Primer group for developing rhesus T cell immune repertoire, high-flux sequencing method and application of method
A technology of cellular immunity and rhesus monkeys, applied in the field of molecular biology, can solve problems such as limited use
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0073] The preparation of T lymphocyte receptor (TCR) genomic DNA sample comprises the following steps:
[0074] (1) Collect 10 milliliters (ml) of fresh peripheral blood samples each, and operate according to the instructions of the LymphoPrep kit (Axis-shield, Cat. No. AS1114544UK) to obtain relatively pure PBMCs;
[0075] (2) Use the PureLink Genomic DNA Mini Kit (Life Technology, Cat.No: K1820-00) kit to extract the genomic DNA of the cells obtained in step (1), and use Nanodrop2000 (Thermo) to measure the concentration and purity of the DNA, and then save the genomic DNA .
Embodiment 2
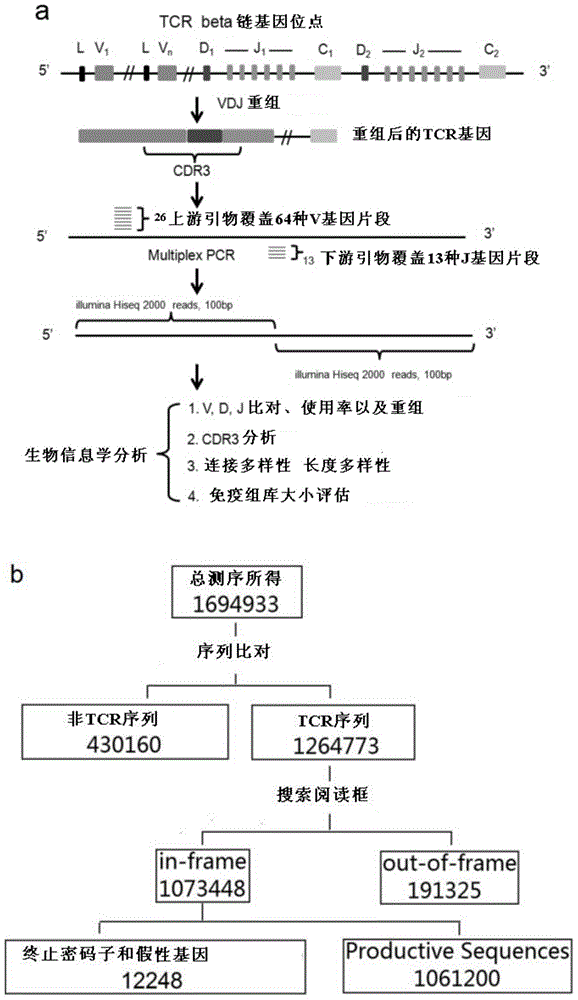
[0077] Using the primer set for developing rhesus monkey T cell immune repertoire to perform multiplex PCR, construct a TCRβ high-throughput sequencing library, and perform high-throughput sequencing, including the following steps:
[0078] (1) Use the genomic DNA obtained in Example 1 as an amplification template, take TCR primers, and then use QIAGEN’s Multiplex PCR kit (article number: 206143) to configure a multiplex PCR system according to the kit instructions, wherein each forward primer, etc. Molar mixing, the total concentration of primers is 10 micromolar, equimolar mixing of each reverse primer, the total concentration of primers is 10 micromolar, the amount of template can be adjusted, 3ug is used in this example, 1ug can also be made, but the diversity will be decline.
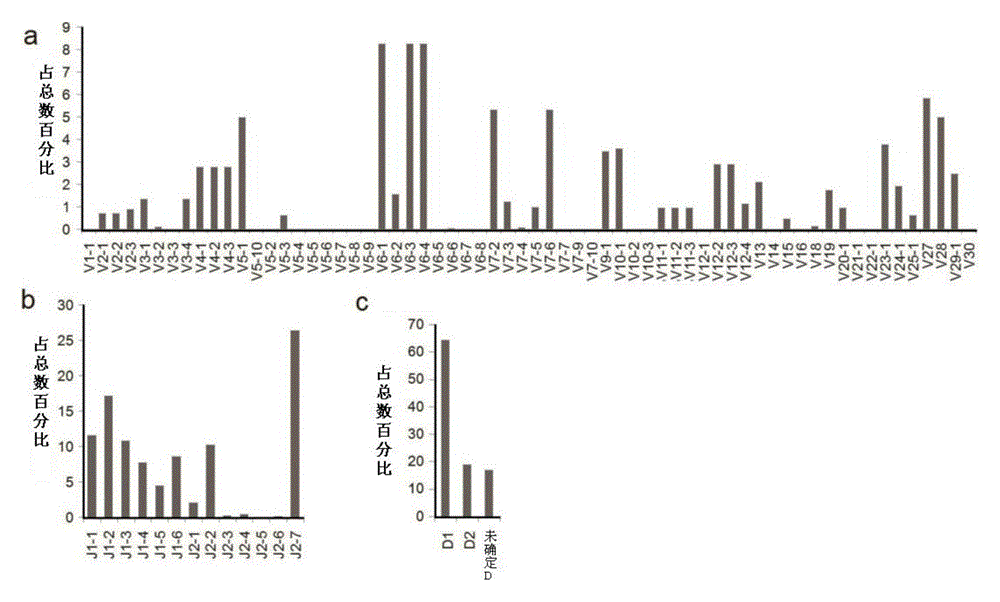
[0079] The upstream primers in this system are equimolar mixtures of primers shown in SEQ ID NO:1 to SEQ ID NO:26; the downstream primers are equimolar mixtures of primers shown in SEQ ID NO:27 t...
Embodiment 3
[0126] Methods for correcting multiplex PCR primer amplification errors include:
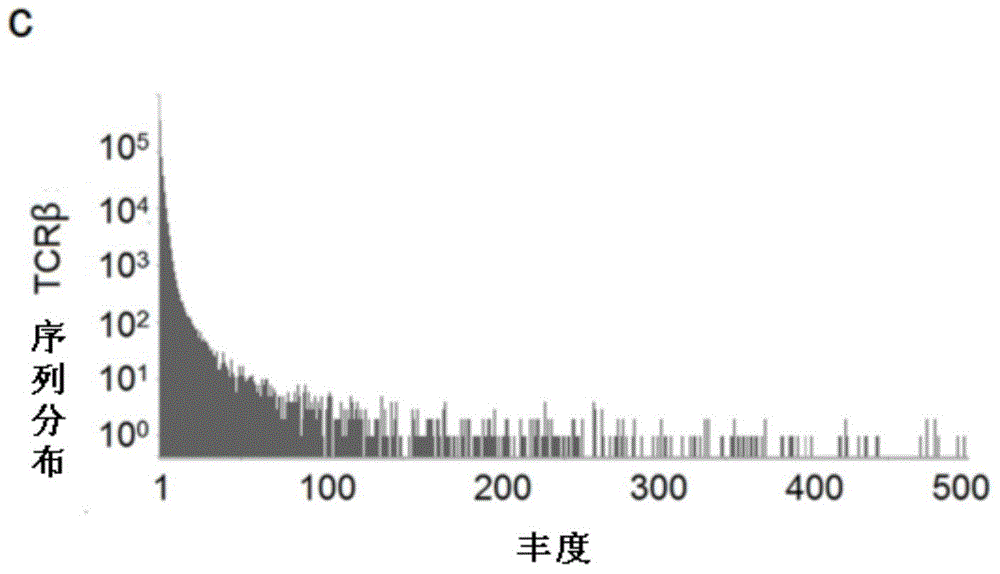
[0127] One of the purposes of the present invention is to analyze the abundance of TCR sequences. However, during the PCR amplification process, due to the amplification preferences of PCR primers for different gene fragments, the real situation of the abundance of TCR sequences will be affected.
[0128] In order to assess this type of systematic error, we adopted the method in Robins et al, we extracted the genome of 1 rhesus monkey according to the method in Example 1, divided into 6 tubes equally, and 3 tubes adopted 25 cycles of multiplex PCR (refer to implementation Example 2), 3 tubes were used for 30 cycles, and the amplified products of each tube were sequenced by Illumina Miseq. In the 25-cycle experimental group, a total of 10,345 specific TCR CDR3 sequences were found, and 15.3% were also found in the 30-cycle PCR experiment.
[0129] We analyzed the 19658 sequencing results found in ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com