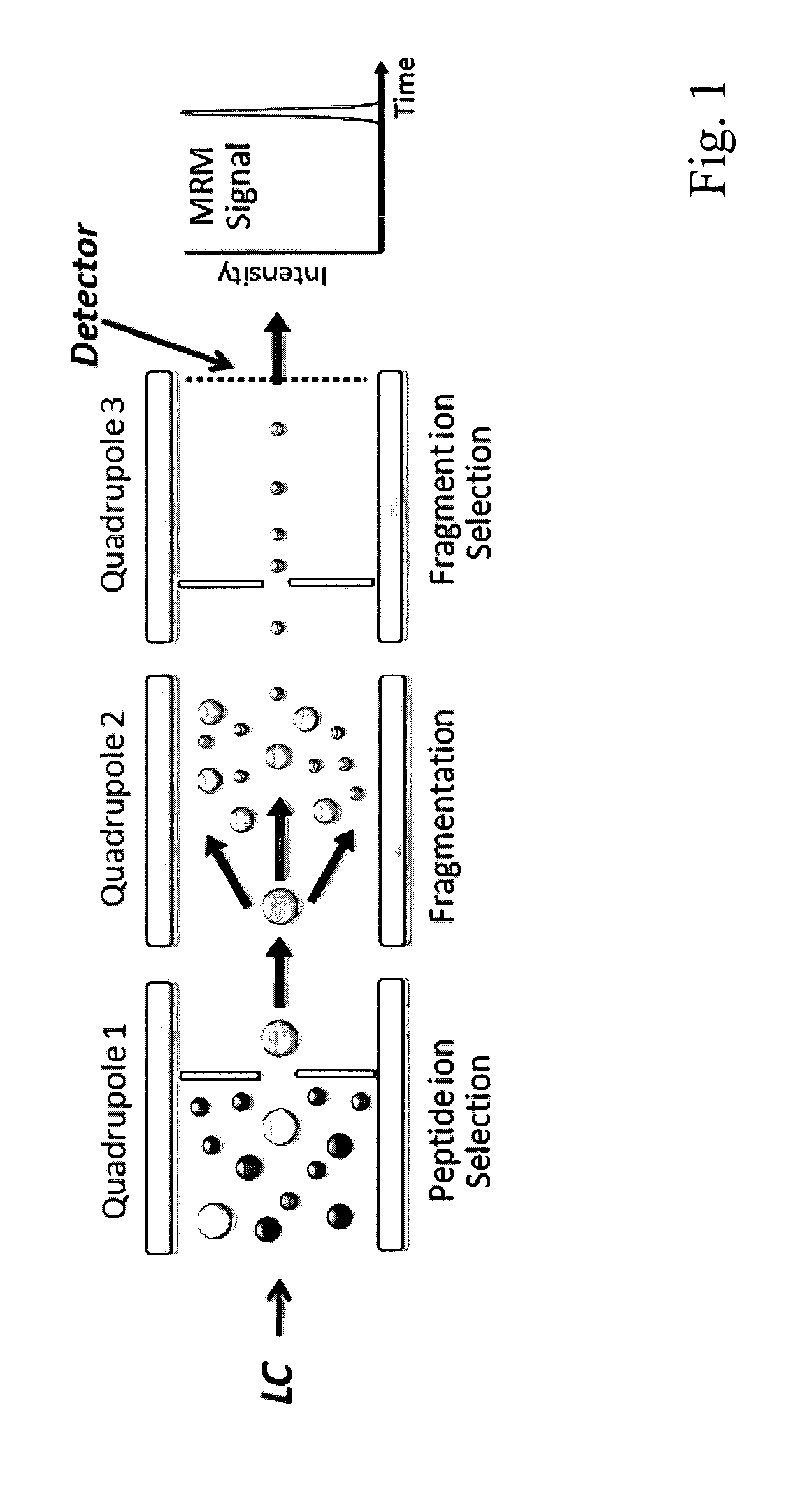
Selected reaction monitoring (SRM) is a method used in tandem mass spectrometry in which an ion of a particular mass is selected in the first stage of a tandem mass spectrometer and an ion product of a fragmentation reaction of the precursor ion is selected in the second mass spectrometer stage for detection.
Selected Reaction Monitoring Assays
ActiveUS20130203096A1Minimal interferenceEarly detectionMicrobiological testing/measurementMass spectrometric analysisMass Spectrometry-Mass SpectrometrySelected reaction monitoring
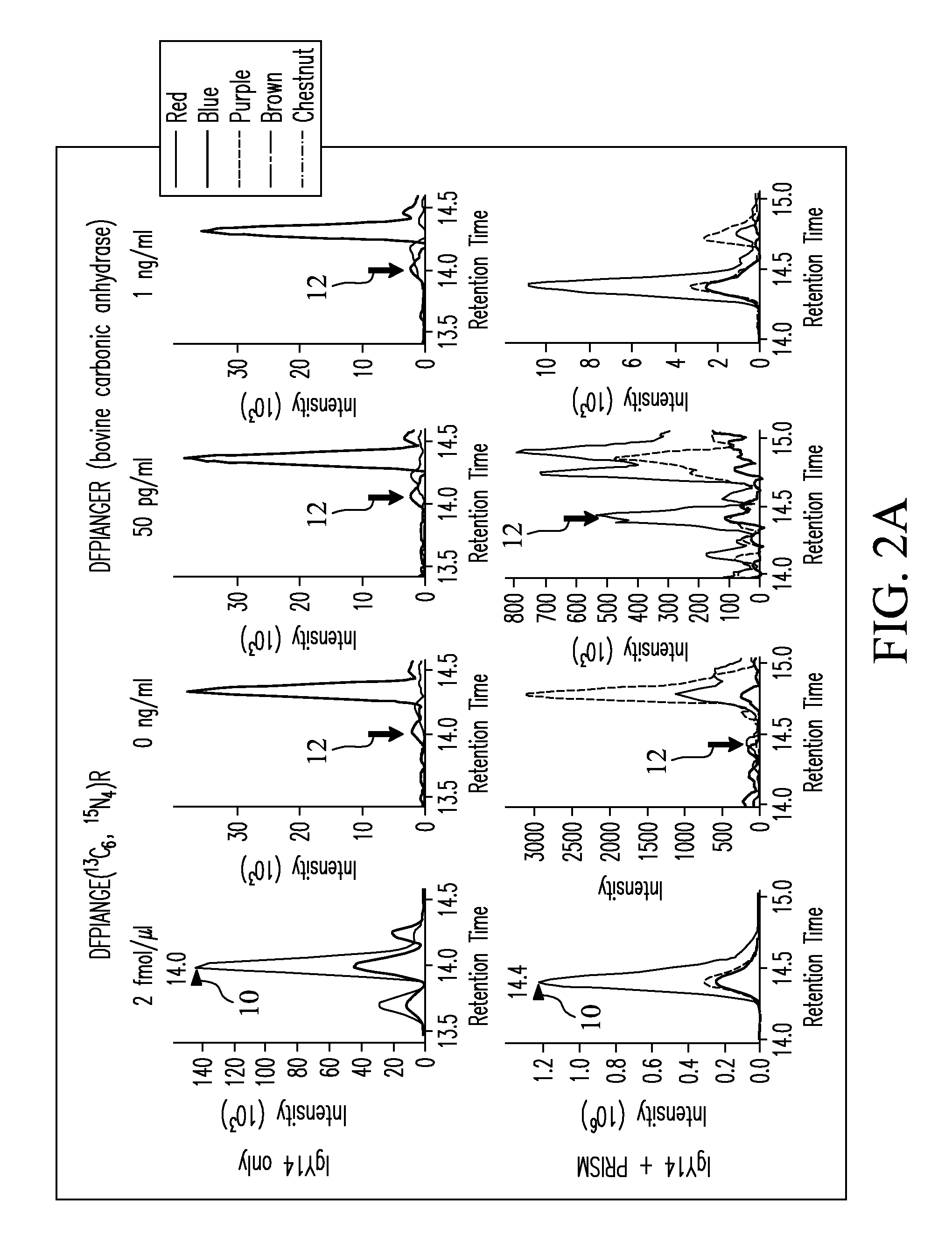
Provided herein are methods for developing selected reaction monitoring mass spectrometry (LC-SRM-MS) assays.
Owner:BIODESIX
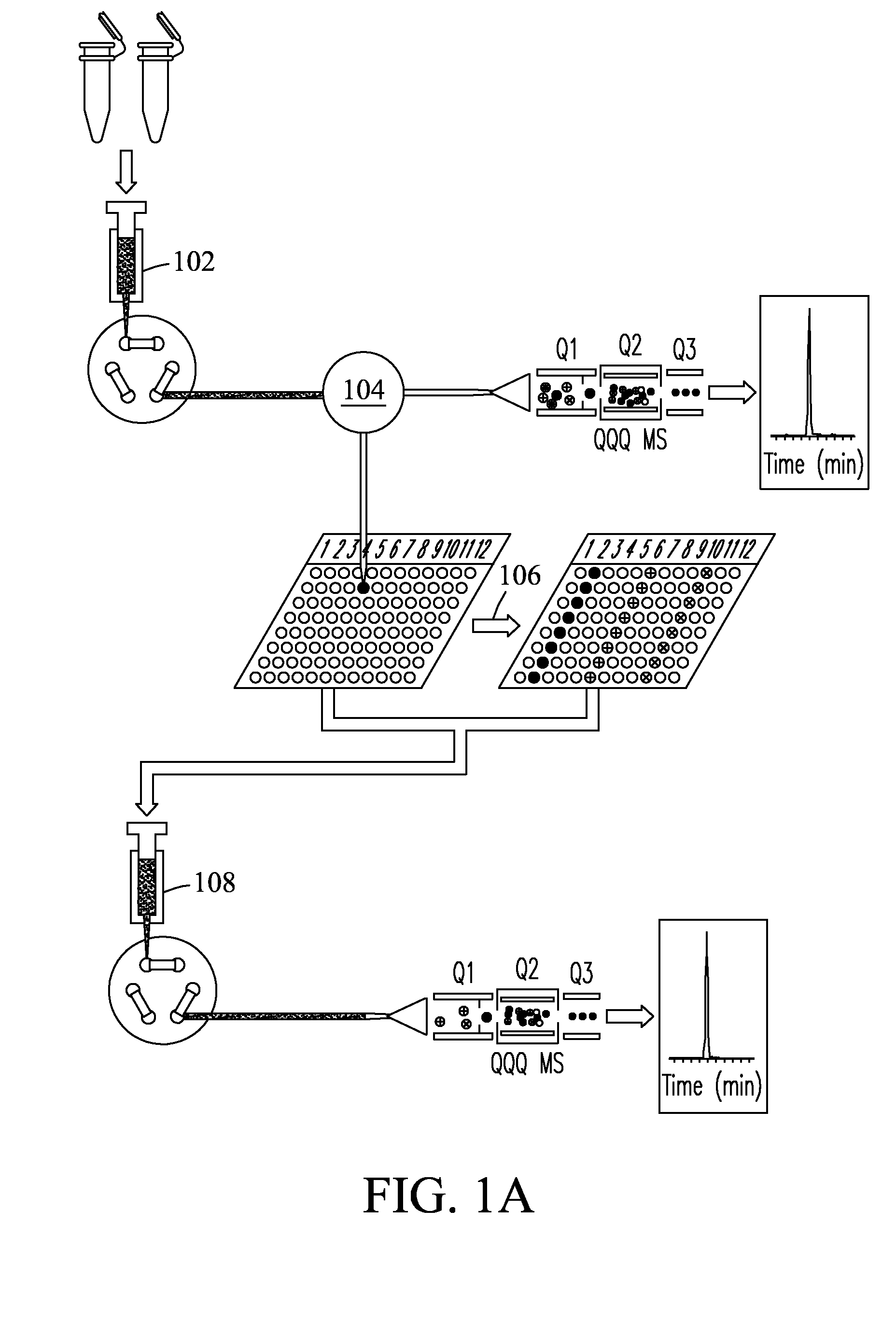
System and process for pulsed multiple reaction monitoring
InactiveUS20110248160A1Reduces background ion noiseIncrease amplitudeStability-of-path spectrometersIsotope separationSelected reaction monitoringPulse injection
A new pulsed multiple reaction monitoring process and system are disclosed that uses a pulsed ion injection mode for use in conjunction with triple-quadrupole instruments. The pulsed injection mode approach reduces background ion noise at the detector, increases amplitude of the ion signal, and includes a unity duty cycle that provides a significant sensitivity increase for reliable quantitation of proteins / peptides present at attomole levels in highly complex biological mixtures.
Owner:BATTELLE MEMORIAL INST
Quantification of vitellogenin
InactiveUS20090011447A1Simple methodMicrobiological testing/measurementPeptidesVitellogeninsGlu-Val-Gly
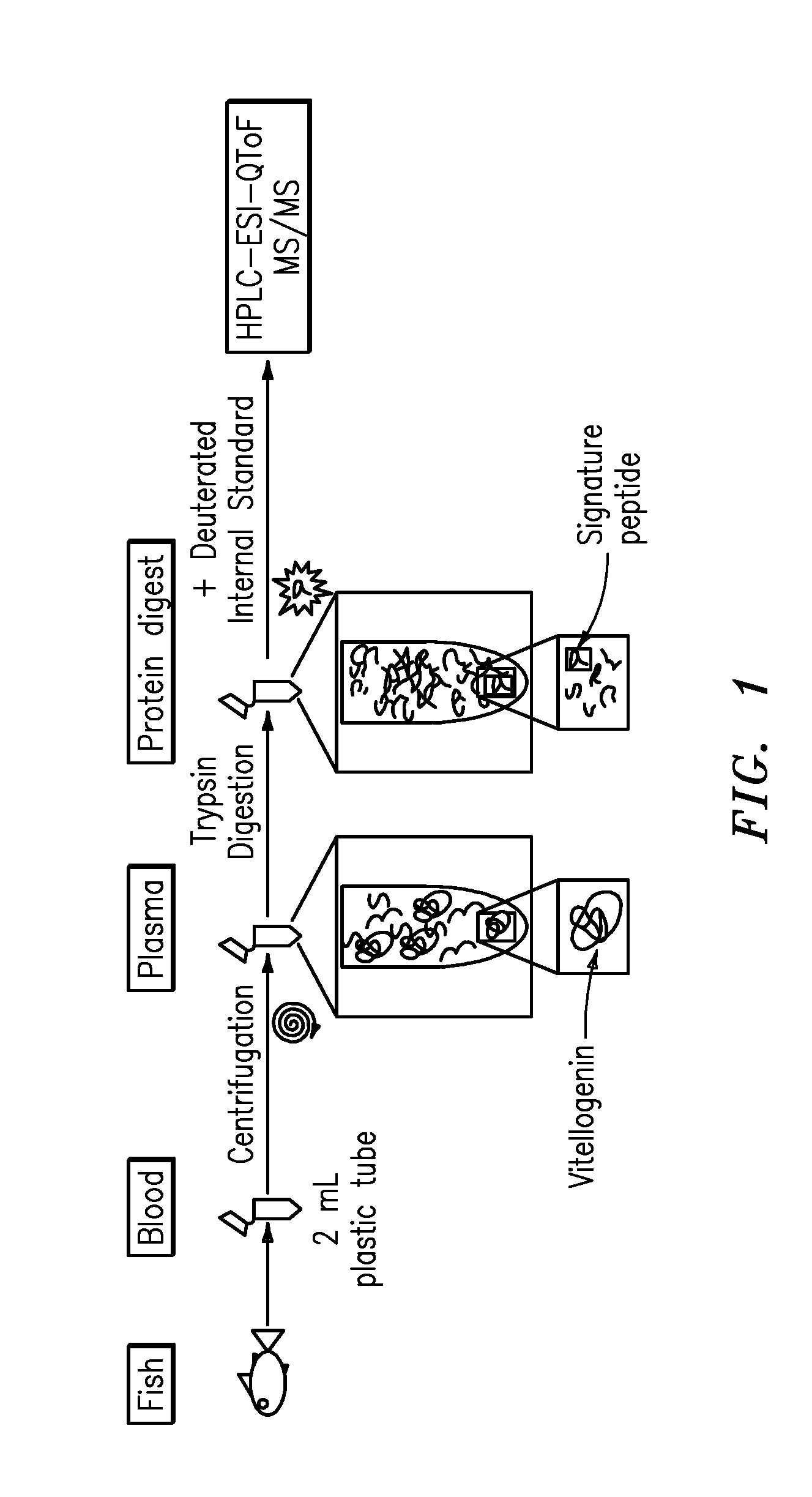
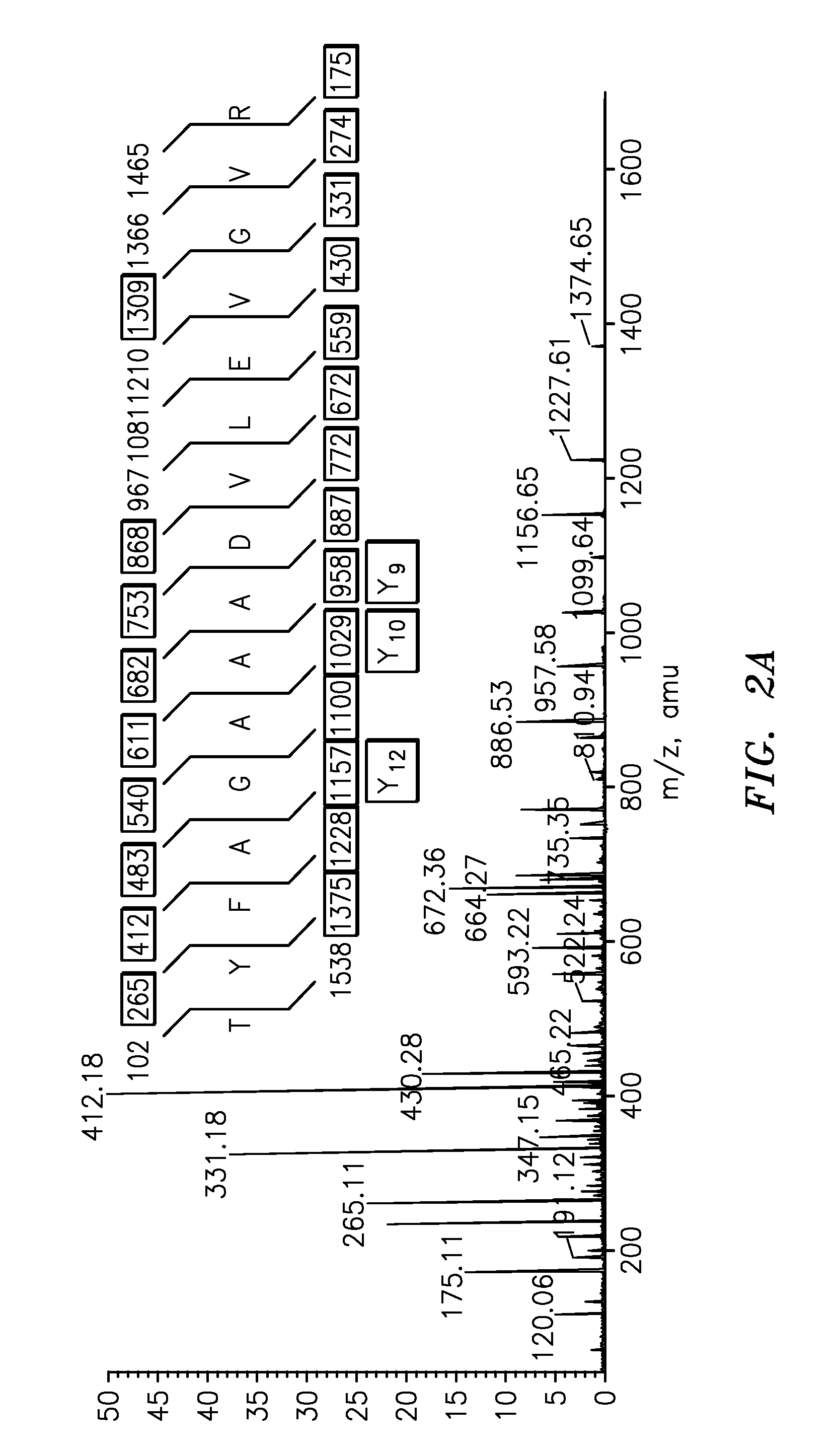
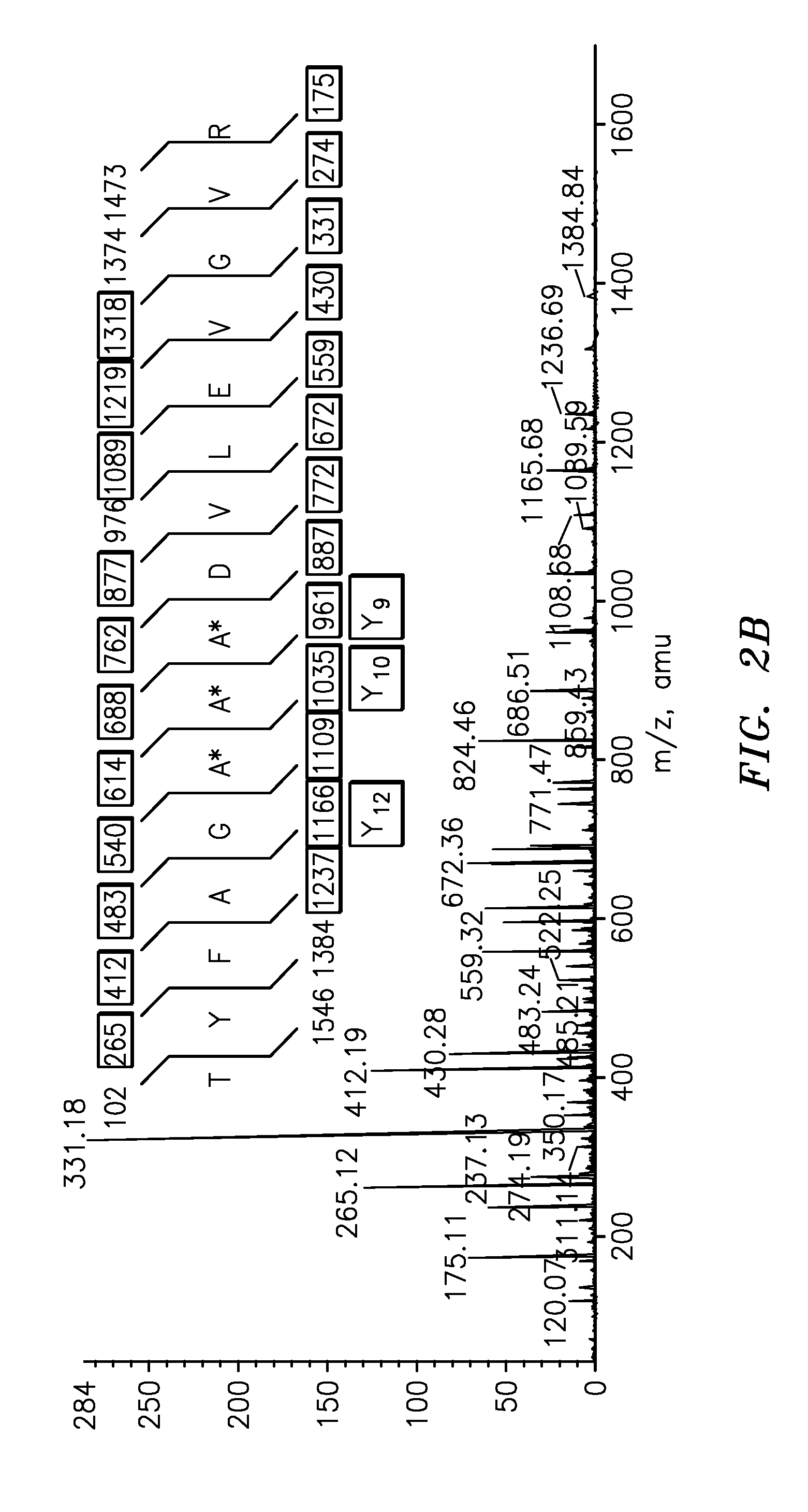
The present invention is directed to a simple method for absolute quantification of plasma vitellogenin from two or more different fish species such as Rainbow trout and Atlantic salmon, or Atlantic cod and haddock. In the case of Rainbow trout and Atlantic salmon, plasma samples obtained from control and β-estradiol induced fish were digested with trypsin. A characteristic ‘signature peptide’ was selected and analyzed by high performance liquid chromatography coupled to an electrospray quadrupole-time-of-flight tandem mass spectrometer, using a deuterated homologue peptide as an internal standard. The hybrid tandem mass spectrometer was operated in a ‘pseudo’ selected reaction monitoring mode by which three diagnostic product ions were monitored for identification and quantification purposes. The reproducibility (coefficient of variation ˜5%) and sensitivity (limit of quantification of 0.009 mg / mL) achieved by this simple assay allow it to be considered as an alternative to immunological assays. In the case of Atlantic cod and haddock, the amino acid sequence of the vitellogenin protein has not yet been determined, but, the Atlantic cod vitellogenin has been characterized using a ‘bottom-up’ mass spectrometric approach. Vitellogenin synthesis was induced ‘in vivo’ with β-Estradiol, and subjected to trypsin digestion for characterization by matrix-assisted laser desorption / ionization-Quadrupole-Time-of-flight tandem mass spectrometry. A peptide mass fingerprint was obtained and ‘de novo’ sequencing of the most abundant tryptic peptides was performed by low energy collision induced dissociation-tandem mass spectrometry. Thus, the sequences of various tryptic peptides have been elucidated. It has also been determined that Atlantic cod vitellogenin shares a series of common peptides with the two different known vitellogenin sequences of Haddock, a closely related species. There are also disclosed novel isolated signature peptides, namely Thr-Tyr-Phe-Ala-Gly-Ala-Ala-Ala-Asp-Val-Leu-Glu-Val-Gly-Val-Arg, Asp Leu Gly Leu Ala Tyr Thr Glu Lys, Phe Phe Gly Gln Glu Ile Ala Asn Ile Asp Lys, Glu Ile Val Leu Leu Gly Tyr Gly Thr Met Ile Ser Lys and Tyr Glu Ser Phe Ala Val Ala Arg.
Owner:BANOUB JOSEPH H +2
Process for ultra-sensitive quantification of target analytes in complex biological systems
InactiveUS20140194304A1Easy to quantifyHigh sensitivityElectrolysis componentsComponent separationChromatographic separationHigh pressure
Antibody-free processes are disclosed that provide accurate quantification of a wide variety of low-abundance target analytes in complex samples. The processes can employ high-pressure, high-resolution chromatographic separations for analyte enrichment. Intelligent selection of target fractions may be performed via on-line Selected Reaction Monitoring (SRM) or off-line rapid screening of internal standards. Quantification may be performed on individual or multiplexed fractions. Applications include analyses of, e.g., very low abundance proteins or candidate biomarkers in plasma, cell, or tissue samples without the need for affinity-specific reagents.
Owner:BATTELLE MEMORIAL INST
Srm assays to chemotherapy targets
InactiveUS20170168057A1Easy to carryControl oxidationMass spectrometric analysisDisease diagnosisERCC1Mass Spectrometry-Mass Spectrometry
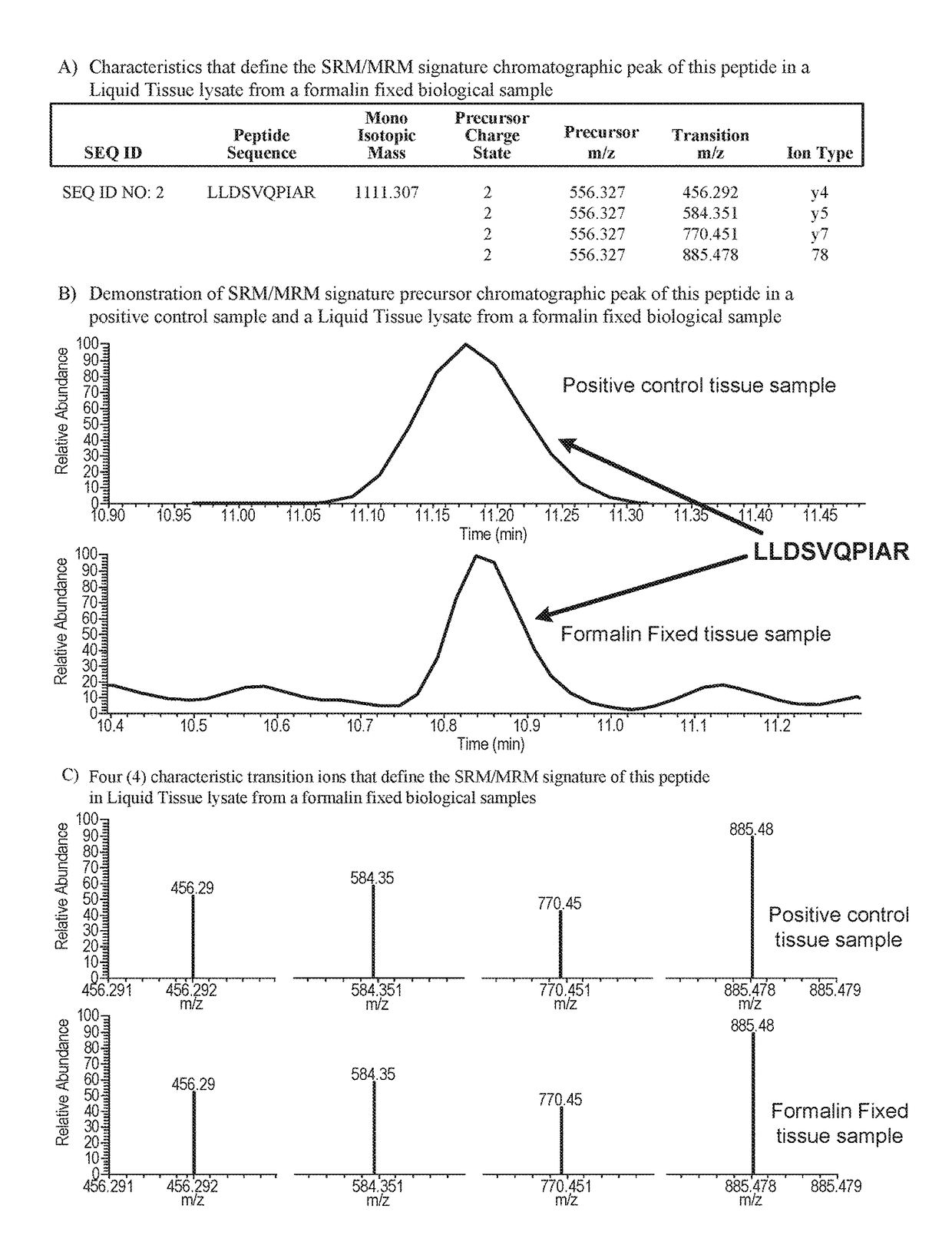
Methods are provided for quantifying the ENT1, ERCC1, FOLR1, RRM1, TUBB3, TOPO1, and / or TOPO2A proteins directly in biological samples that have been fixed in formalin by the method of Selected Reaction Monitoring (SRM) / Multiple Reaction Monitoring (MRM) mass spectrometry. The biological samples are treated with formaldehyde containing agents / fixatives including formalin-fixed tissue / cells, formalin-fixed / paraffin embedded (FFPE) tissue / cells, FFPE tissue blocks and cells from those blocks. A protein digest is prepared from a biological sample and the ENT1, ERCC1, FOLR1, RRM1, TUBB3, TOPO1, and / or TOPO2A proteins are quantitated in the digest by quantitating in the protein sample one or more of the peptides described by the method of SRM / MRM mass spectrometry.
Owner:EXPRESSION PATHOLOGY
Species Detection Using Mass Spectrometry
ActiveUS20160086782A1Reduce generationIsotope separationTesting foodMass Spectrometry-Mass SpectrometryMass spectrometry imaging
Systems and methods are provided for species detection using mass spectrometry. In various embodiments, a multiple reaction monitoring (MRM) experiment is performed on a sample targeting one or more peptide transitions that are unique to one or more species using a tandem mass spectrometer. One or more measured product ion spectra are received from the tandem mass spectrometer using the processor. The one or more measured product ion spectra are compared to product ions of the one or more peptide transitions that are unique to one or more species using the processor. One or more species of the sample are detected by reporting product ions of the one or more peptide transitions that are unique to one or more species that match the one or more measured product ion spectra using the processor.
Owner:DH TECH DEVMENT PTE
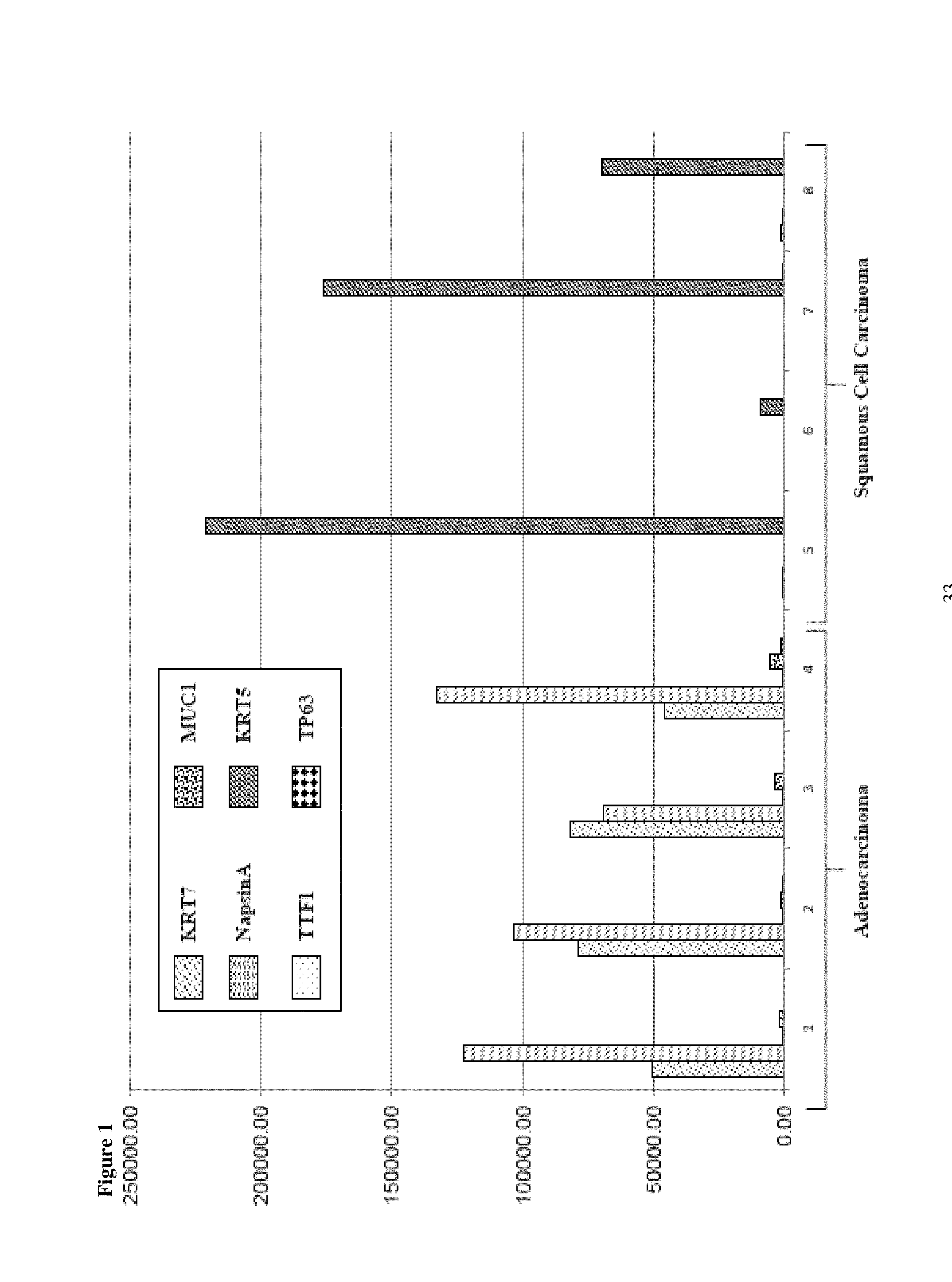
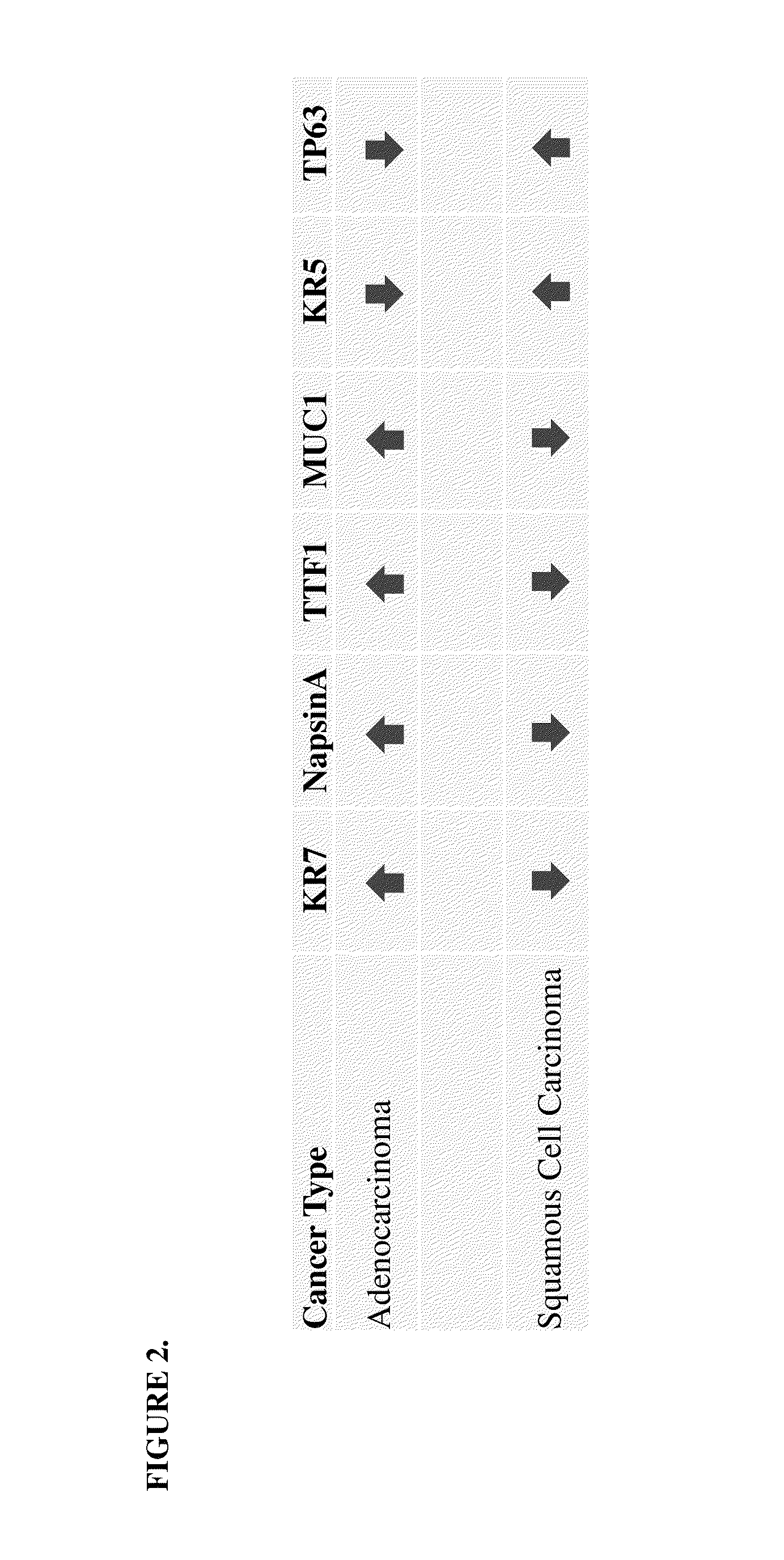
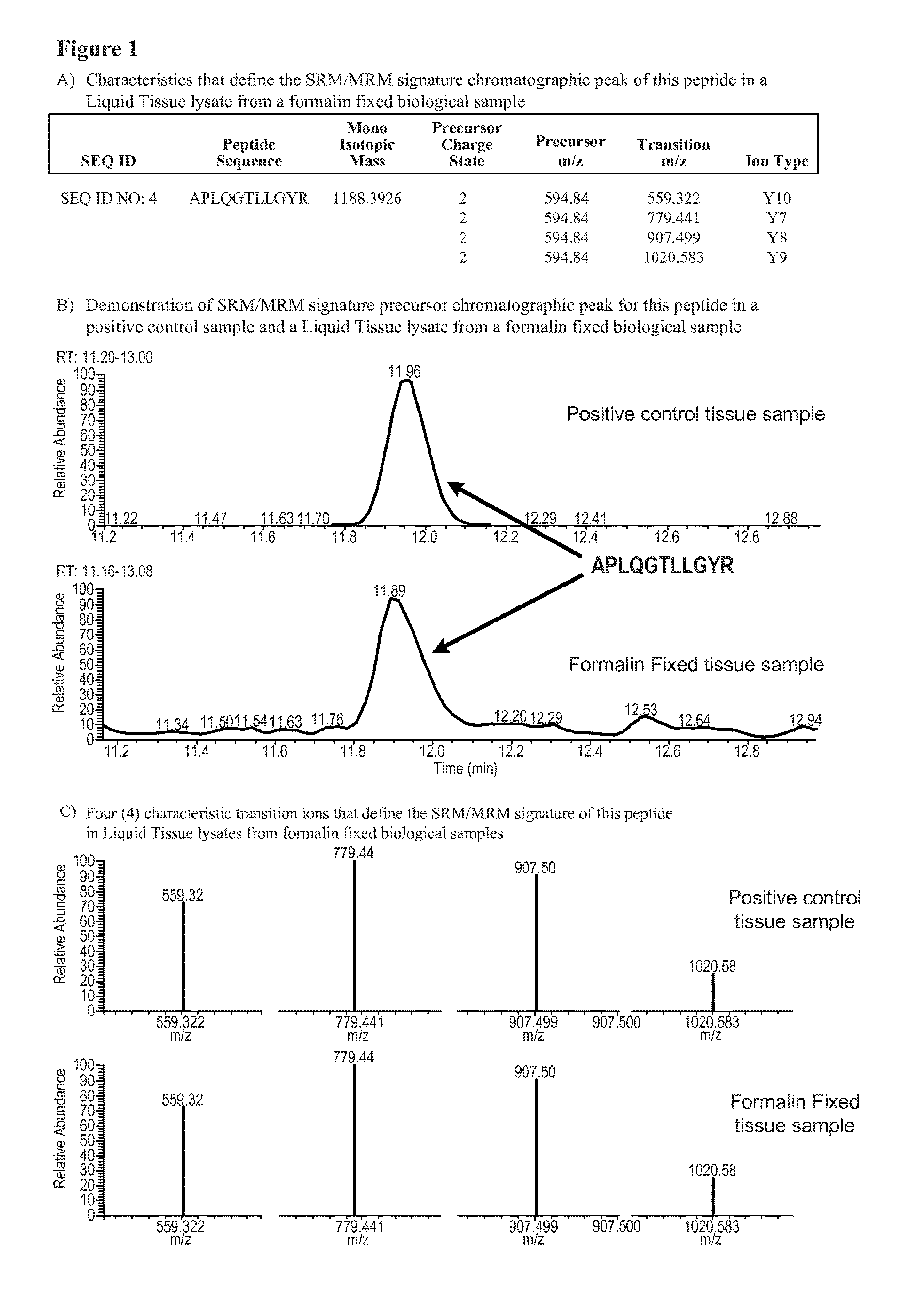
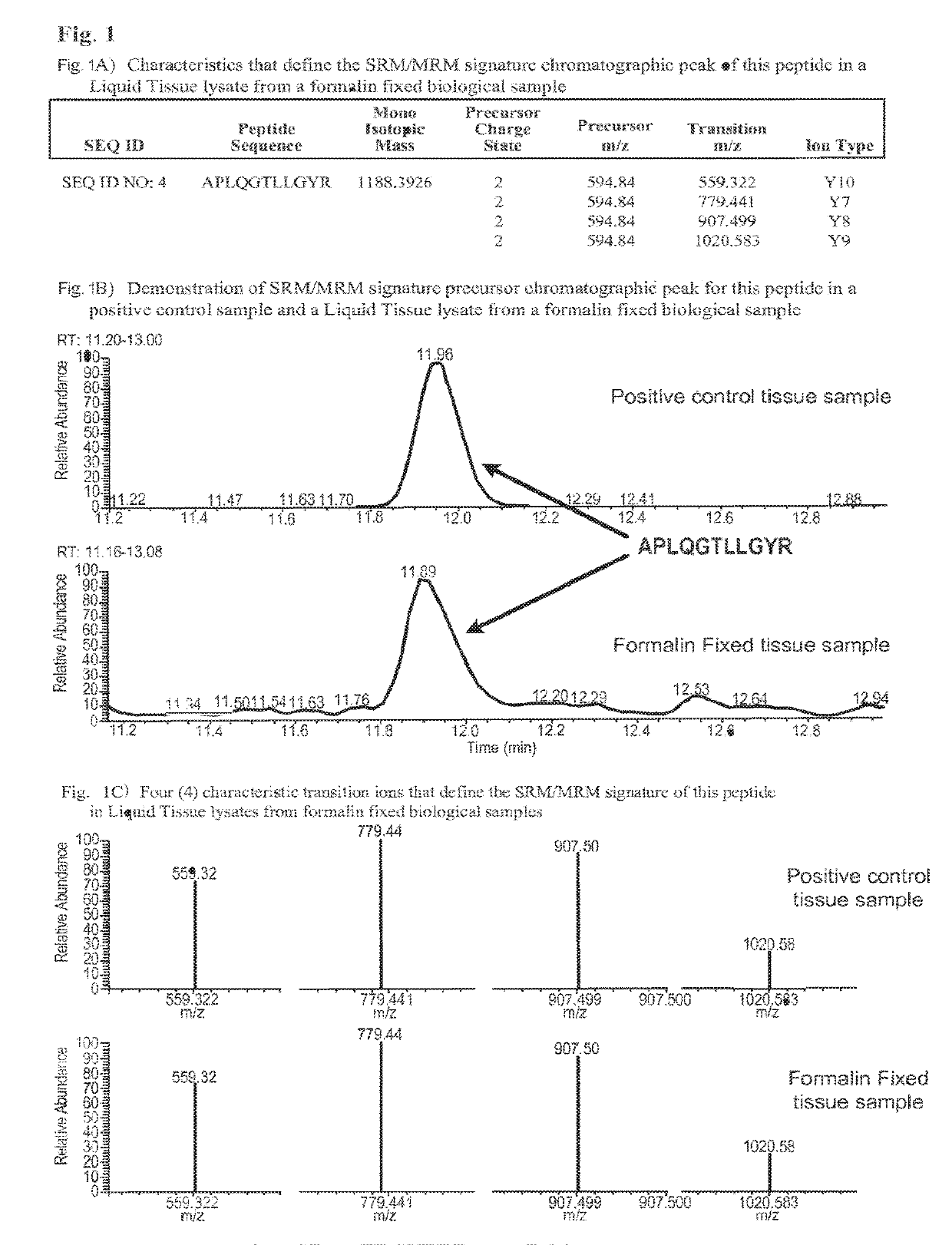
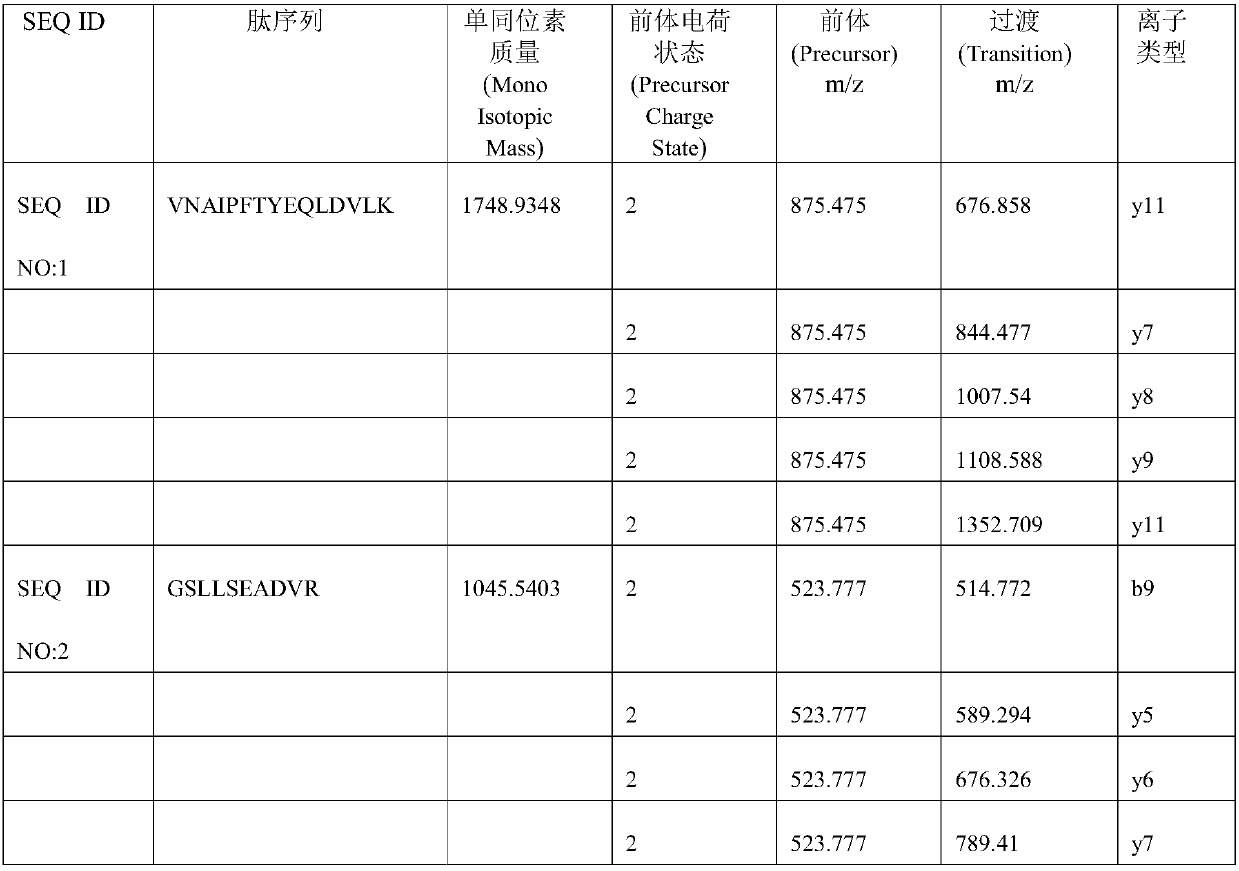
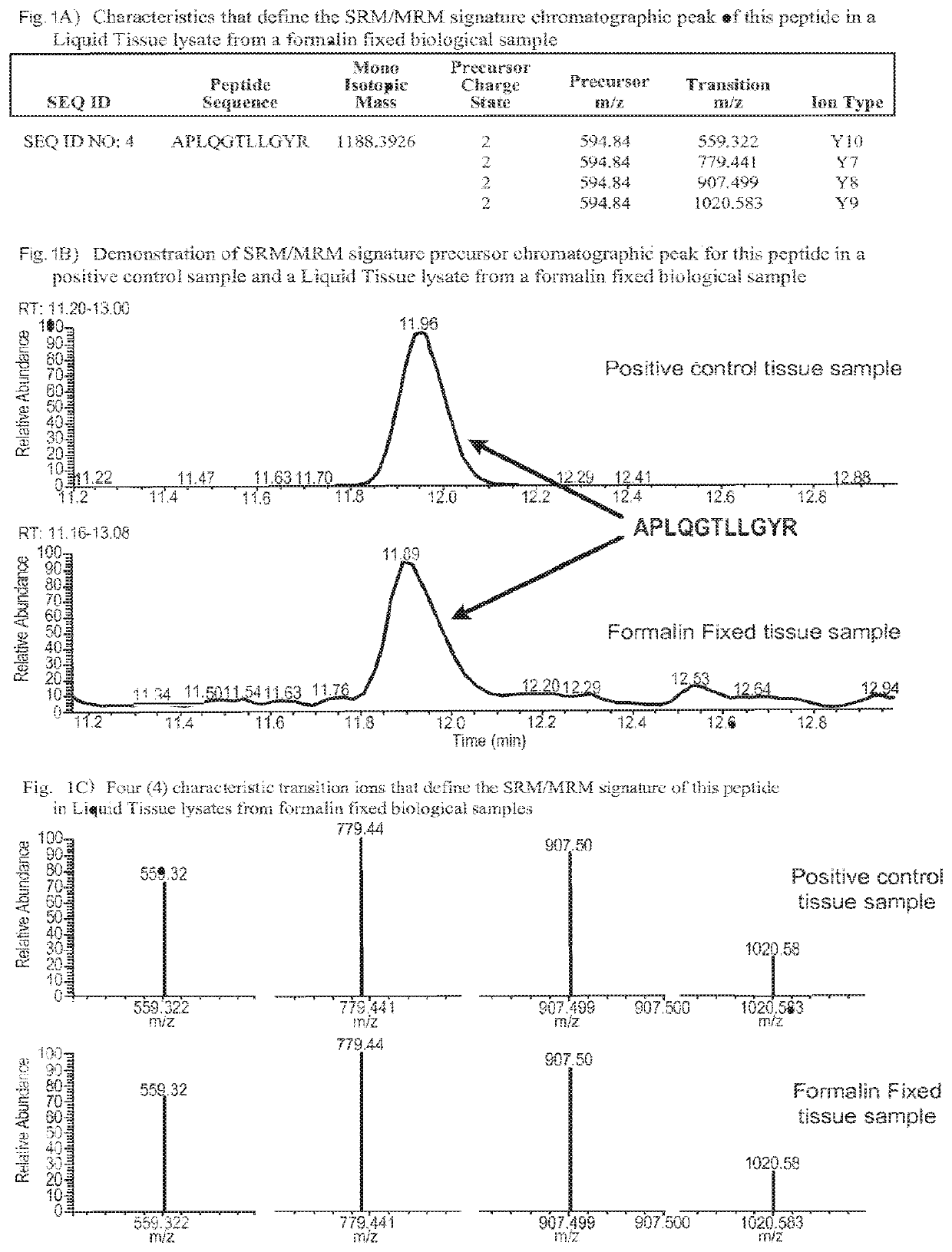
SRM/MRM assay for subtyping lung histology
The current disclosure provides for specific peptides, and derived ionization characteristics of the peptides, from the KRT5, KRT7, NapsinA, TTF1, TP63, and / or MUC1 proteins that are particularly advantageous for quantifying the KRT5, KRT7, NapsinA, TTF1, TP63, and / or MUC1 proteins directly in biological samples that have been fixed in formalin by the method of Selected Reaction Monitoring (SRM) mass spectrometry, or what can also be termed as Multiple Reaction Monitoring (MRM) mass spectrometry. Such biological samples are chemically preserved and fixed wherein said biological sample is selected from tissues and cells treated with formaldehyde containing agents / fixatives including formalin-fixed tissue / cells, formalin-fixed / paraffin embedded (FFPE) tissue / cells, FFPE tissue blocks and cells from those blocks, and tissue culture cells that have been formalin fixed and or paraffin embedded. A protein sample is prepared from said biological sample using the Liquid Tissue™ reagents and protocol and the KRT5, KRT7, NapsinA, TTF1, TP63, and / or MUC1 proteins are quantitated in the Liquid Tissue™ sample by the method of SRM / MRM mass spectrometry by quantitating in the protein sample at least one or more of the peptides described. These peptides can be quantitated if they reside in a modified or an unmodified form. An example of a modified form of a KRT5, KRT7, NapsinA, TTF1, TP63, and MUC1 fragment peptide is phosphorylation of a tyrosine, threonine, serine, and / or other amino acid residues within the peptide sequence.
Owner:EXPRESSION PATHOLOGY
Species detection using mass spectrometry
ActiveUS9373486B2Testing foodSpectrometer combinationsMass Spectrometry-Mass SpectrometryMass spectrometry imaging
Systems and methods are provided for species detection using mass spectrometry. In various embodiments, a multiple reaction monitoring (MRM) experiment is performed on a sample targeting one or more peptide transitions that are unique to one or more species using a tandem mass spectrometer. One or more measured product ion spectra are received from the tandem mass spectrometer using the processor. The one or more measured product ion spectra are compared to product ions of the one or more peptide transitions that are unique to one or more species using the processor. One or more species of the sample are detected by reporting product ions of the one or more peptide transitions that are unique to one or more species that match the one or more measured product ion spectra using the processor.
Owner:DH TECH DEVMENT PTE
Methods for tandem collision-induced dissociation cells
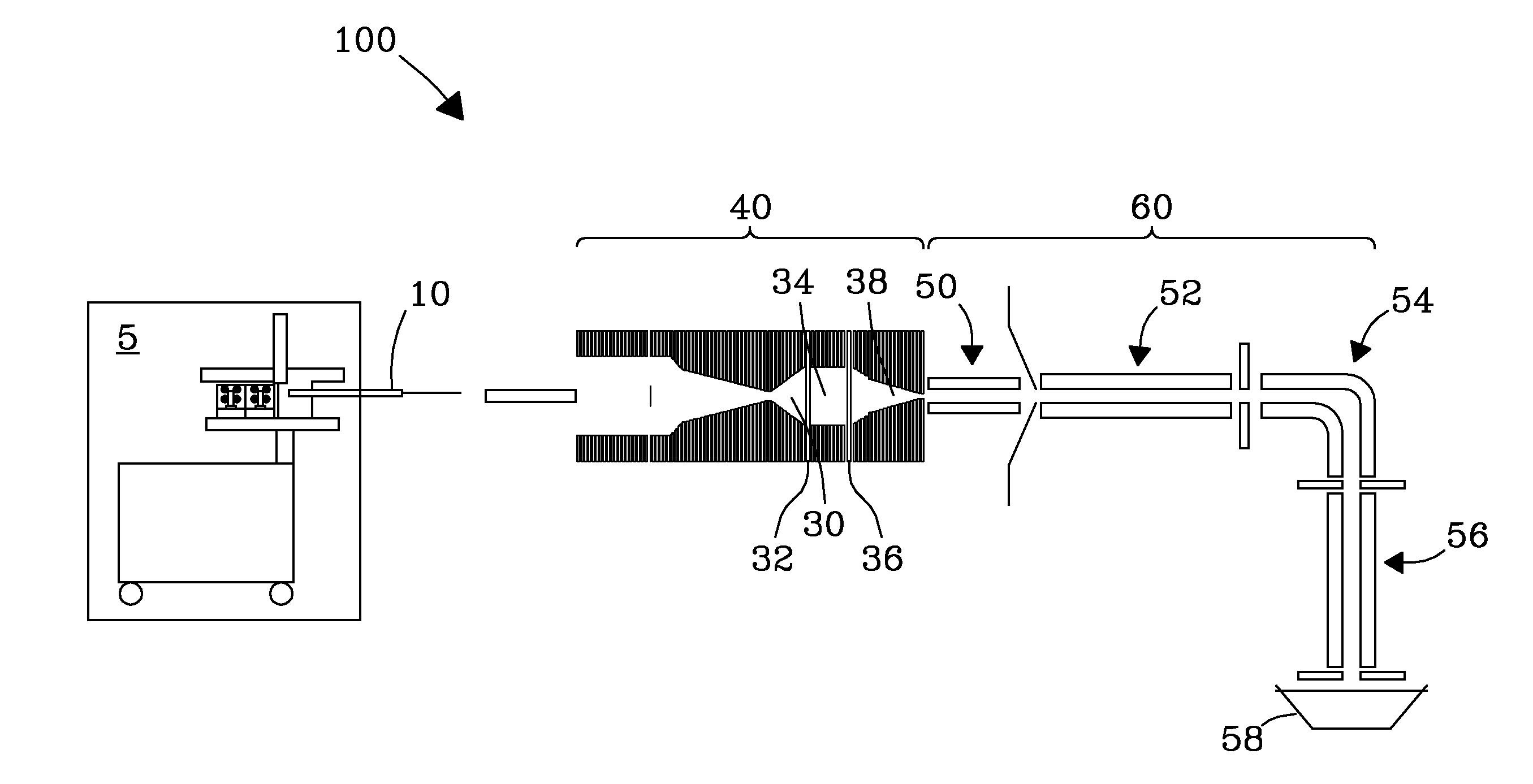
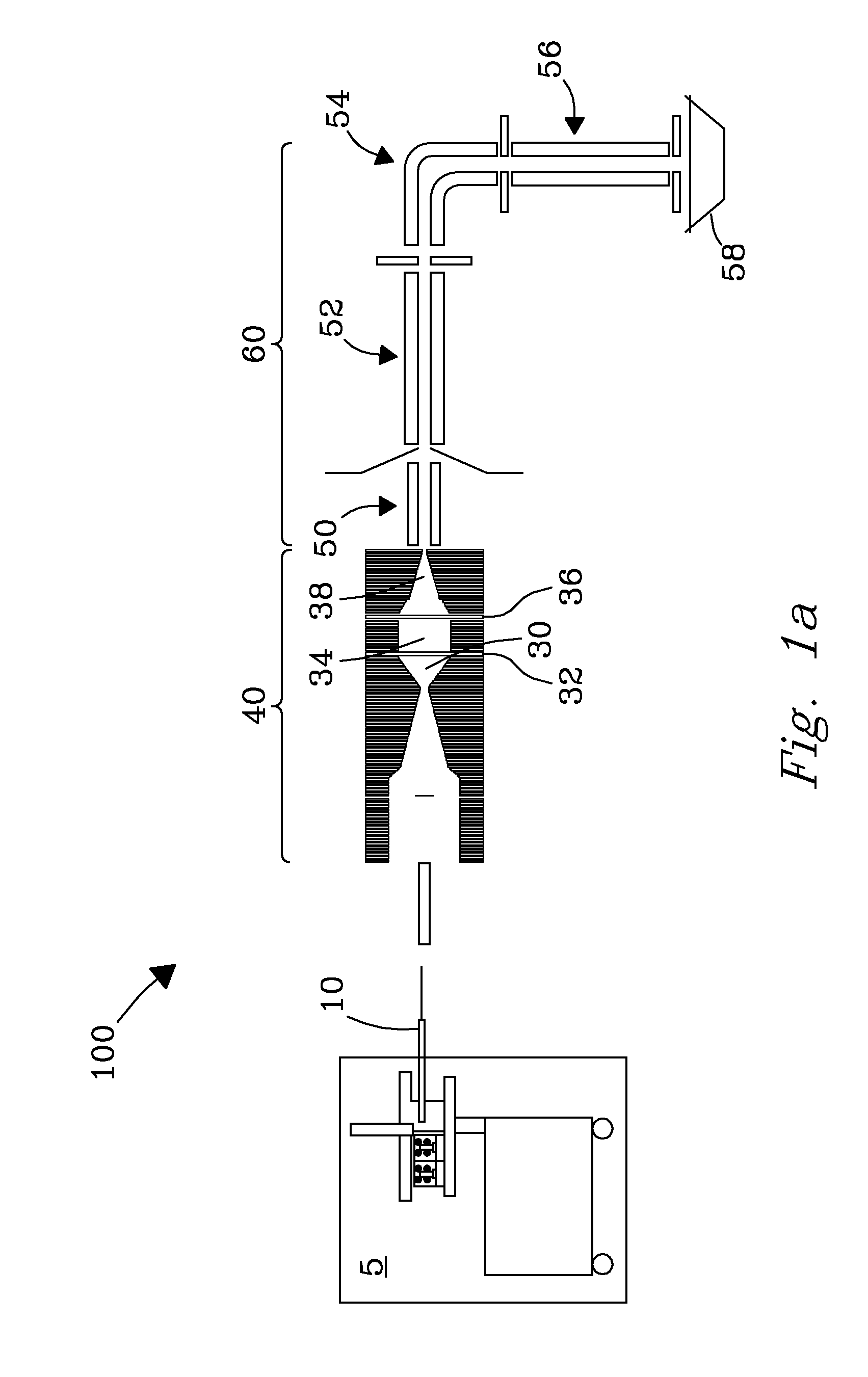
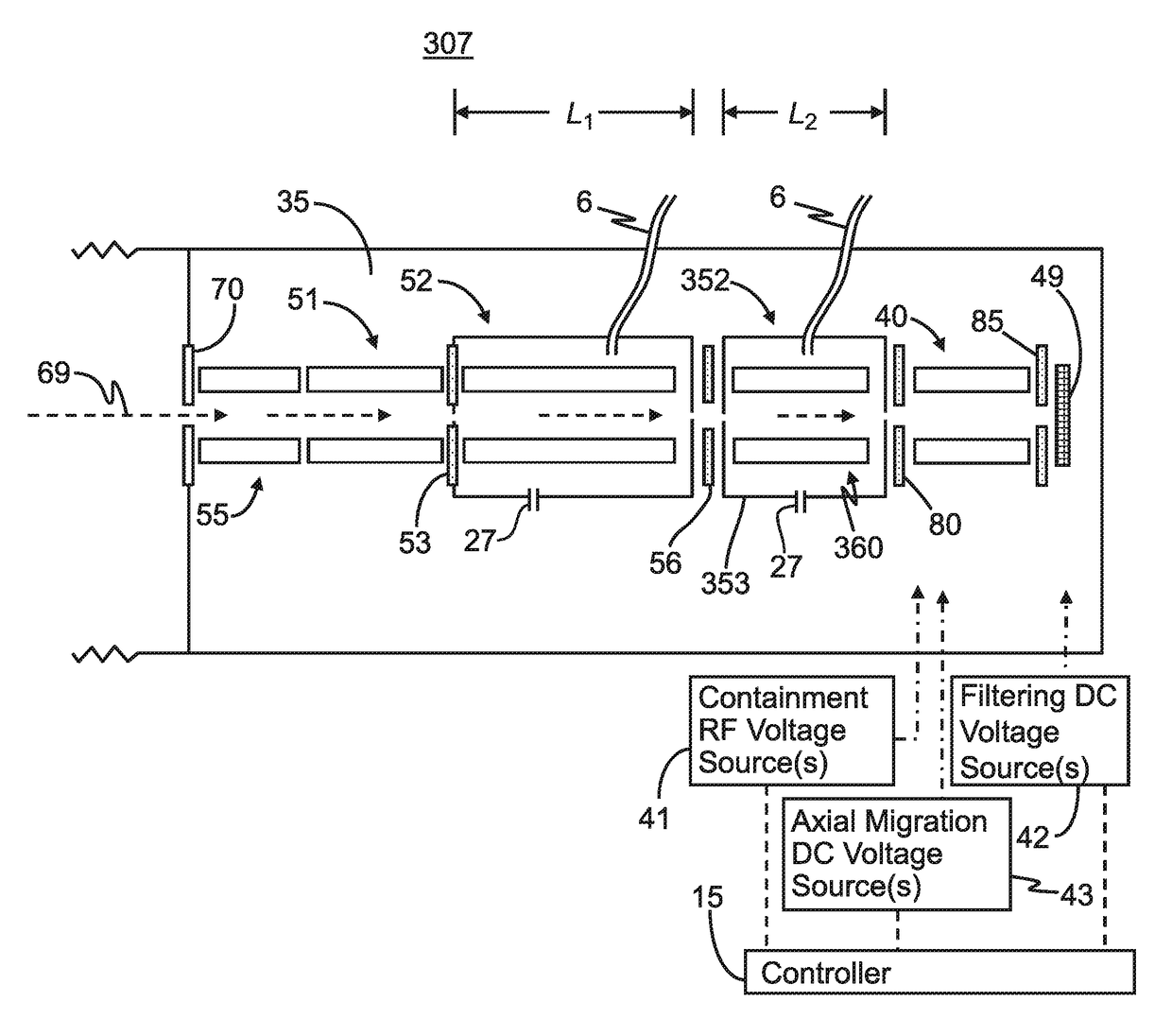
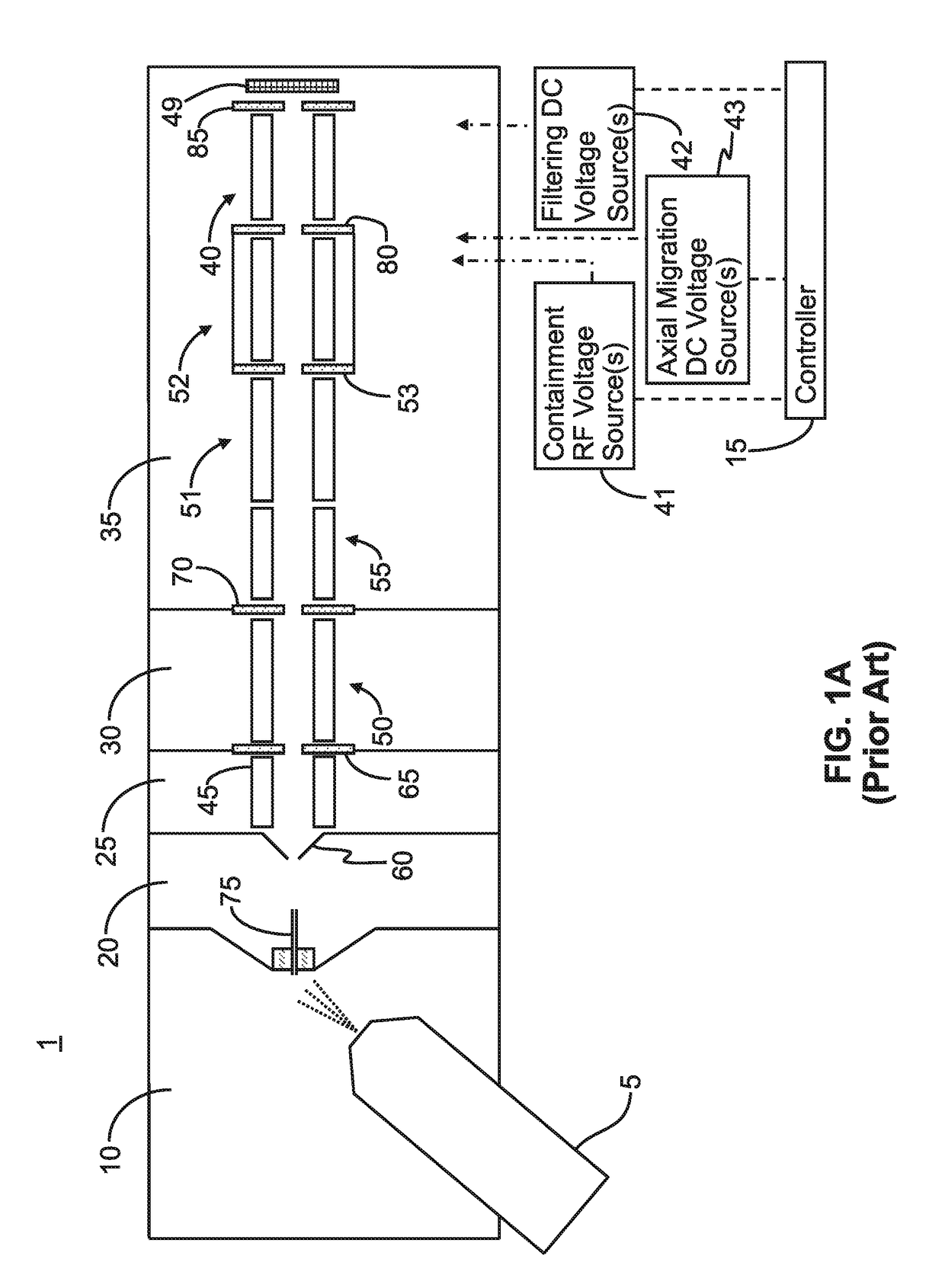
ActiveUS9842730B2Stability-of-path spectrometersSpecific reaction combinationsAnalyteSelected reaction monitoring
A method for operating a mass spectrometer so as to detect or quantify analytes, comprises: (a) identifying a selected-reaction-monitoring (SRM) transition to be used for each respective analyte; (b) determining a time duration required for a fragmentation reaction corresponding to each identified transition to proceed to a threshold percentage of completion; and (c) for each analyte, performing the steps of (i) isolating ions corresponding to a precursor-ion mass-to-charge (m / z) ratio of the respective transition; (ii) fragmenting the respective isolated ions in one of two fragmentation cells or fragmentation cell portions; and (ii) mass analyzing for fragment ions corresponding to a product-ion m / z ratio of the respective transition, wherein, for each analyte, the fragmentation cell or fragmentation cell portion that is used for fragmenting the isolated ions is determined from the time duration determined for the respective analyte.
Owner:THERMO FINNIGAN
SRM Assay for PD-L1
InactiveUS20160313343A1Suppress antitumor activitySuppress presence of PD-LBiological material analysisImmunoglobulins against cell receptors/antigens/surface-determinantsPhosphorylationThreonine
The current disclosure provides for specific peptides, and derived ionization characteristics of the peptides, from the PD-L1 protein that are particularly advantageous for quantifying the PD-L1 protein directly in biological samples that have been fixed in formalin by the method of Selected Reaction Monitoring (SRM) mass spectrometry, or what can also be termed as Multiple Reaction Monitoring (MRM) mass spectrometry. Such biological samples are chemically preserved and fixed wherein said biological sample is selected from tissues and cells treated with formaldehyde containing agents / fixatives including formalin-fixed tissue / cells, formalin-fixed / paraffin embedded (FFPE) tissue / cells, FFPE tissue blocks and cells from those blocks, and tissue culture cells that have been formalin fixed and / or paraffin embedded. PD-L1 peptides having modified or unmodified residues can be quantitated. An example of a modification of a PD-L1 fragment peptide is a phosphorylated tyrosine, threonine, serine, and / or other amino acid residues within the peptide sequence.
Owner:EXPRESSION PATHOLOGY
SRM/MRM assay for the 6-O-methylguanine-DNA methyltransferase (MGMT) protein
ActiveUS10060927B2Improved treatment decision for cancer therapyAccurately determinedPeptide/protein ingredientsMicrobiological testing/measurementMass Spectrometry-Mass Spectrometry6-O-Methylguanine
Owner:EXPRESSION PATHOLOGY
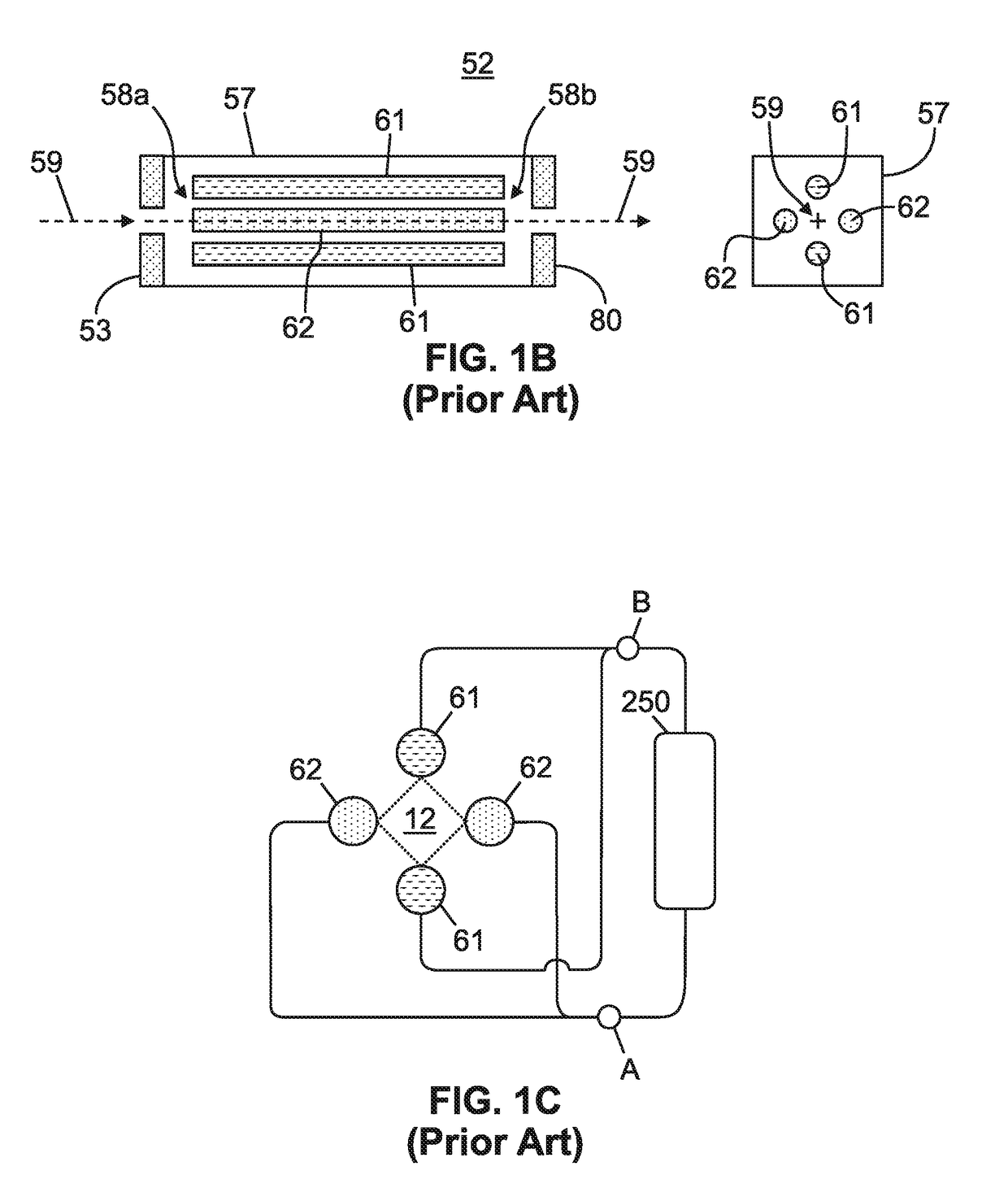
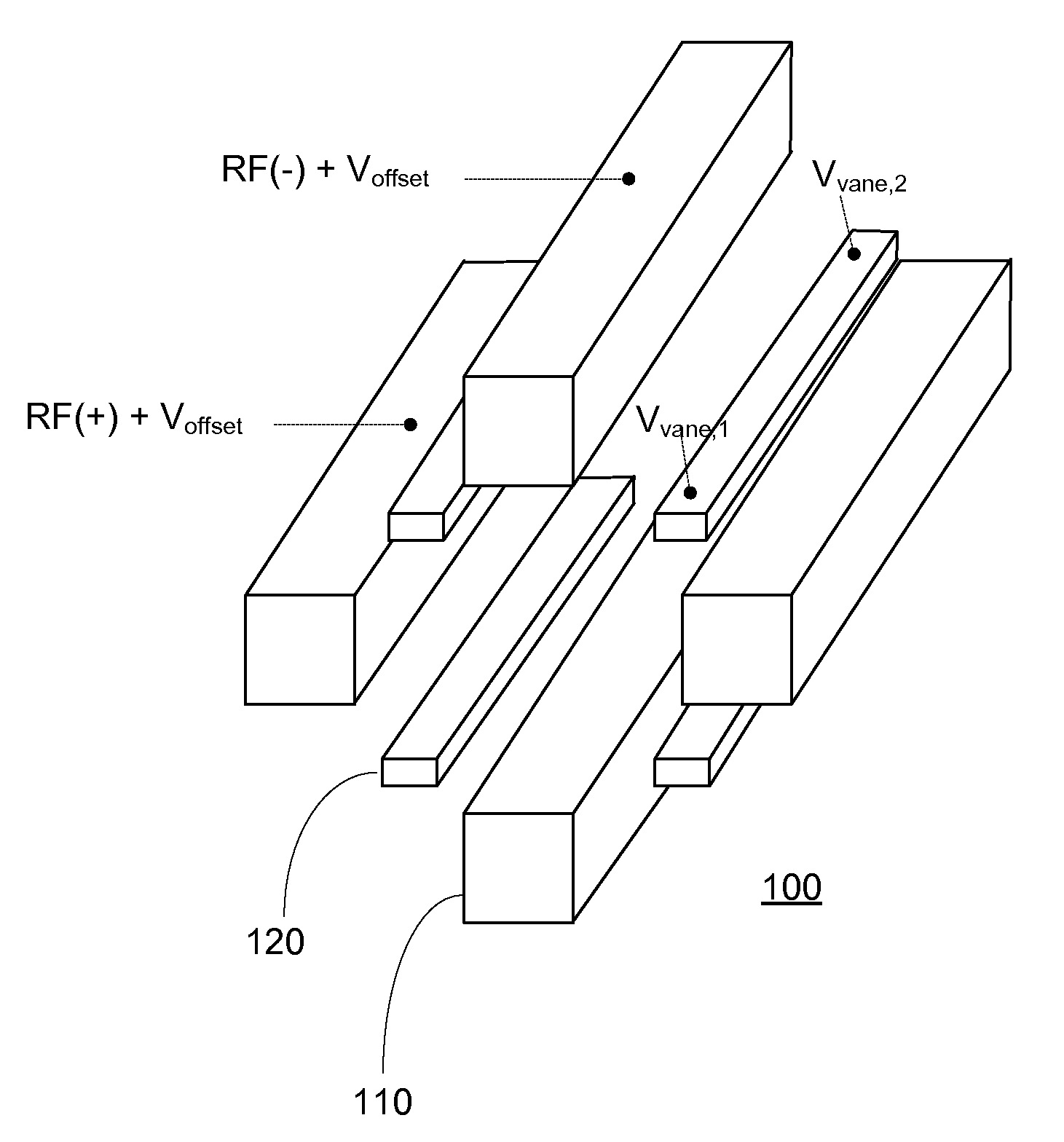
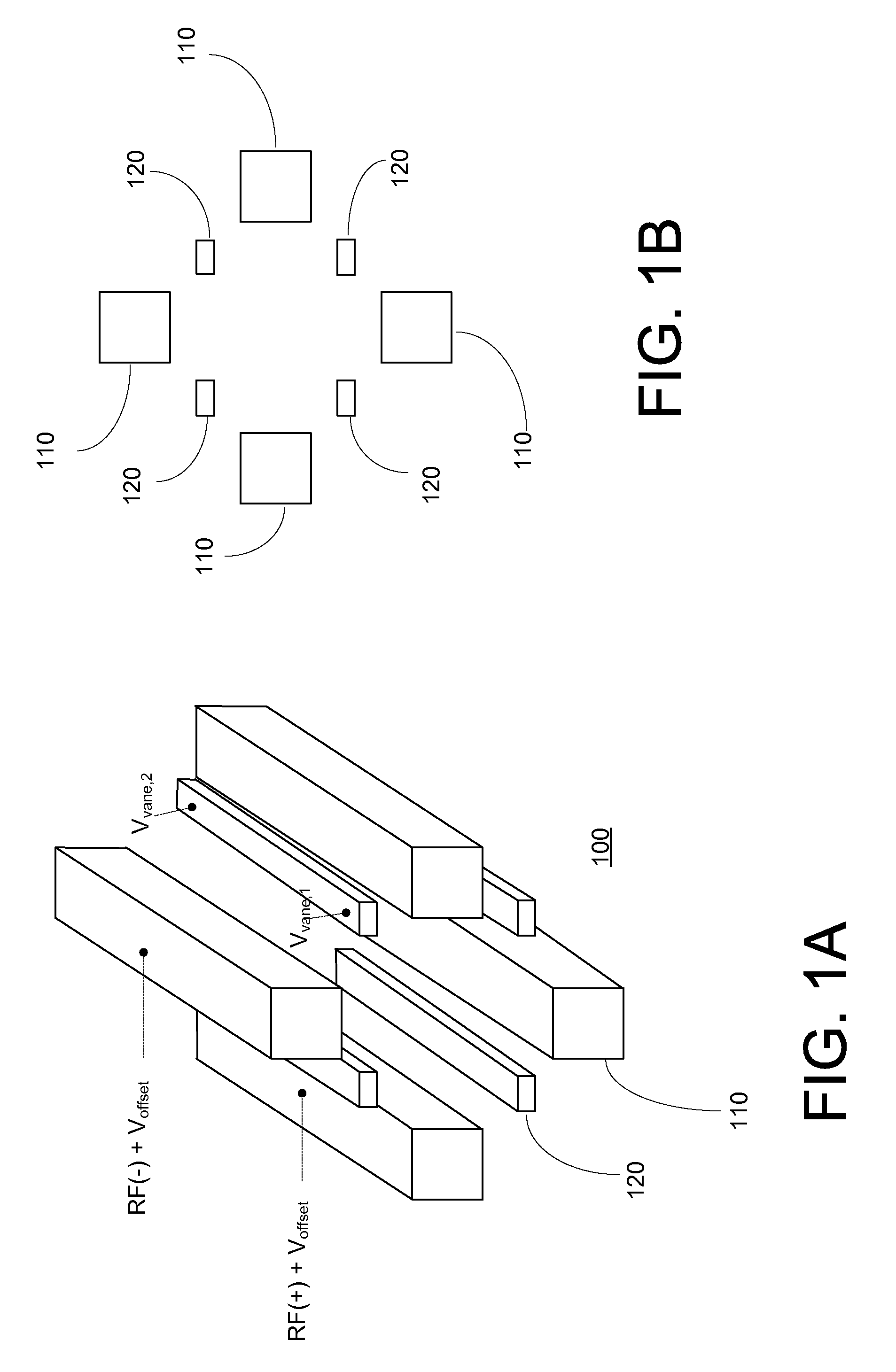
Optimizing drag field voltages in a collision cell for multiple reaction monitoring (MRM) tandem mass spectrometry
ActiveUS9425032B2Stability-of-path spectrometersSpecific reaction combinationsEngineeringSelected reaction monitoring
A collision cell has a plurality of rod electrodes arranged in opposed pairs around an axial centerline and a plurality of drag vanes arranged in the interstitial spaces between the rod electrodes. Operating the collision cell includes, applying a rod offset voltage to the rod electrodes, and varying an offset voltage applied to the drag vanes to identify a vane offset voltage with a maximum intensity for the transition. The method further includes varying a drag field by adjusting the voltages applied to drag vane terminals in opposite directions to identify a drag field value with a cross talk below a cross talk threshold, varying the vane offset voltage by adjusting the voltages applied to the drag vane terminals to maximize the intensity of the transition while preserving the drag field, and operating the collision cell at the vane offset voltage and drag field to monitor the transition.
Owner:THERMO FINNIGAN
Msia-srm assay for biomarker analysis
InactiveUS20160209412A1High binding affinityLarge binding capacityPeptide-nucleic acidsParticle separator tubesMass spectrometric immunoassaySelected reaction monitoring
The present disclosure provides assays, methods and signature peptides for the identification and quantification of biomarkers in a sample. In particular, the present disclosure relates to the development of mass spectrometric immunoassays with selected reaction monitoring mass spectrometry (MSIA-SRM MS) platforms for biomarker analysis. The MSIA-SRM MS platform may specifically be used for discriminating between particular variants of one or more biomarkers. In addition, the MSIA-SRM platform of the present invention may identify and / or quantify a biomarker and / or its variants present at low abundance in a sample.
Owner:NMDX LLC
SRM/MRM Assay for the Fibroblast Growth Factor Receptor 2 (FGFR2) protein
ActiveUS20160334424A1Improve insightsImproved treatment decision for cancer therapyPreparing sample for investigationMass spectrometric analysisFibroblast growth factor receptor 2Mass spectrometry
Methods are provided for quantifying the fibroblast growth factor receptor 2 protein (FGFR2) directly in biological samples that have been fixed in formalin by the method of Selected Reaction Monitoring (SRM) / Multiple Reaction Monitoring (MRM) mass spectrometry. The biological sample may be selected from tissues and cells treated with formaldehyde containing agents / fixatives including formalin-fixed tissue / cells, formalin-fixed / paraffin embedded (FFPE) tissue / cells, FFPE tissue blocks and cells from those blocks, and tissue culture cells. A protein sample is prepared from the biological sample and the FGFR2 protein is quantitated in the sample using the method of SRM / MRM mass spectrometry by quantitating in the protein sample at least one fragment peptide derived from FGFR2.
Owner:EXPRESSION PATHOLOGY
SRM/MRM Assay for the 6-O-methylguanine-DNA methyltransferase (MGMT) protein
ActiveUS20160334408A1Improved treatment decision for cancer therapyAccurately determinedMicrobiological testing/measurementTransferasesMass Spectrometry-Mass Spectrometry6-O-Methylguanine
The current disclosure provides methods for detecting and quantitating the 6-O-methylguanine-DNA methyltransferase protein (MGMT) directly in biological samples that have been fixed in formalin by the method of Selected Reaction Monitoring / Multiple Reaction Monitoring (SRM / MRM) mass spectrometry. Such biological samples are chemically preserved and fixed with formaldehyde containing agents / fixatives and may include formalin-fixed tissue / cells, formalin-fixed / paraffin embedded (FFPE) tissue / cells, FFPE tissue blocks and cells from those blocks, and tissue culture cells that have been formalin fixed and / or paraffin embedded. A protein sample is prepared from the biological sample and the MGMT protein is quantitated in the sample using SRM / MRM mass spectrometry by quantitating one or more fragment peptides.
Owner:EXPRESSION PATHOLOGY
High-resolution time-of-flight mass spectrometer quasi-in-situ reaction monitoring system
PendingCN110361443AFunction increaseAddress reactivityMaterial analysis by electric/magnetic meansChemical reactionMass analyzer
The invention discloses a high-resolution time-of-flight mass spectrometer quasi-in-situ reaction monitoring system, which comprises a high-resolution time-of-flight mass spectrometer, an in-situ reaction device and an automatic flow division dilution transmission device; The in-situ reaction device comprises a reactor, two injection pumps and a heating magnetic stirrer, wherein the two injectionpumps are connected to the reactor through PEEK tubes. The automatic flow division dilution transmission device includes a constant flow pump, a stop valve, a six-way valve and a flow divider, whereinthe stop valve, the constant flow pump and the flow divider are connected with the six-way valve through the PEEK tubes. Reaction liquid to be measured in an in-situ reactor is quickly divided, diluted and transferred to the mass spectrometer under the condition of no contact with air to complete the quasi-in-situ reaction monitoring of the chemical reaction, thereby thoroughly solving the problem that it is difficult to detect chemical intermediates and unstable intermediates under a conventional chemical detection method of the high-resolution time-of-flight mass spectrometer and expandingthe function the high-resolution time-of-flight mass spectrometer in the aspect of reaction mechanism study.
Owner:LANZHOU INST OF CHEM PHYSICS CHINESE ACAD OF SCI
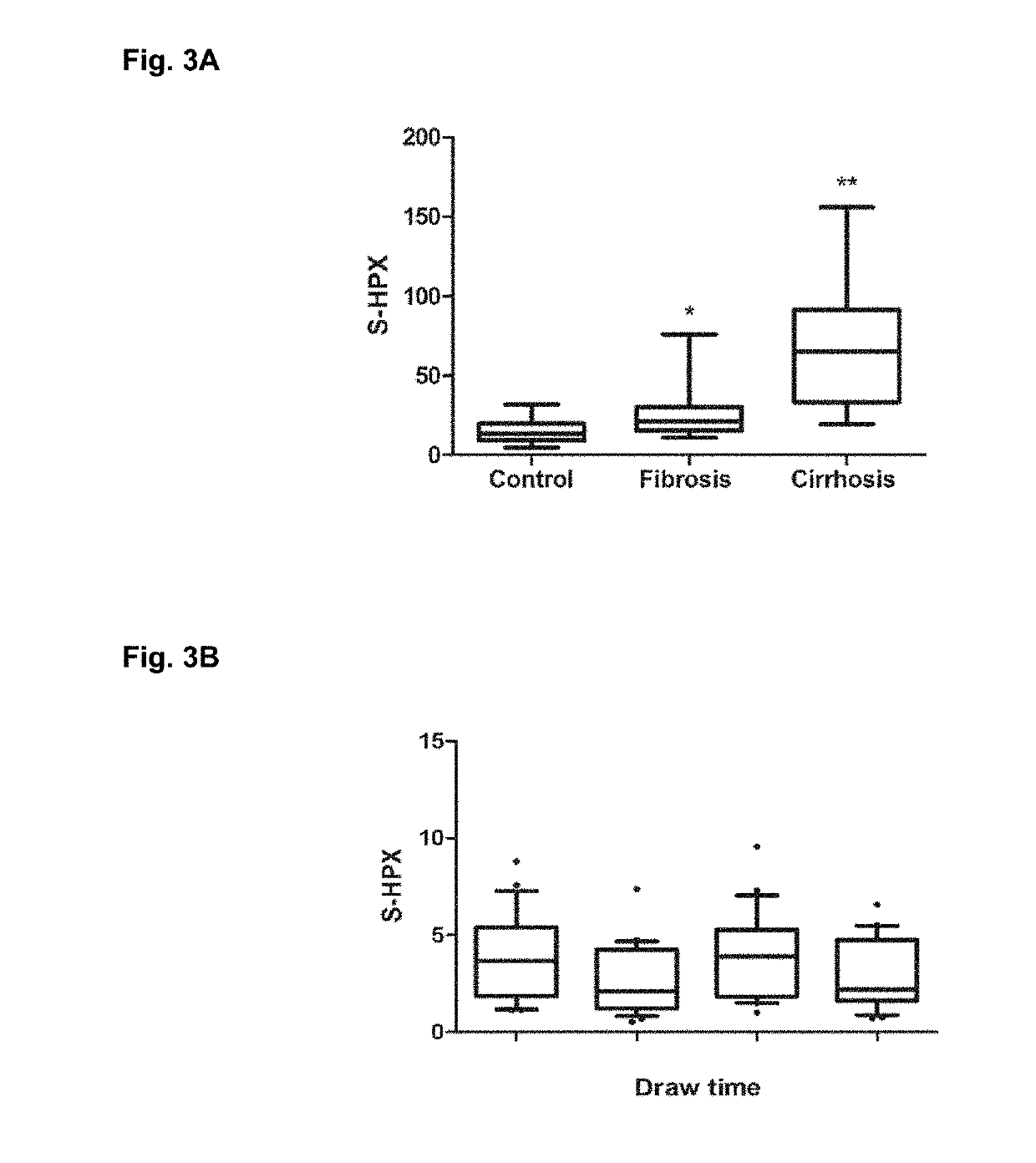
Serologic assay of liver fibrosis
ActiveUS20190242907A1Reduce the amount presentDisease diagnosisBiological testingLiver fibrosisSelected reaction monitoring
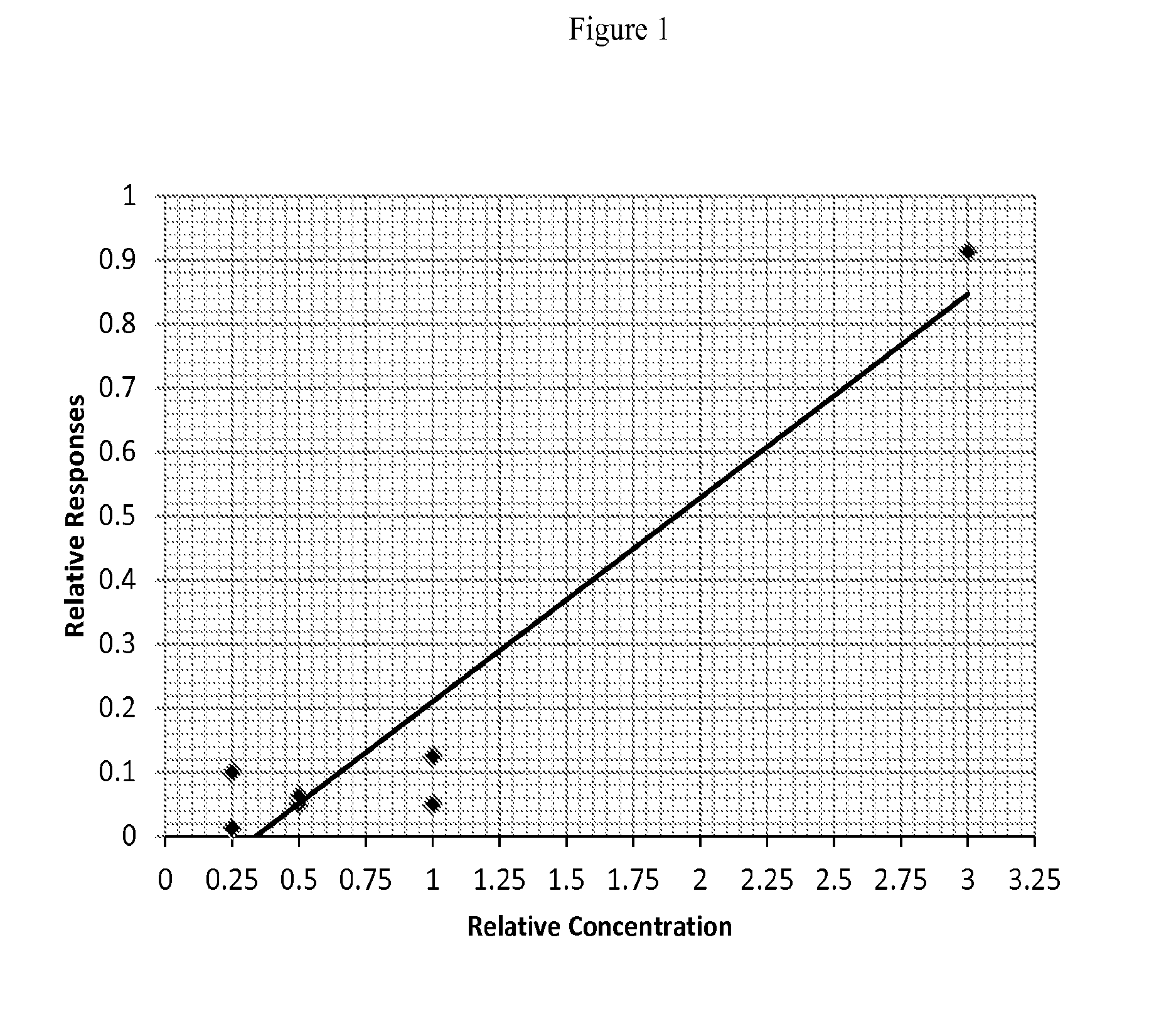
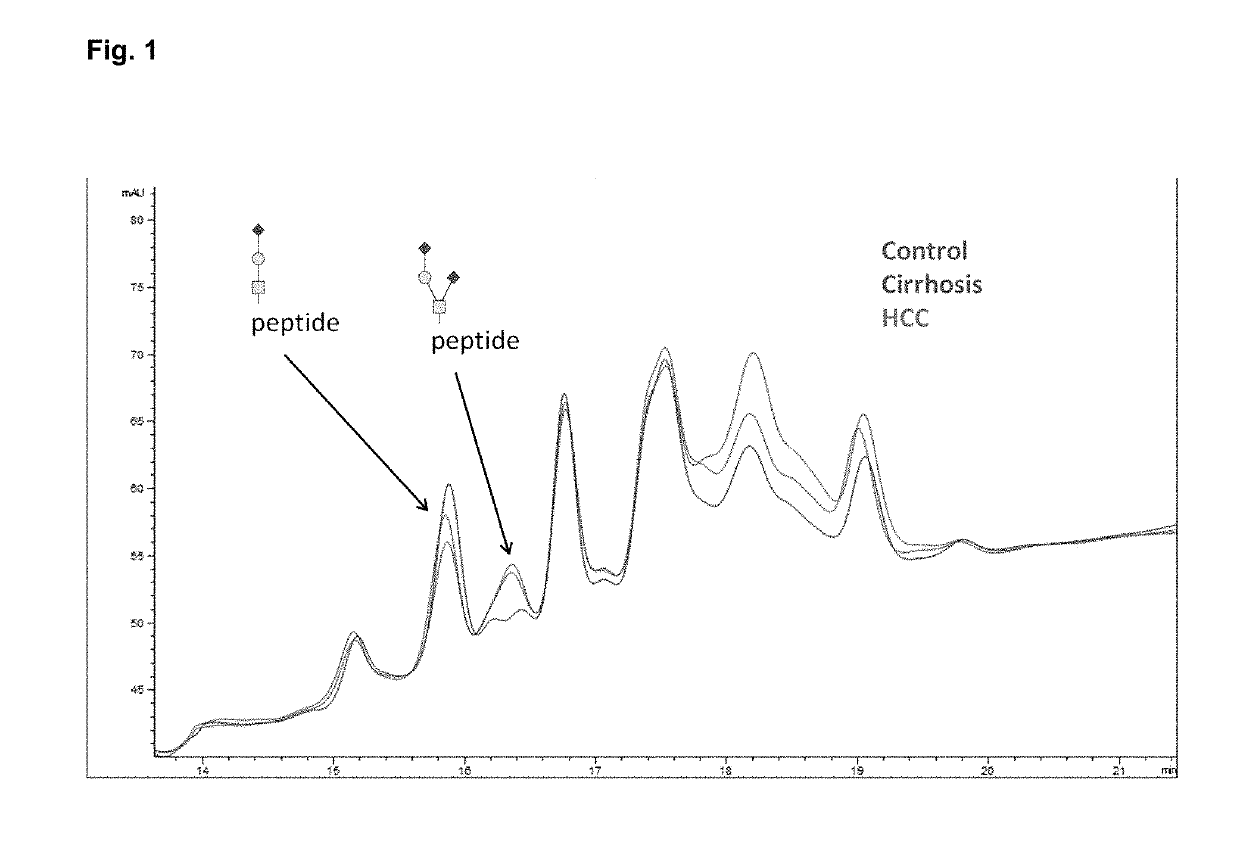
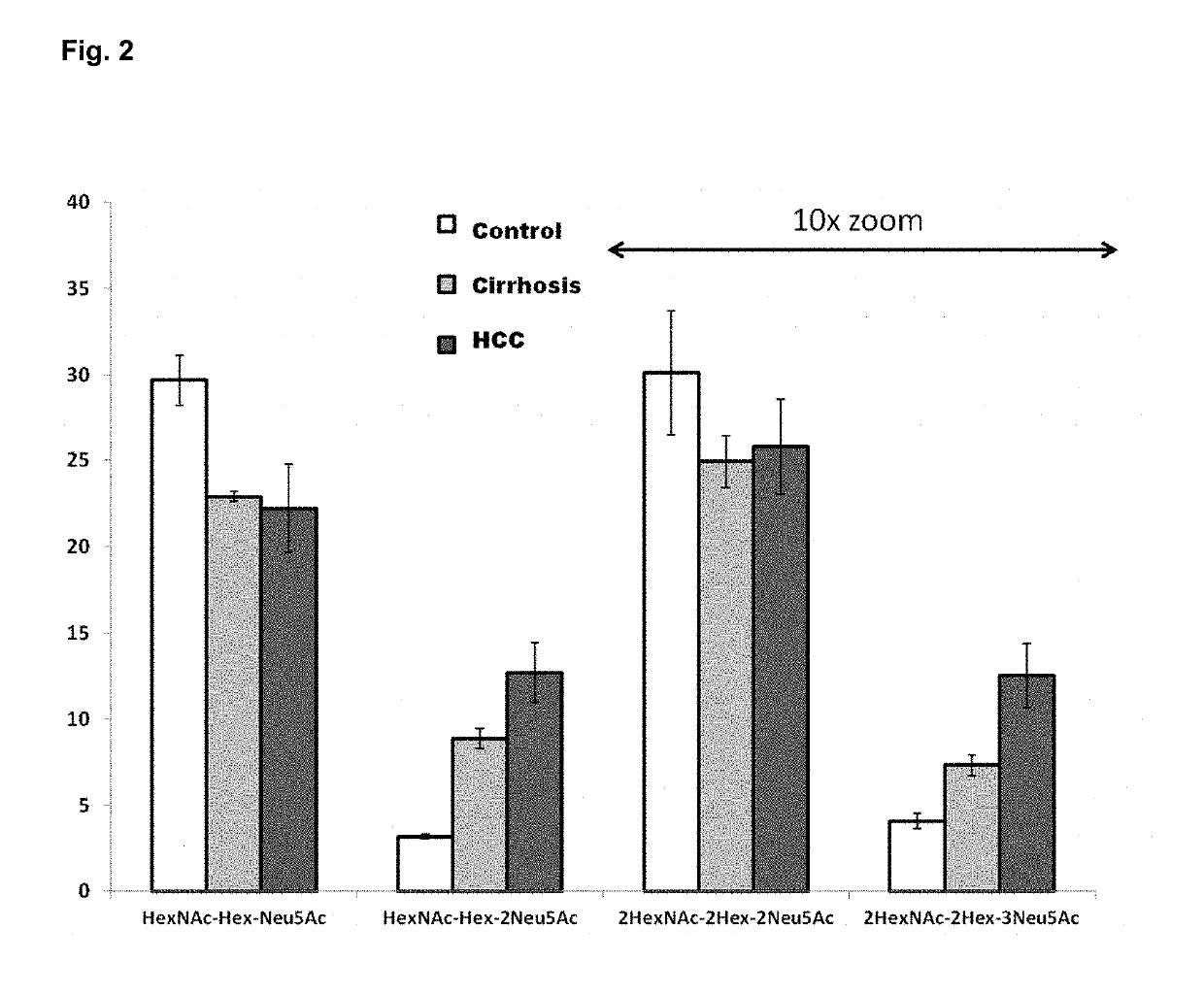
Provided are in vitro serologic methods of assessing the presence of, and assessing the progression of, liver fibrosis in a subject. Also provided are methods of assessing efficacy of an agent for the treatment of liver fibrosis, and methods of treating liver fibrosis. The methods involve quantitatively measuring di-sialylated and mono-sialylated O-glycoforms of a peptide fragment of hemopexin (HPX) in a test serum sample obtained from a test subject, and comparing the measured amounts of di-sialylated and mono-sialylated O-glycoforms of the peptide fragment of hemopexin to a reference amount. In certain embodiments, the measuring is performed using LC-MS / MS-MRM (liquid chromatography / tandem mass spectrometry / multiple reaction monitoring). In certain embodiments, the measuring is performed using LC / MS3 (liquid chromatography with triple-stage mass spectrometric detection).
Owner:GEORGETOWN UNIV
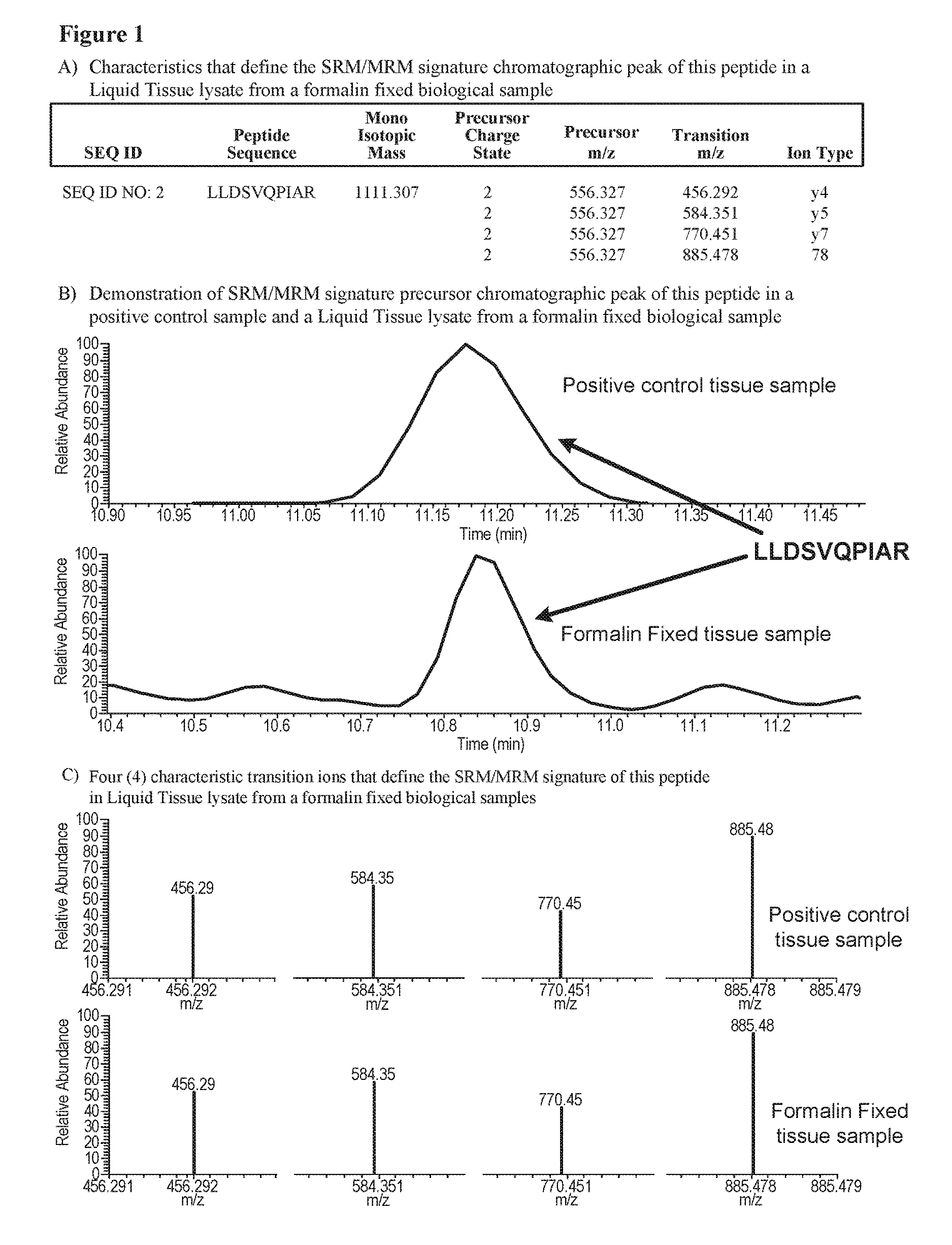
SRM/MRM assay for the androgen receptor (AR) protein
ActiveUS10126307B2Improved treatment decision for cancer therapyMicrobiological testing/measurementDisease diagnosisTissue sampleMass Spectrometry-Mass Spectrometry
Methods are provided for quantifying the Androgen receptor protein (AR) protein directly in biological samples that have been fixed in formalin, using Selected Reaction Monitoring (SRM) / Multiple Reaction Monitoring (MRM) mass spectrometry. The biological samples are chemically preserved and fixed and can be, for example, tissues treated with formaldehyde containing agents / fixatives including formalin-fixed tissue / cells, formalin-fixed / paraffin embedded (FFPE) tissue / cells, FFPE tissue blocks and cells from those blocks. A protein sample is prepared from said biological sample using, for example, the Liquid Tissue protocol and the AR protein is quantitated in the Liquid Tissue sample by the method of SRM / MRM mass spectrometry by quantitating in the protein sample at least one or more of the peptides described.
Owner:EXPRESSION PATHOLOGY
SRM/MRM Assay for the tyrosine-protein kinase receptor UFO (AXL) protein
ActiveUS20170052196A1Improved treatment decision for cancer therapyMass spectrometric analysisMaterial analysis by electric/magnetic meansSelected reaction monitoringSelective reaction
Peptides from the tyrosine-protein kinase receptor UFO protein (AXL) are provided that are particularly advantageous for quantifying the AXL protein directly in biological samples that have been fixed in formalin by the method of Selected Reaction Monitoring (SRM) / Multiple Reaction Monitoring (MRM) mass spectrometry. Such biological samples are chemically preserved and fixed and include formalin-fixed tissue / cells, formalin-fixed / paraffin embedded (FFPE) tissue / cells, FFPE tissue blocks and cells from those blocks. A protein digest is prepared from the biological sample and the AXL protein is quantitated in the Liquid Tissue sample by the method of SRM / MRM mass spectrometry by quantitating in the protein sample at least one or more of the peptides described.
Owner:EXPRESSION PATHOLOGY
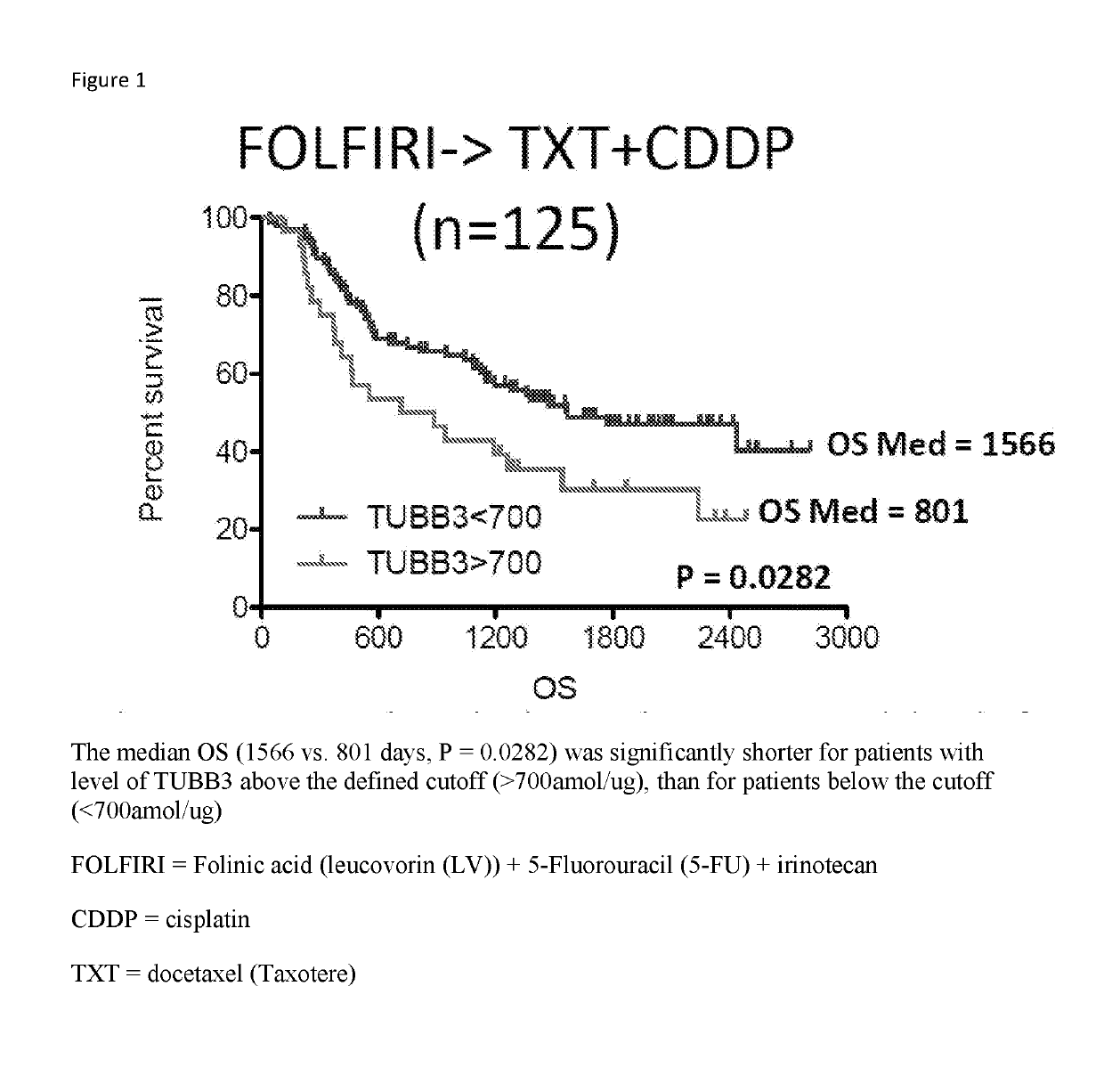
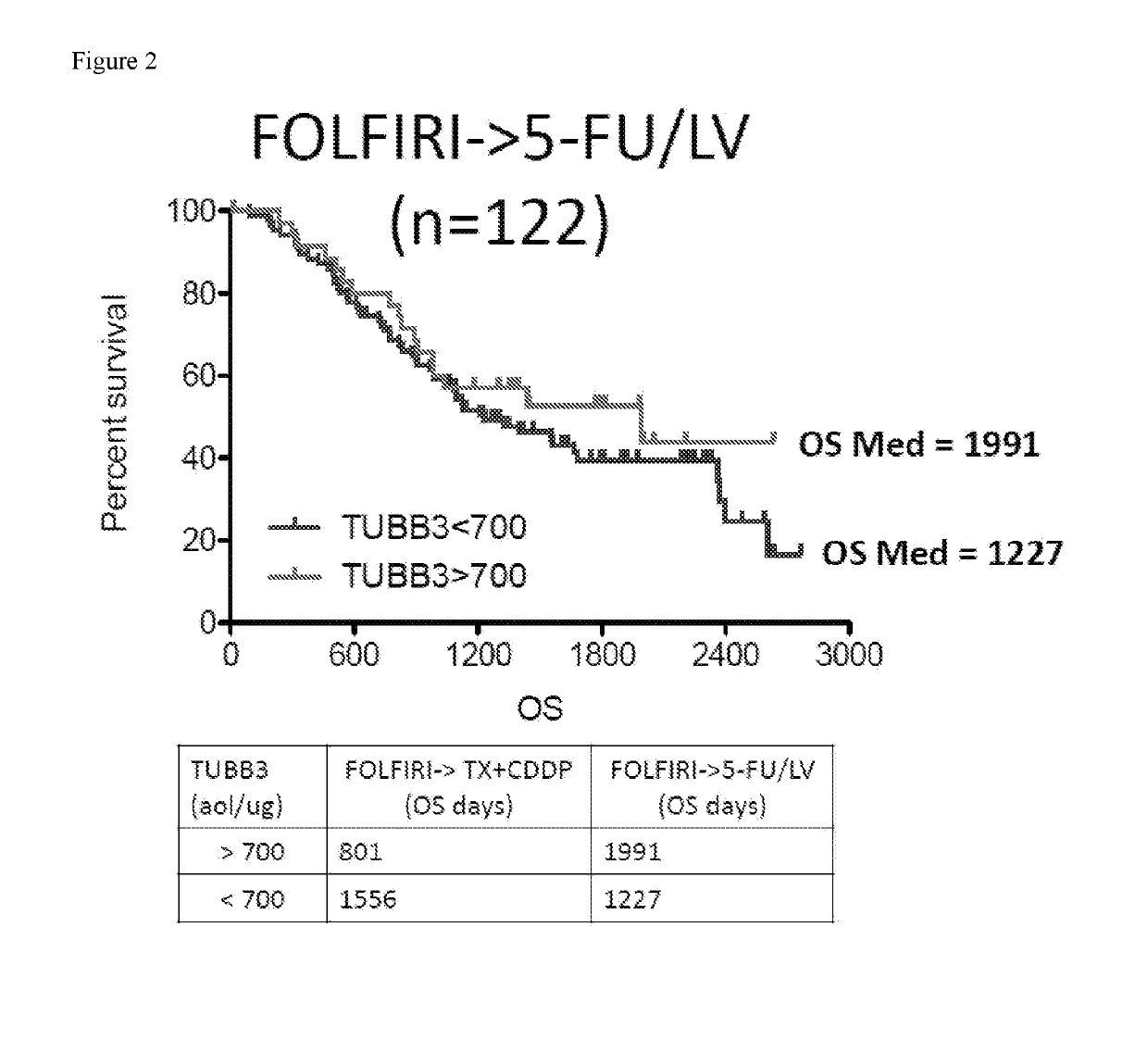
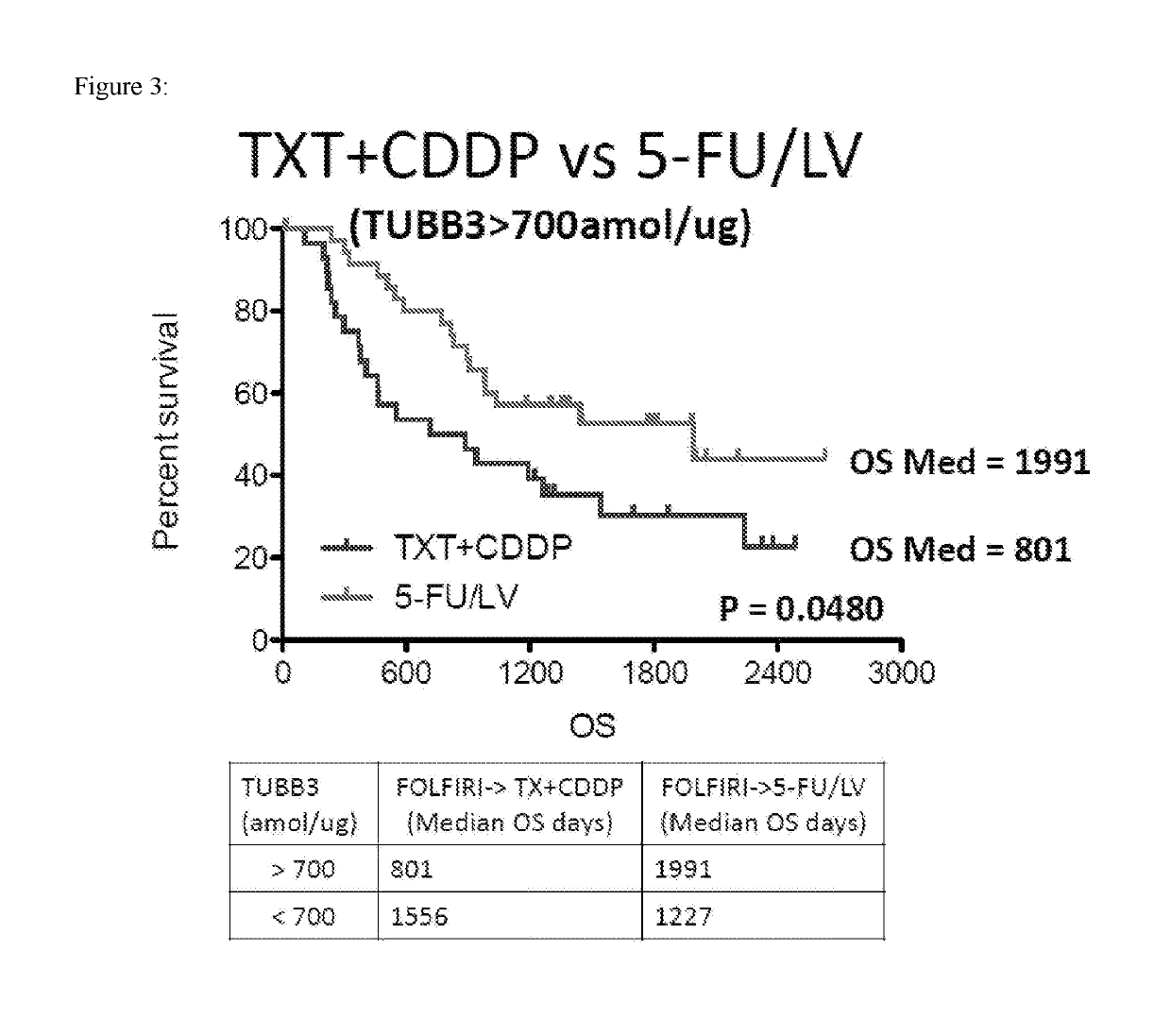
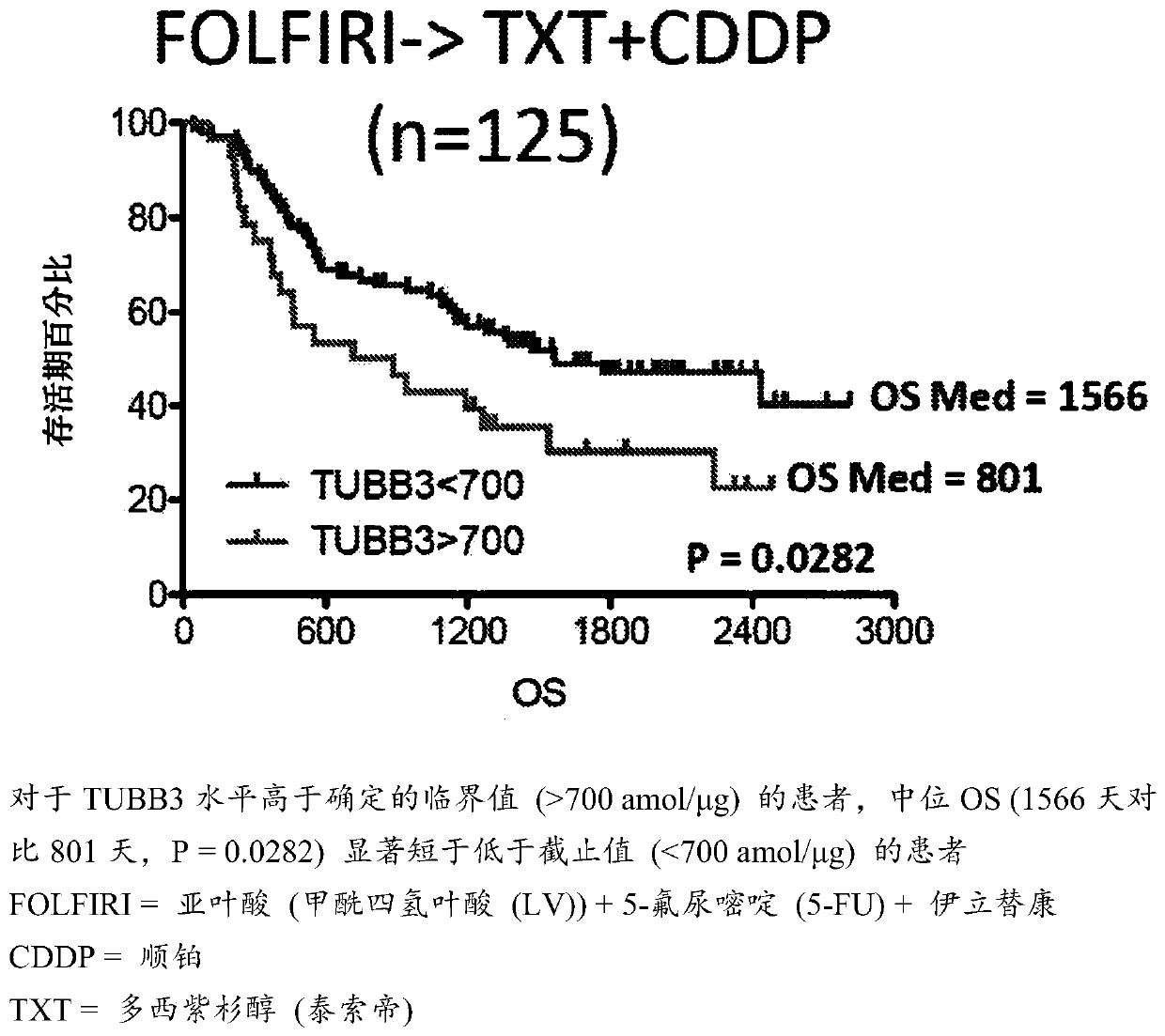
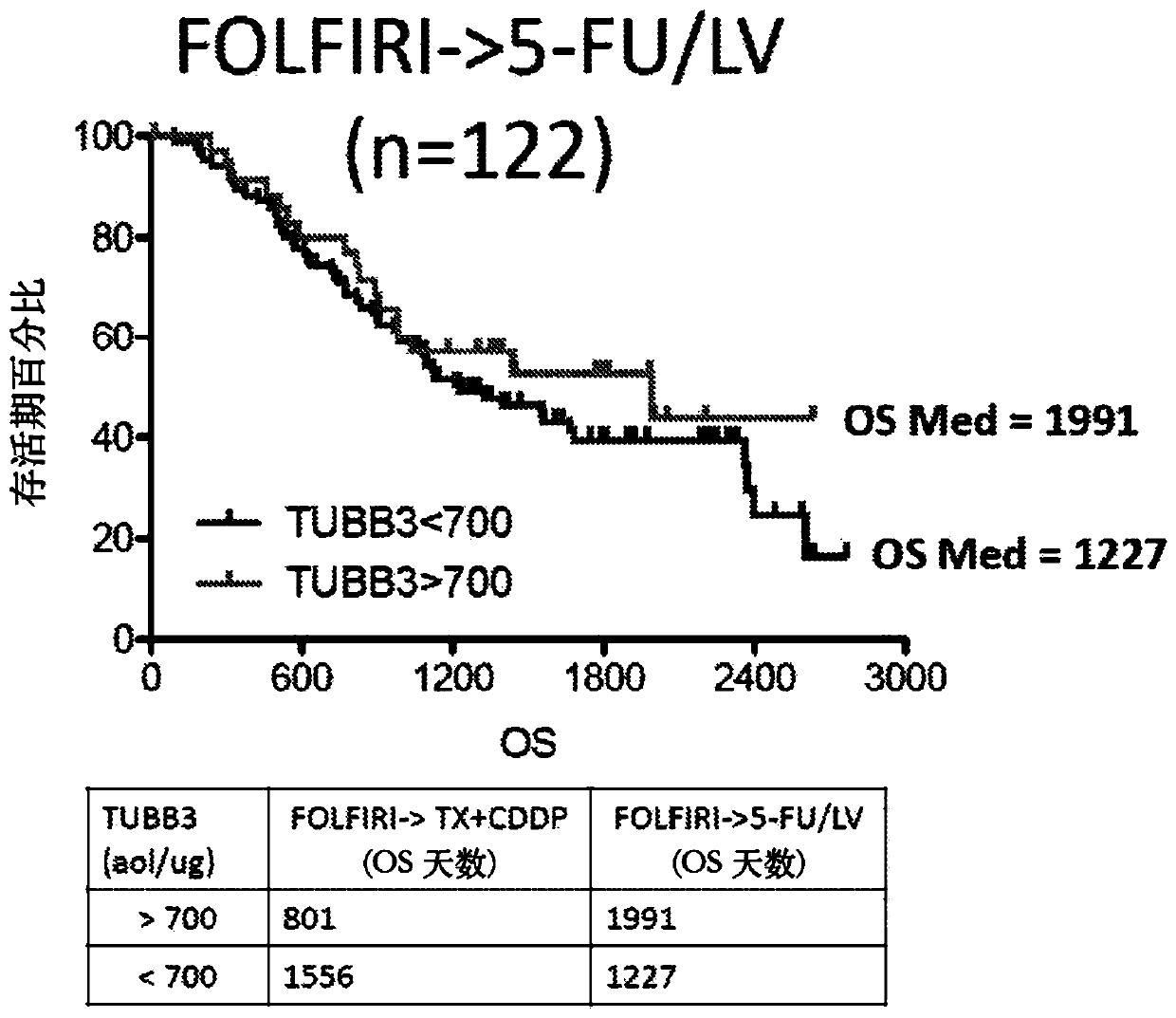
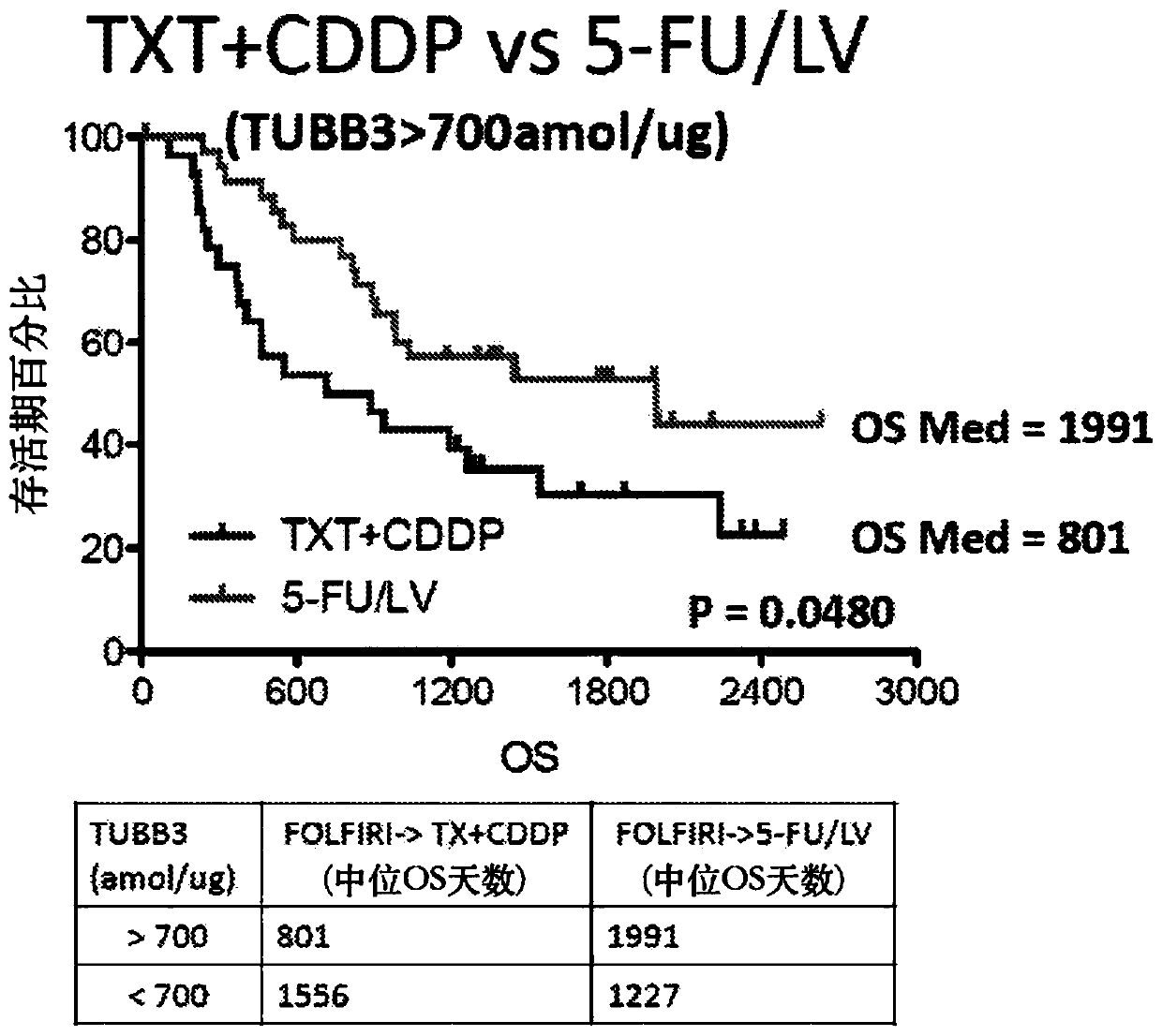
SRM/MRM Assay For The Tubulin Beta-3 Chain (TUBB3) Protein
InactiveUS20190219549A1Improved treatment decision for cancer therapyComponent separationMicrobiological testing/measurementWilms' tumorSelected reaction monitoring
The current disclosure provides for a specific peptide, and derived ionization characteristics of a peptide from the tubulin beta-3 chain protein (TUBB3) that is particularly advantageous for quantifying the TUBB3 protein directly in biological samples that have been fixed in formalin by the method of Selected Reaction Monitoring (SRM) mass spectrometry. A protein sample is prepared from a biological sample using the Liquid Tissue reagents and protocol and the TUBB3 protein is quantitated by SRM / MRM mass spectrometry analysis of the sample, where the specific peptide is quantitated. Methods of treatment are provided in which the measured level of TUBB3 in a patient tumor sample is compared with a reference level and the patient is treated with a taxane-based treatment regimen when the measured TUBB3 level is lower than the reference level. A suitable reference level is, for example, about 700 amol / μg tissue.
Owner:EXPRESSION PATHOLOGY
Process for ultra-sensitive quantification of target analytes in complex biological systems
ActiveUS20160238614A1Easy to quantifyHigh sensitivityComponent separationMass spectrometric analysisChromatographic separationHigh pressure
Owner:BATTELLE MEMORIAL INST
Srm/mrm assay for the tubulin beta-3 chain (TUBB3) protein
InactiveCN109863405AComponent separationMicrobiological testing/measurementSelected reaction monitoringMass spectrography
Owner:EXPRESSION PATHOLOGY
SRM/MRM Assay For The Tyrosine-Protein Kinase Receptor UFO (AXL) Protein
ActiveUS20190265252A1Improved treatment decision for cancer therapyMass spectrometric analysisMaterial analysis by electric/magnetic meansPhosphorylationTyrosine
The current disclosure provides for specific peptides, and derived ionization characteristics of the peptides, from the tyrosine-protein kinase receptor UFO protein (AXL) that are particularly advantageous for quantifying the AXL protein directly in biological samples that have been fixed in formalin by the method of Selected Reaction Monitoring (SRM) mass spectrometry, or what can also be termed as Multiple Reaction Monitoring (MRM) mass spectrometry. Such biological samples are chemically preserved and fixed wherein said biological sample is selected from tissues and cells treated with formaldehyde containing agents / fixatives including formalin-fixed tissue / cells, formalin-fixed / paraffin embedded (FFPE) tissue / cells, FFPE tissue blocks and cells from those blocks, and tissue culture cells that have been formalin fixed and or paraffin embedded. A protein sample is prepared from said biological sample using the Liquid Tissue reagents and protocol and the AXL protein is quantitated in the Liquid Tissue sample by the method of SRM / MRM mass spectrometry by quantitating in the protein sample at least one or more of the peptides described. These peptides can be quantitated if they reside in a modified or an unmodified form. An example of a modified form of an AXL peptide is phosphorylation of a tyrosine, threonine, serine, and / or other amino acid residues within the peptide sequence.
Owner:EXPRESSION PATHOLOGY
SRM/MRM assay for the mesothelin (MSLN) protein
ActiveCN107850605ADisease diagnosisBiological testingMass Spectrometry-Mass SpectrometrySelected reaction monitoring
The current disclosure provides methods for quantifying the MSLN protein directly in biological samples, including samples that have been fixed in formalin by Selected Reaction Monitoring / Multiple Reaction Monitoring (SRM / MRM) mass spectrometry. Samples that can be assayed include tissues and cells treated with formaldehyde containing agents / fixatives including formalin-fixed tissue / cells, formalin-fixed / paraffin embedded (FFPE) tissue / cells, FFPE tissue blocks and cells from those blocks, and tissue culture cells that have been formalin fixed and or paraffin embedded. Methods to prepare a protein sample from such a biological sample are provided and MSLN protein is quantitated by SRM / MRM mass spectrometry by quantitating in the protein sample at least one or more of the peptides described.
Owner:EXPRESSION PATHOLOGY
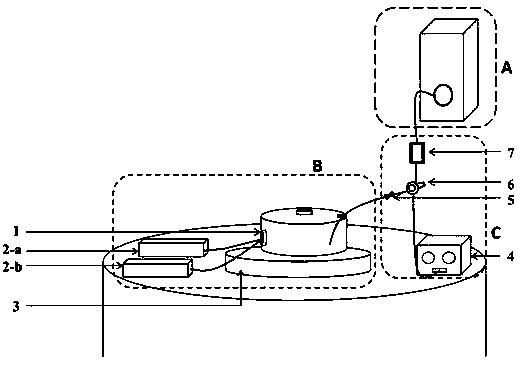
Detection device of immune multiple-reaction mass spectrum
InactiveCN104101642APreparing sample for investigationMaterial analysis by electric/magnetic meansDiseaseOperation mode
The invention discloses a detection device of immune multiple-reaction mass spectrum. The device comprises a multiple-reaction mass spectrum detection part and a protein biomarker immunization enrichment part. Multiple-reaction mass-spectrometric technique is to use triple quadrupole instrument, and an operation mode of is called multiple-reaction monitoring (MRM). Combined with isotope labeled polypeptide reference substance and specific ion transition, the multiple-reaction mass detection has the characteristic of high sensitivity detection. Combined with antibody immunization enrichment technology, low concentration biomarkers in biological fluids (such as blood and saliva) can be quantified according to the standard, so as to provide practical basis for early diagnosis of disease.
Owner:江苏金太生命科技有限公司
SRM/MRM assay for the tyrosine-protein kinase receptor UFO(AXL) protein
ActiveUS10718780B2Improved treatment decision for cancer therapyMass spectrometric analysisMaterial analysis by electric/magnetic meansPhosphorylationSelected reaction monitoring
The current disclosure provides for specific peptides, and derived ionization characteristics of the peptides, from the tyrosine-protein kinase receptor UFO protein (AXL) that are particularly advantageous for quantifying the AXL protein directly in biological samples that have been fixed in formalin by the method of Selected Reaction Monitoring (SRM) mass spectrometry, or what can also be termed as Multiple Reaction Monitoring (MRM) mass spectrometry. Such biological samples are chemically preserved and fixed wherein said biological sample is selected from tissues and cells treated with formaldehyde containing agents / fixatives including formalin-fixed tissue / cells, formalin-fixed / paraffin embedded (FFPE) tissue / cells, FFPE tissue blocks and cells from those blocks, and tissue culture cells that have been formalin fixed and or paraffin embedded. A protein sample is prepared from said biological sample using the Liquid Tissue reagents and protocol and the AXL protein is quantitated in the Liquid Tissue sample by the method of SRM / MRM mass spectrometry by quantitating in the protein sample at least one or more of the peptides described. These peptides can be quantitated if they reside in a modified or an unmodified form. An example of a modified form of an AXL peptide is phosphorylation of a tyrosine, threonine, serine, and / or other amino acid residues within the peptide sequence.
Owner:EXPRESSION PATHOLOGY
SRM/MRM assay for the fibroblast growth factor receptor 2 (FGFR2) protein
ActiveUS10620223B2Improved treatment decision for cancer therapyAccurately determinedPreparing sample for investigationMass spectrometric analysisFibroblast growth factor receptor 2Histiocyte
Owner:EXPRESSION PATHOLOGY
SRM/MRM Assay for the Androgen Receptor (AR) Protein
ActiveUS20170052197A1Improve insightsImproved treatment decision for cancer therapyMicrobiological testing/measurementDisease diagnosisTissue sampleMass Spectrometry-Mass Spectrometry
Methods are provided for quantifying the Androgen receptor protein (AR) protein directly in biological samples that have been fixed in formalin, using Selected Reaction Monitoring (SRM) / Multiple Reaction Monitoring (MRM) mass spectrometry. The biological samples are chemically preserved and fixed and can be, for example, tissues treated with formaldehyde containing agents / fixatives including formalin-fixed tissue / cells, formalin-fixed / paraffin embedded (FFPE) tissue / cells, FFPE tissue blocks and cells from those blocks. A protein sample is prepared from said biological sample using, for example, the Liquid Tissue protocol and the AR protein is quantitated in the Liquid Tissue sample by the method of SRM / MRM mass spectrometry by quantitating in the protein sample at least one or more of the peptides described.
Owner:EXPRESSION PATHOLOGY
SRM/MRM assay for the mesothelin (MSLN) protein
ActiveUS10078084B2Improved treatment decision for cancer therapyAccurately determinedPeptide/protein ingredientsDisease diagnosisMass Spectrometry-Mass SpectrometryProtein methods
Methods are provided for detecting and quantifying the Mesothelin protein (MSLN) in biological samples that have been fixed in formalin using Selected Reaction Monitoring (SRM) / Multiple Reaction Monitoring (MRM) mass spectrometry. The biological sample may be, for example, tissues and cells treated with formaldehyde containing agents / fixatives including formalin-fixed tissue / cells, formalin-fixed / paraffin embedded (FFPE) tissue / cells, FFPE tissue blocks and cells from those blocks, and tissue culture cells that have been formalin fixed and or paraffin embedded. A protein sample is prepared from the biological sample and the MSLN protein is quantitated by SRM / MRM mass spectrometry by quantitating one or more MSLN fragment peptides in the protein sample.
Owner:EXPRESSION PATHOLOGY
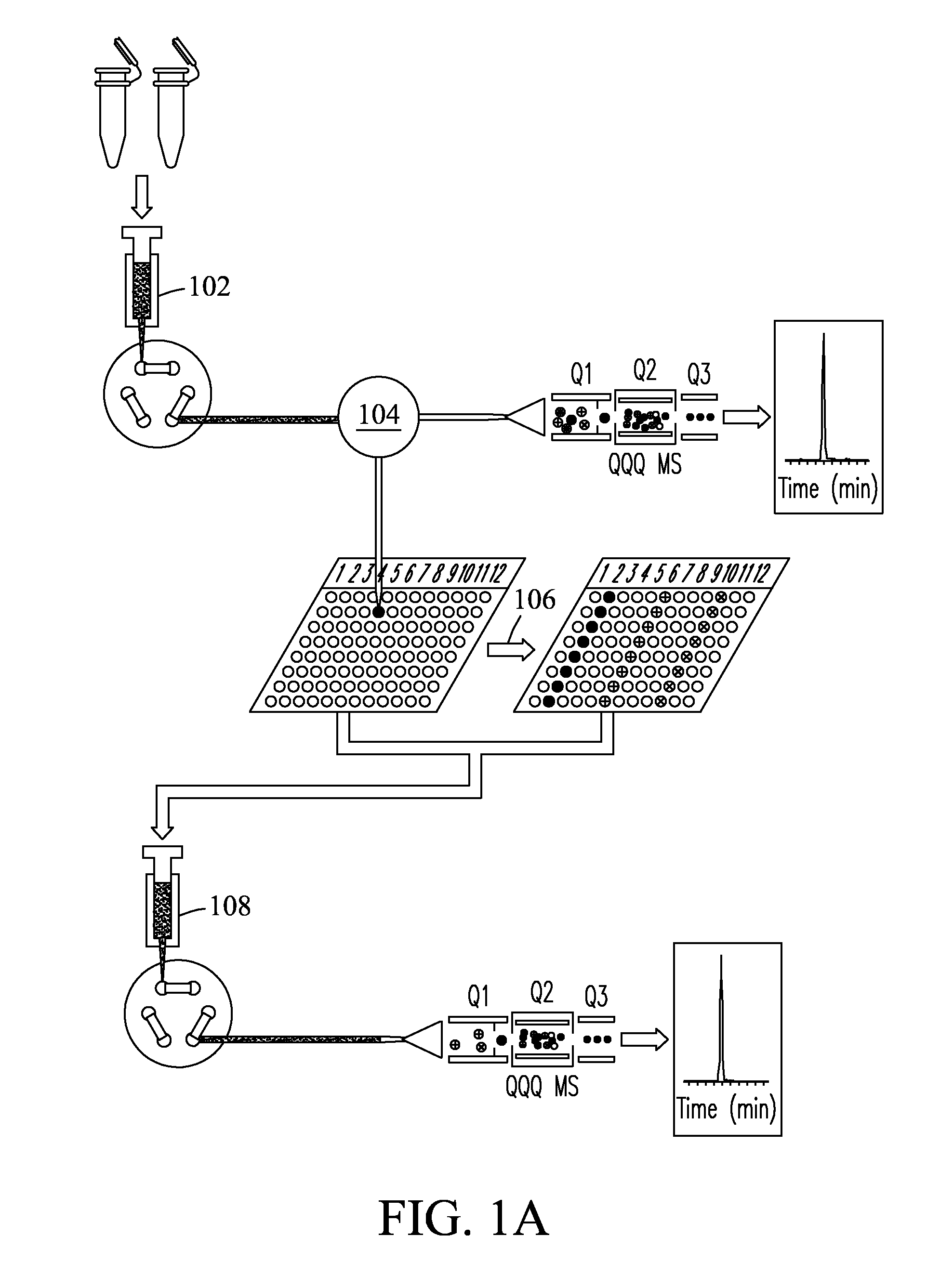
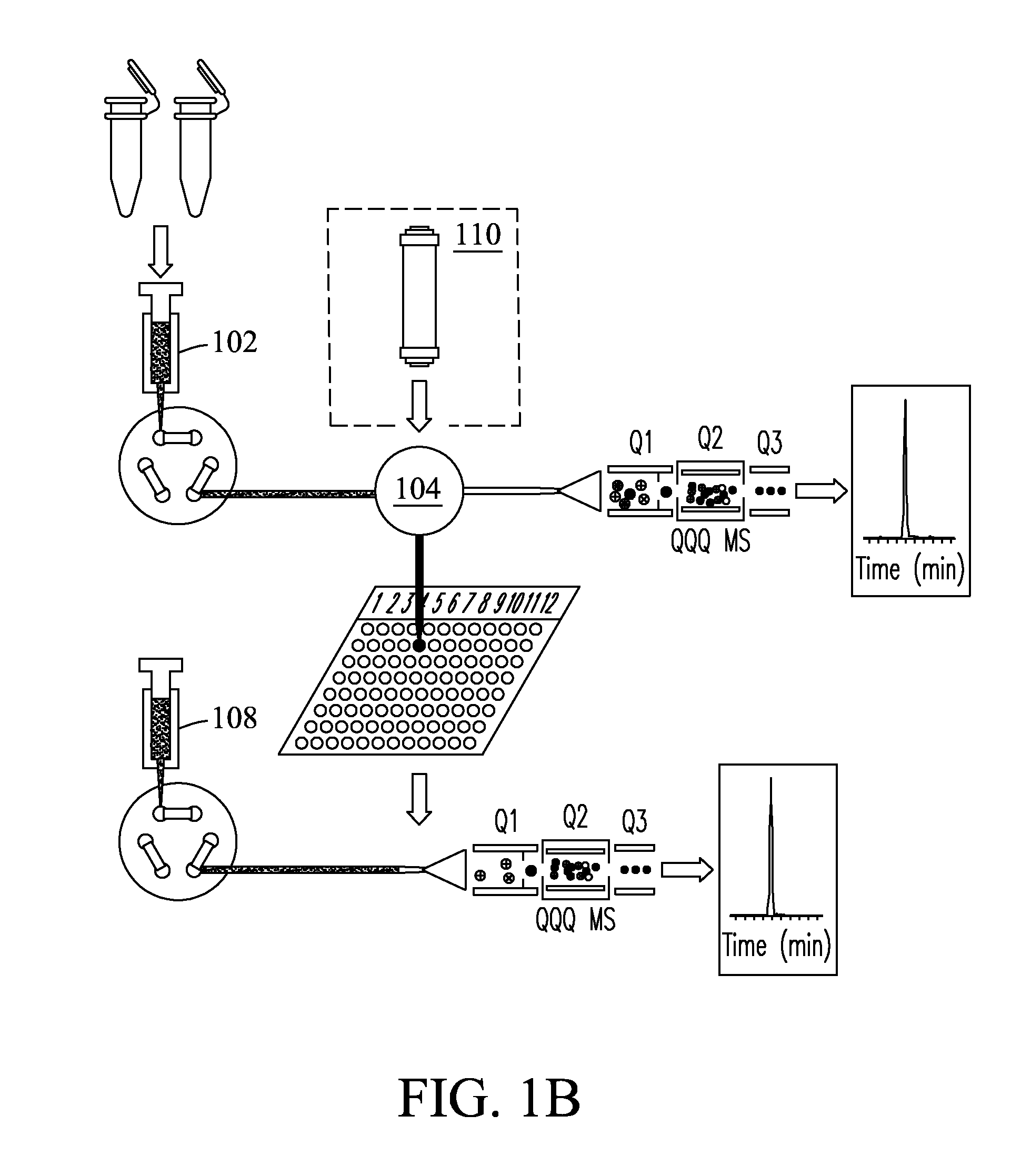
Method for high throughput peptide/protein assay generation and assays generated therewith
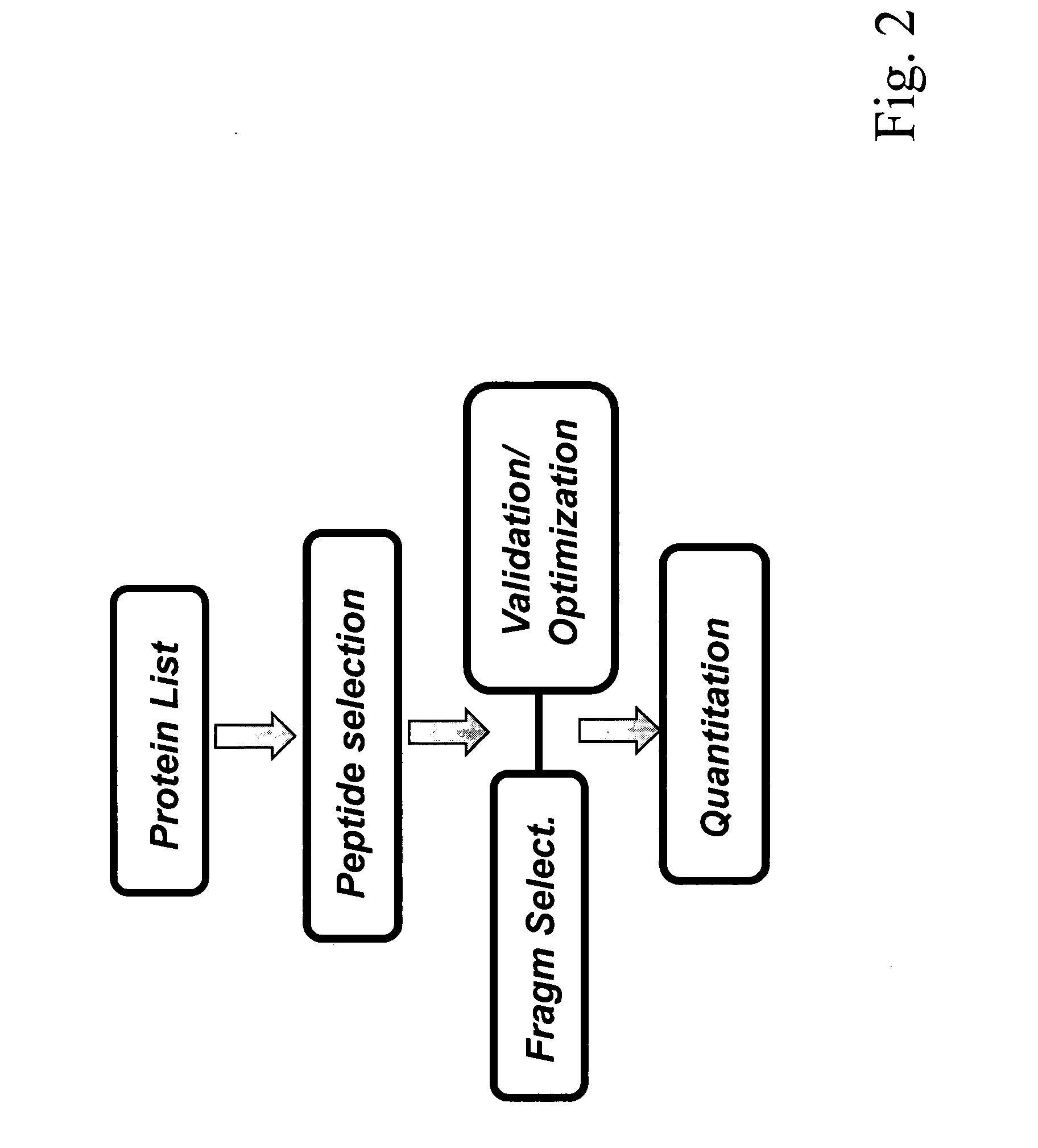
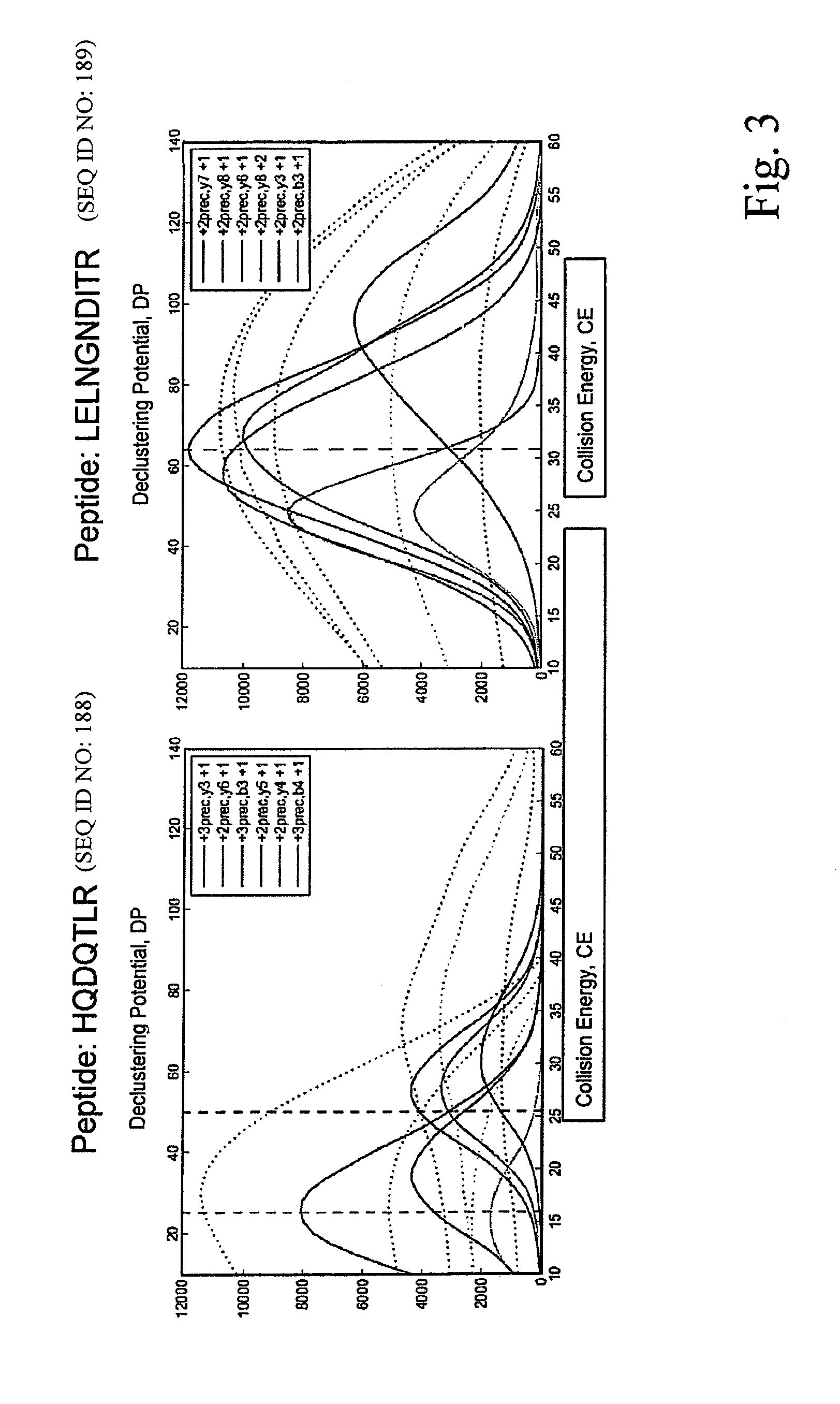
ActiveUS8501421B2Improve throughputStrong specificityAnalysis using chemical indicatorsPeptide/protein ingredientsElutionSelected reaction monitoring
The invention relates to a method for the determination of an MRM or SRM assay for a protein of interest, a peptide of interest, or a group of proteins / peptides of interest or a whole proteome. It essentially includes the following steps: (1) a list of proteins of interest is selected and for each member at least one or a list of candidate proteotypic peptides is derived (2) this at least one peptide is synthesized / generated essentially without subsequent purification; (3) this at least one unpurified peptide is analyzed by selected reaction monitoring (SRM) preferably coupled to liquid chromatography (LC-SRM) or analogous techniques; (4) validation and / or optimisation of the corresponding assay of the at least one peptide with determination of the SRM coordinates for a peptide / protein of interest and / or of a regulator of interest is achieved. A protein sample of interest is enzymatically digested and can then be analyzed in SRM mode or time-constrained SRM mode, using elution times to trigger acquisition of the set of selected SRM traces, thus drastically increasing the throughput. The analysis allows to detect and quantify the set of peptides / proteins of interest. The method additionally relates to a tagging strategy to achieve absolute quantification of the peptides / proteins of interest at low-budget and high-throughput.
Owner:ETH ZZURICH
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com