High-throughput nucleotide library sequencing
A technology of nucleotides and polynucleotides, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA preparation, etc., and can solve problems such as unrelated
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1a
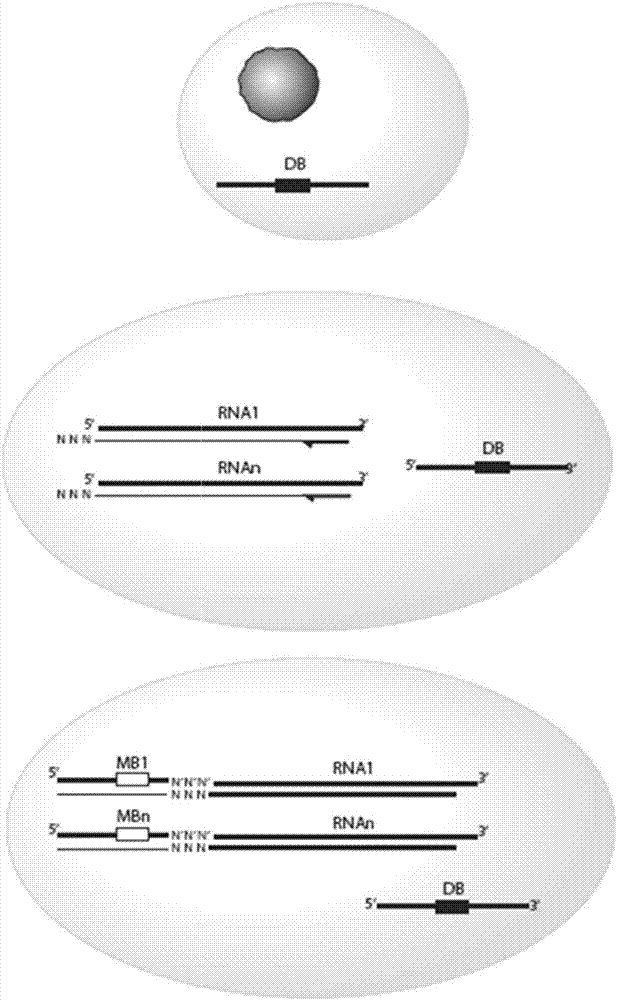
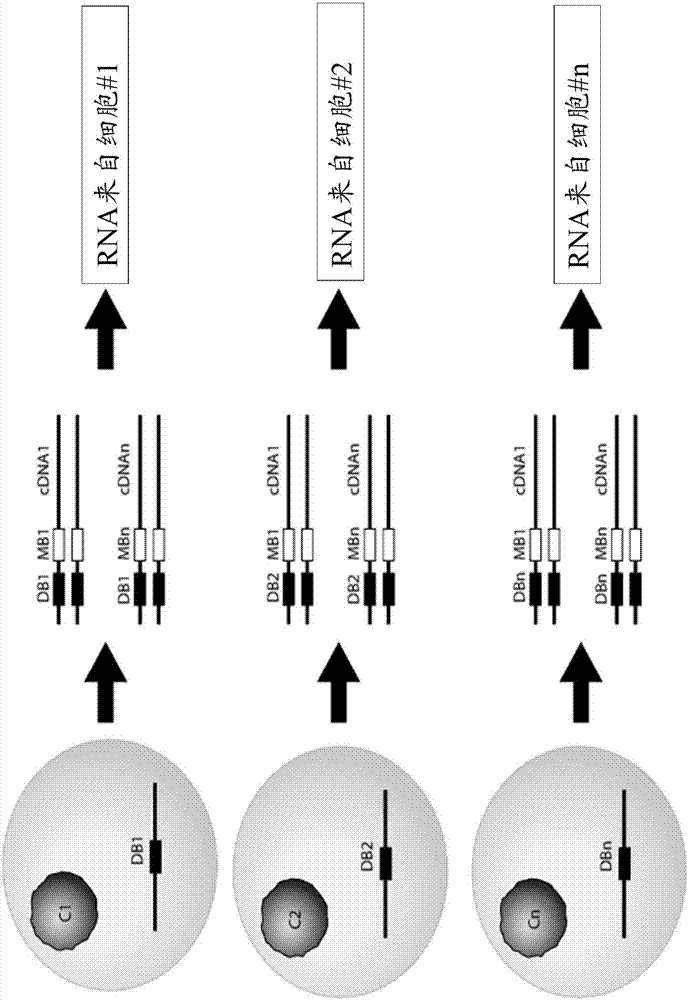
[0342] Example 1a - Protocol for preparing cells for emulsion-based large-scale high-throughput single-cell polynucleotide sequencing.
[0343] Obtain the cell population of interest. These include total PBMC, sorted cells, antibody-enriched B or T cells, or other cell types. Cells have an intact plasma membrane, so they do not leak excessive amounts of mRNA into the surrounding medium. Cells need not be alive.
[0344] Cells were washed twice by centrifugation (200 g, 10 min for T cells or B cells) in cell buffer 1 x Dulbecco's phosphate buffered saline (PBS). Cells were then diluted to 3.5x10 in cell buffer 6 / mL cell concentration. The suspension was then pipetted through a 20 μm cell strainer.
Embodiment 1b
[0345] Example 1b - Protocol for preparation of solid tissues for emulsion-based large-scale high-throughput single-cell polynucleotide sequencing.
[0346] Solid tissues (e.g., tumor or non-tumor biopsies) were treated with various proteases including collagenase III (200 U / mL), DNase I (200 U / mL), and trypsin (5 mg / mL) and NEDB (Invitrogen) ) to produce mixtures of individual cells and aggregates containing more than one cell. Briefly, tumors removed from mice were added to cold medium, and the surrounding mouse mammary gland tissue and fat were removed. Tumors were cut into 2-4 mm pieces, which were then incubated with appropriate dissociation solutions or enzymes for 30 min at 37°C. Tumor fragments were then mixed up and down every 10 min using a 1,000 mL micropipette tip cut to the appropriate diameter for the tissue fragment size. After each incubation period, debris was filtered through a 40 mm nylon mesh cell strainer. The released cells were centrifuged at 1200 r...
Embodiment 2
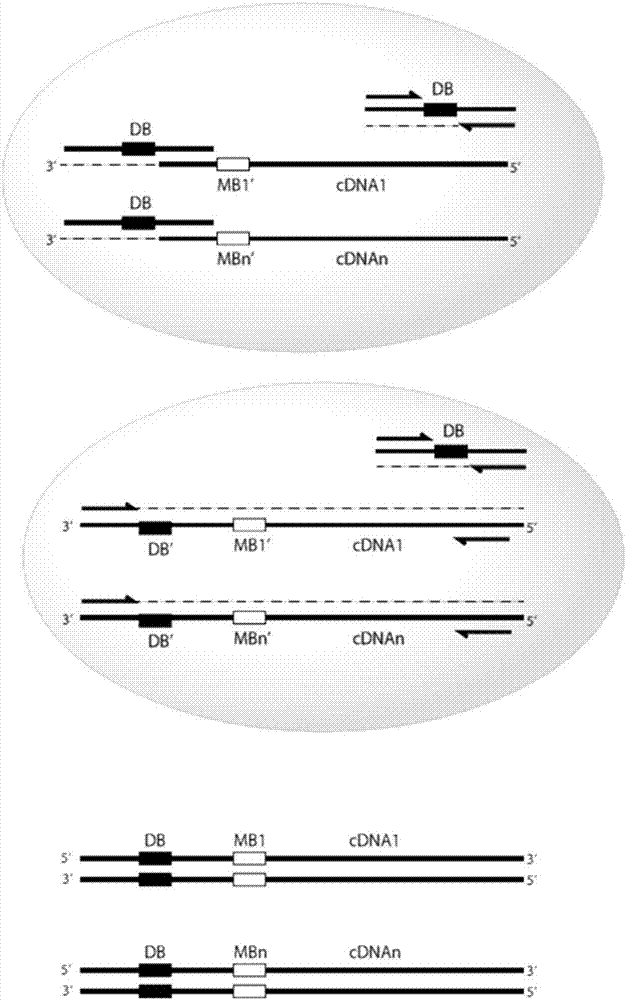
[0348] Example 2 -For large-scale, high-throughput, single-cell polynucleotide sequencing based on emulsions
[0349] Scheme for the preparation of the emulsion reaction mixture.
[0350] Mix the emulsion reaction mixture containing the reagents and oligonucleotides in Table 1 below in a PCR-clean cabinet at room temperature.
[0351] Table 1
[0352]
[0353]
[0354] / 5Biosg / = 5' biotin modification; / iSp18 / = 18-carbon spacer; V = A, C or G; N = any base; rG = riboguanosine; W = A or T.
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com