Authentication method of cattle CAPN1 gene as longisimus dorsi tenderness molecular marker and application
A CAPN1E14R, Longissimus muscle technology, applied in application, genetic engineering, plant genetic improvement, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
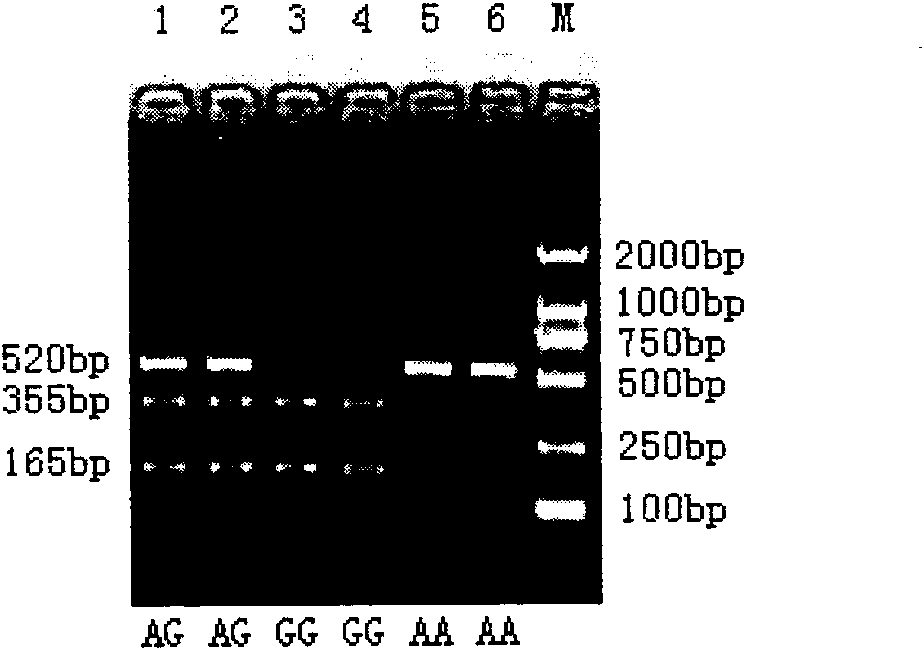
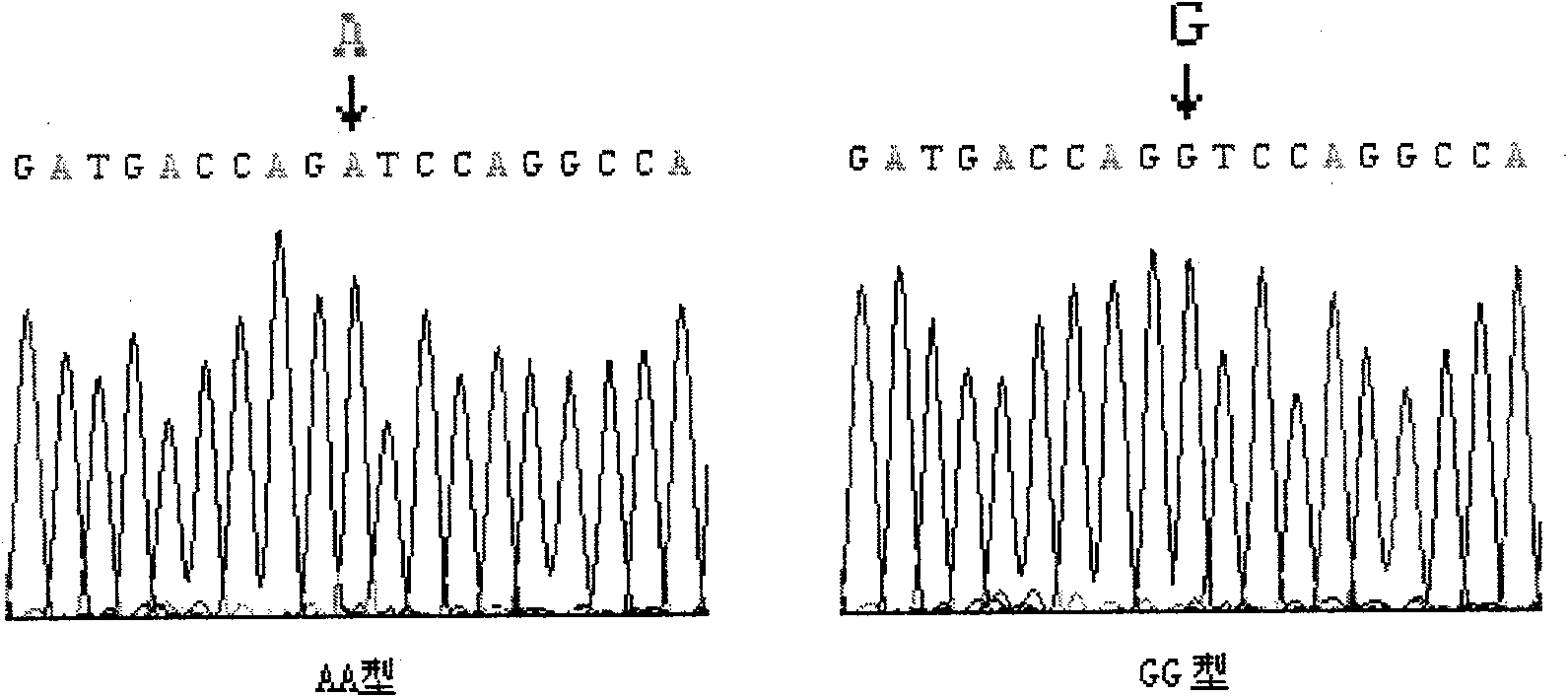
[0088] Six individuals to be slaughtered were randomly selected from the beef cattle group of Haoyue Halal Meat Group Co., Ltd. in Changchun City, Jilin Province, China. Blood was collected during slaughter for genomic DNA extraction and the shear force of the longissimus dorsi muscle was measured. The extracted genomic DNA was determined by a spectrophotometer, the concentration was 36.54ug / ml, and the OD260 / OD280Ratio was 1.824. The primers, PCR reaction system and conditions designed by the present invention are used for PCR amplification. The PCR reaction system is: 10×PCR Buffer (purchased from Tiangen Biochemical Technology Co., Ltd.) 3.0 μl, dNTPs (purchased from Beijing Dingguo Biotechnology Company, the concentration is 2.5mM) 0.8 μl, forward primer and reverse primer (concentration 0.4 μl each of 10 pmol / μl), 0.4 μl of Taq enzyme (purchased from Tiangen Biochemical Technology Co., Ltd., at a concentration of 2.5 U / μl), 0.5 μl of genomic DNA (containing 18.27 ng DNA),...
Embodiment 2
[0102] Ten individuals to be slaughtered were randomly selected from the beef cattle group of Haoyue Halal Meat Industry Group Co., Ltd. in Changchun City, Jilin Province, China. Blood was collected during slaughter for genomic DNA extraction and the shear force of the longissimus dorsi muscle was measured. The extracted genomic DNA was determined by a spectrophotometer, the concentration was 34.54ug / ml, and the OD260 / OD280Ratio was 1.794. The primers, PCR reaction system and conditions designed by the present invention are used for PCR amplification. The PCR reaction system is: 10×PCR Buffer (purchased from Tiangen Biochemical Technology Co., Ltd.) 3.0 μl, dNTPs (purchased from Beijing Dingguo Biotechnology Company, the concentration is 2.5mM) 0.8 μl, forward primer and reverse primer (concentration 0.4 μl each of 10 pmol / μl), 0.4 μl of Taq enzyme (purchased from Tiangen Biochemical Technology Co., Ltd., at a concentration of 2.5 U / μl), 0.5 μl of genomic DNA (containing 17.27...
Embodiment 3
[0115] Nine individuals to be slaughtered were randomly selected from the beef cattle group of Haoyue Halal Meat Industry Group Co., Ltd. in Changchun City, Jilin Province, China. Blood was collected during slaughter for genomic DNA extraction and the shear force of the longissimus dorsi muscle was measured. The extracted genomic DNA was determined by a spectrophotometer, the concentration was 37.34ug / ml, and the OD26O / OD280 Ratio was 1.803. The primers, PCR reaction system and conditions designed by the present invention are used for PCR amplification. The PCR reaction system is: 10×PCR Buffer (purchased from Tiangen Biochemical Technology Co., Ltd.) 3.0 μl, dNTPs (purchased from Beijing Dingguo Biotechnology Company, the concentration is 2.5mM) 0.8 μl, forward primer and reverse primer (concentration 0.4 μl each of 10 pmol / μl), 0.4 μl of Taq enzyme (purchased from Tiangen Biochemical Technology Co., Ltd., at a concentration of 2.5 U / μl), 0.5 μl of genomic DNA (containing 18....
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com